Abstract
Exosomes are the small vesicles that are secreted by different types of normal and tumour cells and can incorporate and transfer their cargo to the recipient cells. The main goal of the present work was to study the tumour exosomes’ ability to accumulate the parent mutant DNA or RNA transcripts with their following transfer to the surrounding cells. The experiments were performed on the MCF7 breast cancer cells that are characterized by the unique coding mutation in the PIK3CA gene. Using two independent methods, Sanger sequencing and allele-specific real-time PCR, we revealed the presence of the fragments of the mutant DNA and RNA transcripts in the exosomes secreted by the MCF7 cells. Furthermore, we demonstrated the MCF7 exosomes’ ability to incorporate into the heterologous MDA-MB-231 breast cancer cells supporting the possible transferring of the exosomal cargo into the recipient cells. Sanger sequencing of the DNA from MDA-MB-231 cells (originally bearing a wild type of PIK3CA) treated with MCF7 exosomes showed no detectable amount of mutant DNA or RNA; however, using allele-specific real-time PCR, we revealed a minor signal from amplification of a mutant allele, showing a slight increase of mutant DNA in the exosome-treated MDA-MB-231 cells. The results demonstrate the exosome-mediated secretion of the fragments of mutant DNA and mRNA by the cancer cells and the exosomes’ ability to transfer their cargo into the heterologous cells.
1. Introduction
Exosomes are among the most popular and perspective objects of recent molecular oncology studies. Exosomes represent the 30–100 nm-sized microvesicles that are generated in the cells, released into the extracellular space, and carry a wide spectrum of bioactive molecules. The most intriguing property of exosomes represents their ability to incorporate into the recipient cells resulting in the cascade of both genomic and nongenomic changes. Among the exosomal cargo, proteins preserving their biological activity after transferring such as ABC transporters, receptor tyrosine kinases, growth factors, etc., and small stable RNAs, primarily microRNAs were of great interest for research. Basing on the different experimental models and clinical data the ability of exosome-transferred molecules to modulate the main properties of the recipient cells that are growth and survival, drug resistance was found to occur [1,2,3,4,5].
In addition, the exosomes can transfer the fragments of the parent DNA or mRNA, but their role in the regulation of the recipient cells is unclear and requires further investigation. Concerning cancer cells, certain observations demonstrate the presence of the mutant tumour DNA in the native exosomes [6,7,8,9]; however, the efficiency of mutant DNA accumulation and the possible presence of the mutant RNA transcripts in the exosomes are open questions—as well as the possible involvement of the DNA-enriched exosomes in the gene exchange between the cells.
Here, the analysis of MCF7 breast cancer cells revealed the presence of the DNA and RNA transcripts with the native PIK3CA mutation in the secreted exosomes. We demonstrated the effect of MCF7 exosomes’ incorporation into the heterologous recipient cells supporting the possible transferring of the exosomal cargo into the recipient cells.
2. Results
The experiments were performed on the MCF7 breast cancer cells characterized by the unique coding mutation in the PIK3CA gene. Exosomes were prepared from the MCF7 conditioned medium by the differential ultracentrifugation, and exosome imaging was carried out by transmission electron microscope as described in Methods (Figure 1a). The exosome functional studies were performed as recommended by ISEV in [10] and fully described in [11]. The quality of purified DNA and RNA samples was controlled by separation on the 0.8% and 1.2% agarose gel, respectively (Figure 1b,c). Most DNA was found inside the exosomes and did not degrade after treatment with DNase.
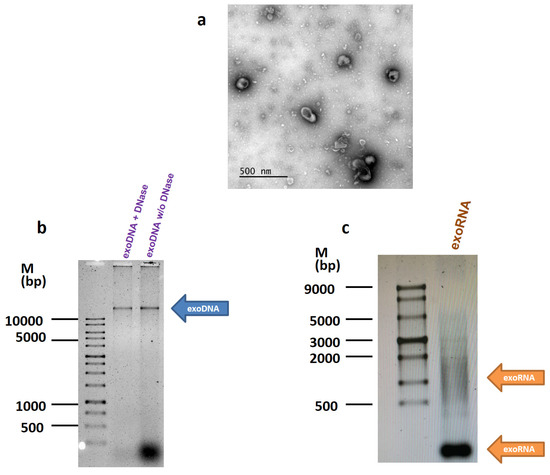
Figure 1.
