Abstract
Due to its high sensitivity and resolving power, gas chromatography-ion mobility spectrometry (GC-IMS) is a powerful technique for the separation and sensitive detection of volatile organic compounds. It is a robust and easy-to-handle technique, which has recently gained attention for non-targeted screening (NTS) approaches. In this article, the general working principles of GC-IMS are presented. Next, the workflow for NTS using GC-IMS is described, including data acquisition, data processing and model building, model interpretation and complementary data analysis. A detailed overview of recent studies for NTS using GC-IMS is included, including several examples which have demonstrated GC-IMS to be an effective technique for various classification and quantification tasks. Lastly, a comparison of targeted and non-targeted strategies using GC-IMS are provided, highlighting the potential of GC-IMS in combination with NTS.
1. Introduction
Quality control and early detection of hazard chemicals, allergens, or biological contaminants are critical to ensure product safety. Environmental pollutants, pesticides, or toxins, among others, can compromise food safety and pose a public health risk [1]. Furthermore, food adulteration and food fraud, accelerated by globalization, continue to cause economic losses and customer dissatisfaction and emphasize the need for robust, inexpensive, and fast analytical methods [2]. While new scientific findings continuously identify potential hazardous or allergenic compounds [3], commonly employed methods, which focus on the detection and identification of a particular compound or class of compounds, lack the ability to identify new or unknown compounds. Due to the inherent diversity of biogenic samples, as observed in food analysis, and the chemical complexity of the sample matrices, analysis often requires advanced sample preparation strategies [4]. For systematic monitoring of product quality, it is therefore desirable to develop analytical methods capable of discovering unknown or non-targeted compounds from the complex sample matrices. This approach, also referred to as NTS, requires comprehensive extraction and analysis of compounds of interest. Analysis of the volatile organic compounds (VOCs) of samples, also known as VOC profiling, allows for the detection of compounds in complex sample matrices without the need for detailed a priori knowledge of the molecular composition. Due to its high sensitivity and resolving power on the one hand and its simplicity and robustness on the other, ion mobility spectrometry (IMS) has gained popularity for the analysis of VOCs [5]. Moreover, gas chromatography coupled to ion mobility spectroscopy (GC-IMS) has been shown to be an easy-to-handle and yet highly effective tool for VOC profiling [6]. As a result, non-targeted VOC profiling based on GC-IMS in combination with machine learning has emerged as a promising method for sample monitoring.
Since the 1970s, when IMS was first known as ‘plasma chromatography’, IMS has developed into a highly sensitive technique for the analysis of VOCs at ultratrace concentration levels, which accounts for additional information regarding the ion’s mobility [7,8,9]. Due to the robust and easy-to-handle instrumentation, a wide range of application fields have been found for IMS today, such as food flavor analysis [5], process monitoring [10,11], and quality control [12], as well as detection and quantification of warfare agents [13] and explosives [14,15].
With IMS, analytes are first ionized in the ionization region of the instrument. The most common ionization method is the atmospheric pressure chemical ionization [16] by beta emitters, which frequently use nickel-63 (Ni-63) [11,15,17,18] or the less hazardous beta-emitting tritium (H-3) [19] or alpha-emitting americium-241 (Am-241) [20,21]. Other ionization methods are atmospheric pressure photo ionization (APPI) [22], which uses ultraviolet light (UV) [23,24] or corona discharge (CD) atmospheric pressure chemical ionization [17,25,26,27], where a high electric field between a needle and a metal plate or discharge electrode is used. Yet another method is the laser desorption/ionization technique (LDI), which employs a laser pulse as ion source [28].
According to the European Union directive, the exemption limit for the total activity for tritium was set to 1 GBq [29]. Therefore, the usage of a low-radiation tritium ion source with an activity of 300 MBq is not subject to authorization, hence leading to a broad adoption of tritium ion sources in a number of commercially available systems on the market [30,31,32,33,34]. Beta particles, which are emitted by the tritium source, initiate a gas-phase reaction cascade of the drift gas (nitrogen or air), resulting in predominant proton-water clusters H+[H2O]n, which are commonly referred to as ‘reactant ions’ [35]. The number of water molecules (n) depends on the gas temperature and the moisture content of the gas atmosphere [8]. Depending on the proton affinity, molecules entering the ionization region react with the reactant ions to protonated monomers MH+[H2O]n−x, while decreasing the intensity of the reactant ion peak (RIP). At higher analyte concentrations, proton-bound dimers M2H+[H2O]m−x are formed by the attachment of additional analyte molecules. When the concentration further increases, the formation of higher molecular cluster ions, such as trimers or tetramers, is possible; however, due to their low stability and short lifetime, higher molecular cluster ions are rarely observed [36]. In general, nonlinear behaviors are observed for the ratio of the RIP and the distribution between the protonated monomer and the proton-bound dimer [36,37]. The principles of a drift-time IMS including a H-3 ionization source are shown in Figure 1.