Detection of DNA and RNA in the exosomes. (a) The transmission electron microscopy of the exosomes. Exosomes were prepared from the MCF7 conditioned medium by the differential ultracentrifugation, and imaged as described in Methods. (b,c) DNA and RNA were isolated from the MCF7 exosomes and separated on 0.8% and 1.2% agarose gel, respectively.
Sanger sequencing of exosomal DNA and RNA revealed the PIK3CA mutation E545K (c.1633G > A) in both DNA and RNA samples (Figure 2).
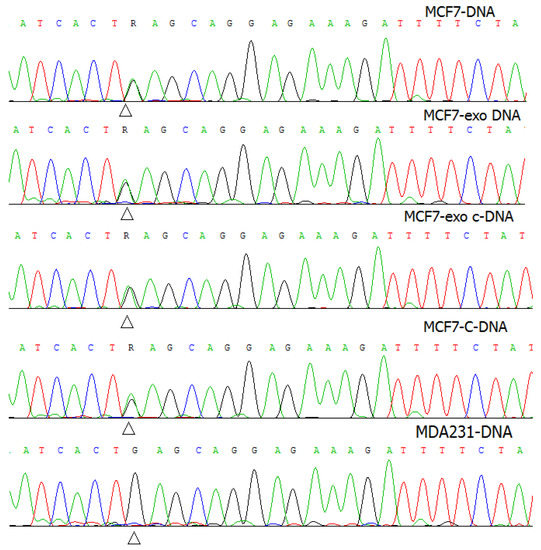
Figure 2.
Mutation validation by Sanger sequence analysis. The sequence electrophoregrams of the MCF7-DNA, exosomal DNA from MCF7, cDNA from MCF7, and exosomal cDNA from MCF7 contained the PIK3CA mutation c.1633G > A (p.E545K) (R), compared to the MDA-231 DNA.
Are the exosomes capable to transfer the mutant nucleic acids to the recipient cells? To answer this question, the subsequent experiments were performed using MDA-MB-231 breast cancer cells characterized by the wild type of the PIK3CA gene. The MDA-MB-231 cells were treated with the exosomes of MCF7 cells, and cytoplasmic DNA or RNA of MDA-MB-231 cells was isolated and sequenced. There was no detectable amount of mutant DNA or RNA found in the treated MDA-MB-231 cells (Figure 3).
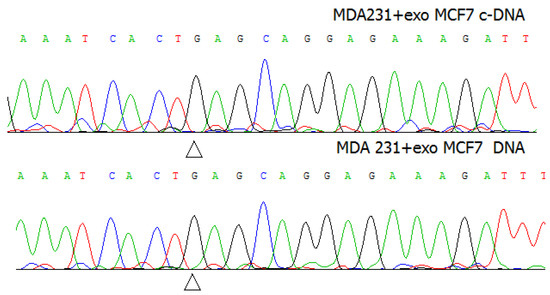
Figure 3.
The sequence electrophoregrams of DNA and c-DNA of MDA-MB-231 cells after treatment with the exosomes of MCF7 cells.
Furthermore, the PIK3CA mutation was detected by an independent method—allele-specific real-time PCR. The threshold cycle (Ct value) was calculated as the cycle when the fluorescence of the sample exceeded a threshold level corresponding to 10 standard deviations from the mean of the baseline fluorescence. We provided three technical replications of the reaction per sample and calculated average Ct, standard deviation, and indexes associated with them according to the Guide for TaqMan® Mutation Detection Assay (Thermo Fisher Scientific, Waltham, MA, USA). The average Ct for all reactions was determined, and the delta Ct of the mutant and reference (normal) alleles was calculated. Then, the cut-off for delta Ct was determined as the total average Ct of the negative control minus three standard deviations. As shown, the delta Ct of MDA-MB-231 cells treated with MCF7 exosomes was less than the cut-off delta Ct of the negative control (MDA-MB-231 cells without PIK3CA mutation) in absolute values but did not differ significantly revealing the slight tendency to accumulation of mutant DNA in the exosome-treated cells. Probably, the amount of mutant DNA in the treated cells was under the threshold of analytical sensitivity of the used real-time PCR protocol in this case (Table 1).

Table 1.
Allele-specific real-time PCR of the DNA samples of exosome-treated cells.
To demonstrate the possibility of MCF7 exosomes to incorporate into the MDA-MB-231 cells, the vesicles were labelled by fluorescent dye CellTracker™ Red CMPTX Dye (Thermo Fisher Scientific, Waltham, MA, USA). Then thoroughly stained MCF7 exosomes were washed in PBS twice by the ultracentrifugation 100 000× g, and precipitates were dissolved in PBS. One portion of the exosomes was destructed by sonication (used as a negative control), and native and sonicated exosomes were incubated with MDA-MB-231 cells for 1 h. As shown, the MCF7 exosomes were able to accumulate the fluorescent dye and transfer it to MDA-MB-231 recipient cells (Figure 4).
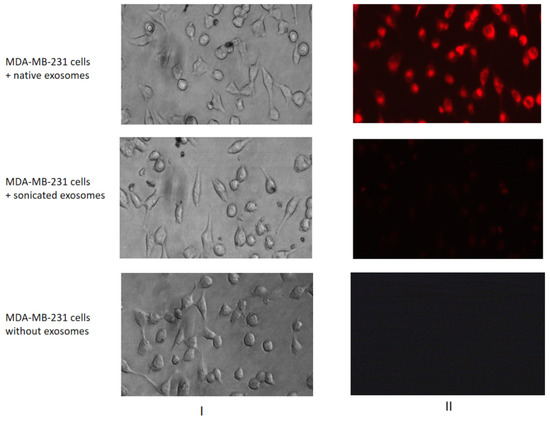
Figure 4.
Transferring of fluorescent-labelled compounds by exosomes. MDA-MB-231 cells were cultured without labelled exosomes or with the native or sonicated MCF7 exosomes stained by a fluorescent dye. The efficiency of dyeing exosome incorporation was checked with a fluorescent microscope Nikon Eclipse Ti-E. The images of light (I) and fluorescent (II) microscopy are presented.
We showed the accumulation of the fragments of mutant DNA or RNA transcripts in the exosomes of the cancer cells. Based on our data and data from other researchers showing the exosomes’ ability to incorporate into the recipient cells, we can propose the possible involvement of the exosomal tumour DNA in the recipient cell rearrangement. The latter one agrees with the earlier observations showing the exosomes’ capability to transfer the fragments of tumour DNA to recipient cells, including the microenvironment cells, and to induce in those cancer-associated changes [12,13,14,15]. We did not find a detectable amount of the mutant DNA in the exosome-treated recipient cells—probably because of the extremely low amount of the incorporated DNA. A similar problem was pointed out by other authors who noted the low efficiency of the detection of heterologous exosomal DNA in the recipient cells [13].
The origin of the exosomal DNA is still unclear; it may be the micronuclei resulting from chromosome aberrations during mitosis [16], cytoplasmic fragments of the damaged DNA [17], and DNA transposons or cDNA reversed from retrotransposons [18]. Another important problem is whether the possibility of the integration of the heterologous exosomal DNA into the genomic DNA of the recipient cells exists. Single publications demonstrate the transfer of exosomal DNA into the recipient cells accompanied by stable integration of the DNA and transmission to daughter cells after some passaging [13,19]. In general, the presence of the mutant DNA in the exosomes does not seem to be sufficient to induce the malignant transformation of the recipient cells, and the combination of tumour exosomes with the additional cancer-initiating agents is required.
3. Materials and Methods
The human breast cancer cell lines MCF7 and MDA-MB-231 were purchased from ATCC. The cells were authenticated by morphology and STR profiling provided by Gordiz (http://gordiz.ru/, accessed on 12 August 2020). The cells were cultured in standard DMEM medium (Gibco) supplemented with 10% foetal bovine serum (FBS) (HyClone) at 37 °C and 5% CO2. Exosomes were prepared from the MCF7 conditioned medium by the differential ultracentrifugation and were characterized as described in our recent paper [11]. Transmission Electron Microscopy (TEM) was used to visualize the exosome samples. They were deposited onto the carbon-coated grids (Ted Pella, Redding, USA) pre-treated using a K100X glow discharge unit (Quorum Technologies Ltd, Lewes, Great Britain) and stained with 1% uranyl acetate (Electron Microscopy Sciences, Hatfield, UK). Imaging was carried out using a JEM-1011 transmission electron microscope (JEOL Ltd., Tokyo, Japan) at 80 kV.
To reduce external DNA contamination, before DNA extraction, exosomes were treated with DNase I, and exosomal DNA was extracted by phenol/chloroform mixture; total exosomal RNA was extracted in Trisol.
For cell treatments, MCF7 exosomes were added to the MDA-MB-231 cells for 3 days at the final concentration of 1.7 μg/mL of exosomal protein or (5.5 ± 0.3) × 109 vesicles/mL; then the cells were washed with PBS, and cytoplasmic DNA and total RNA were extracted and subjected for sequencing. For exosome labelling, the MCF7 vesicles were incubated with fluorescent dye CellTracker™ Red CMPTX Dye (Thermo Fisher Scientific, Waltham, MA, USA) and washed in PBS by the ultracentrifugation for 2 h at 100,000× g. Precipitates were dissolved in PBS and incubated with MDA-MB-231 cells for 1 h before fluorescent microscopy analysis.
cDNA conversion: cDNA synthesis was carried out with 1 µg total RNA by using the MiScript Reverse Transcription Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
Sanger sequencing: The fragments with PIK3CA (p.Glu545Lys) mutation were amplified using specific primers from nuclear and exosomal DNA and cDNA (Table 2).

Table 2.
Sequence primers PIK3CA detection.
The direct sequencing of individual PCR products from primers that flank areas of specific mutations was performed on the automatic genetic analyser 3500 (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocols.
Real-time PCR. The PIK3CA mutation c.1633G > A (p.E545K) was detected by allele-specific real-time PCR with TaqMan probes using TaqMan® Mutation Detection Assay (Hs00000824_mu/Hs00001025_rf assay, recommended reagents, consumables, and protocols; Thermo Fisher Scientific, Waltham, MA, USA) and DT-Prime thermocycler (DNA-Technology, Moscow, Russia). The mutant and normal (reference) alleles of the same site of the PIK3CA gene were amplificated; DNA from MDA-MB-231 cells (originally contained a wild type of PIK3CA) was used as a negative control.
4. Conclusions
The results demonstrate the exosome-mediated secretion of the fragments of mutant DNA and mRNA by the cancer cells and demonstrate the exosomes’ ability to incorporate into the heterologous cells. We propose that further investigations will delineate the role of the exosome-transferred mutant DNA in the tumour progression as well as in the formation of the cancer-associated phenotype of the cells of the tumour microenvironment.
Author Contributions
Conceptualization, M.A.K.; Data curation, M.A.K. and M.V.N.; Formal analysis, M.A.K., M.V.N., A.M.S., and O.E.A.; Investigation, O.E.A., Y.Y.S., E.I.M., D.V.S., I.V.B., E.B.K., D.S.M., D.V.B. and M.A.K.; Methodology, O.E.A., M.V.G., M.V.N., D.V.B. and M.A.K.; Project administration, M.A.K.; Supervision, M.A.K. and M.V.N.; Writing—original draft, M.A.K. and M.V.N.; Writing—review and editing, A.M.S., M.V.G., and M.V.N. All authors have read and agreed to the published version of the manuscript.
Funding
The research is supported by the Russian Science Foundation (project 19-15-00245, experiments with fluorescent-labelled compounds) and the Russian Foundation for Basic Research (project 18-29-09016, experiments with exosomes).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Datas are available from the authors.
Acknowledgments
The authors would like to thank Svetlana Semina for her kind help with exosome purification protocols. The TEM measurements were performed at the User Facilities Center “Electron microscopy in life sciences” of Lomonosov Moscow State University.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Sample Availability
Samples are available from the authors.
References
- Kim, J.; Morley, S.; Le, M.; Bedoret, D.; Umetsu, D.T.; Di Vizio, D.; Freeman, M.R. Enhanced shedding of extracellular vesicles from amoeboid prostate cancer cells: Potential effects on the tumor microenvironment. Cancer Biol. Ther. 2014, 15, 409–418. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal. Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, C.; Harikumar, K.B. The Origin and Functions of Exosomes in Cancer. Front. Oncol. 2018, 8, 66. [Google Scholar] [CrossRef]
- Tamkovich, S.N.; Yunusova, N.V.; Tugutova, E.; Somov, A.K.; Proskura, K.V.; Kolomiets, L.A.; Stakheeva, M.N.; Grigor’eva, A.E.; Laktionov, P.P.; Kondakova, I.V. Protease Cargo in Circulating Exosomes of Breast Cancer and Ovarian Cancer Patients. Asian Pac. J. Cancer Prev. APJCP 2019, 20, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Konoshenko, M.; Sagaradze, G.; Orlova, E.; Shtam, T.; Proskura, K.; Kamyshinsky, R.; Yunusova, N.; Alexandrova, A.; Efimenko, A.; Tamkovich, S. Total Blood Exosomes in Breast Cancer: Potential Role in Crucial Steps of Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 7341. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Shin, S.; Kim, B.; Lee, K.A. Selecting short length nucleic acids localized in exosomes improves plasma EGFR mutation detection in NSCLC patients. Cancer Cell Int. 2019, 19, 251. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.X.; Li, Y.M.; Ye, M.; Guo, Y.Y.; Li, Q.W.; Peng, X.M.; Wang, Q.; Zhang, S.F.; Zhao, H.X.; Zhang, H.; et al. KRAS and BRAF mutations in serum exosomes from patients with colorectal cancer in a Chinese population. Oncol. Lett. 2017, 13, 3608–3616. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. Discovery of Double-Stranded Genomic DNA in Circulating Exosomes. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef] [PubMed]
- Semina, S.E.; Scherbakov, A.M.; Vnukova, A.A.; Bagrov, D.V.; Evtushenko, E.G.; Safronova, V.M.; Golovina, D.A.; Lyubchenko, L.N.; Gudkova, M.V.; Krasil’nikov, M.A. Exosome-Mediated Transfer of Cancer Cell Resistance to Antiestrogen Drugs. Molecules 2018, 23, 829. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Cornils, K.; Speiseder, T.; Badbaran, A.; Reimer, R.; Indenbirken, D.; Grundhoff, A.; Brunswig-Spickenheier, B.; Alawi, M.; Lange, C. Indication of Horizontal DNA Gene Transfer by Extracellular Vesicles. PLoS ONE 2016, 11, e0163665. [Google Scholar] [CrossRef] [PubMed]
- Stefanius, K.; Servage, K.; de Souza Santos, M.; Gray, H.F.; Toombs, J.E.; Chimalapati, S.; Kim, M.S.; Malladi, V.S.; Brekken, R.; Orth, K. Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation. Elife 2019, 8, e40226. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Han, Y.; Ren, H.; Chen, C.; He, D.; Zhou, L.; Eisner, G.M.; Asico, L.D.; Jose, P.A.; Zeng, C. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J. Mol. Cell Biol. 2013, 5, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Knasmueller, S.; Bolognesi, C.; Holland, N.; Bonassi, S.; Kirsch-Volders, M. Micronuclei as biomarkers of DNA damage, aneuploidy, inducers of chromosomal hypermutation and as sources of pro-inflammatory DNA in humans. Mutat. Res. 2020, 786, 108342. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Loo, T.M.; Okada, R.; Kamachi, F.; Watanabe, Y.; Wakita, M.; Watanabe, S.; Kawamoto, S.; Miyata, K.; Barber, G.N.; et al. Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat. Commun. 2018, 9, 1249. [Google Scholar] [CrossRef] [PubMed]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Guan, W.; Tan, X.; Chen, C.; Li, L.; Wang, N.; Zou, X.; Zhou, F.; Wang, J.; Pei, F.; et al. SRY gene transferred by extracellular vesicles accelerates atherosclerosis by promotion of leucocyte adherence to endothelial cells. Clin. Sci. 2015, 129, 259–269. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).