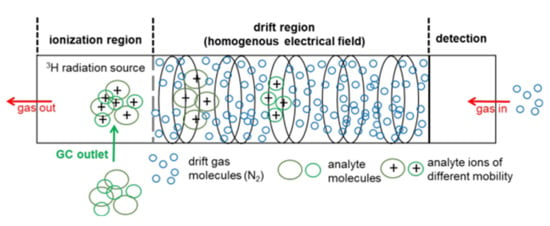
Figure 1.
Setup of a drift-time IMS with a tritium (H-3) ionization source, adopted from [38] with permission (ID5138730886281).
Subsequent to ionization, the analyte ions enter the drift region, where they are accelerated towards the detector, typically a Faraday plate, and are separated by their drift time (or mobility) in an electrical field at ambient pressure. The ions are slowed down by the collision with counterflowing drift gas molecules in the collision cross-section (CCS). Due to an equilibrium between the acceleration by an electric field and deceleration by the collision with the drift gas molecules, the ions move with a constant velocity to the detector. Depending on the characteristic mass, charge, and structure, the ions are separated in the drift tube and reach the detector at different drift times [39]. For identification of the analyte, the inverse of the measured drift time is normalized to the drift length and the electric field resulting in the spectrum of ion mobility. The reduced ion mobility K0 (see Equation (1)), which is independent of ambient conditions and experimental setup, is obtained after further normalization to pressure and temperature.
With
- K0 = reduced ion mobility in cm²V−1s−1
- L = drift length in cm
- E = electric field strength in Vcm−1
- tD = drift time in s
- p = pressure of the drift gas in hPa
- p0 = ambient pressure: p0 = 1013.2 hPa
- T = temperature of the drift gas in K
- T0-ambient temperature: T0 = 273.2 K
Instead of measuring temperature and pressure, the normalization is often carried out using the known mobility of the ions produced in the pure drift gas or by adding a reference analyte [40]. The signal intensity is proportional to the concentration and enables the quantification in ppbv (for some compounds even pptv) levels within a few milliseconds.
The state-of-the-art IMS technologies can be classified into time-dispersive, space-dispersive, and trapping technologies [41]. Time-dispersive IMS separates ions as a function of their mobility in a neutral gas, whereas space-dispersive IMS separates ions by the ratio of low-field to high-field mobilities [42]. Examples of time-dispersive IMS are drift tube ion mobility spectrometry (DTIMS) and travelling tube ion mobility spectrometry (TWIMS). High-field asymmetric waveform ion mobility spectrometry (FAIMS) and differential ion mobility spectrometry (DIMS or DMS) are examples for space-dispersive techniques [43]. The third class is represented by trapped ion mobility spectrometry (TIMS), which contains a trapping technology able to confine and release ions.
IMS alone has been applied for quantification [11,15,17,27] and classification [5] tasks in controlled environments. However, due to the inherent diversity of biogenic samples, the applications of IMS with direct sample introduction are often not sufficient, requiring prior purification or separation. The commonly used purification methods for VOC profiling in combination with IMS are solvent extraction [20,26,44] and solid-phase microextraction (SPME) [13]. SPME devices are constructed of a silica fiber coated with a thin layer of a suitable polymeric sorbent or immobilized liquid, used for the direct extraction of analytes from gaseous and liquid media [45]. While SPME coupled to IMS has been successfully used for quantification tasks, such as the detection and quantification of precursors and degradation products of chemical warfare agents [13], SPME is commonly extended by column separation techniques [18,46,47].
To avoid clustering in the ionization or drift region, IMS devices are commonly coupled to column separation techniques, such as liquid chromatography (LC) or gas chromatography (GC). Column separation coupled to drift-time IMS separates analytes into two orthogonal features, first the retention time through chromatography and second, the drift time or mobility through IMS, resulting in a two-dimensional (2D) highly resolved GC-IMS spectrum [6,38]. In LC analysis, any soluble compound can be separated, but sample preparation is a critical step for the data quality [48]. A comprehensive extraction method which enables the extraction of a wide range of compounds with minimized potentially interfering coextractives is needed for NTS approaches, since unspecific compounds are being targeted [49]. LC-MS in combination with NTS has been applied for the detection of food contaminants and environmental hazards [50,51].
In GC analysis, the volatility of a sample is a prerequisite. Headspace (HS)-based techniques allow for the analysis of untreated samples, avoiding the time-consuming sample pretreatment steps [52]. The analysis of non-volatile samples may be achieved through the derivatization with a functional group onto the molecule of interest. Although the modification of the functional group enables the analysis of compounds that otherwise could not be easily monitored by GC, NTS approaches usually do not incorporate derivatization, in particular due to the high level of variance.
The advantages of GC-IMS in comparison to established techniques, such as mass spectrometry, are its simple and inexpensive design primarily due to being operated at atmospheric pressure and hence not requiring vacuum pumps [8]. Furthermore, the use of radioactive ionization sources allows for portability, miniaturization, and mechanical robustness and therefore is suitable for field and benchtop applications [52]. Due to efficient ionization, in combination with its fast and sensitive detection, IMS is a universal technique for the analysis of organic and inorganic molecules, atoms, or particles [38]. One potential challenge of IMS analysis is that spectra may contain interference due to widespread ionization, which results in low selectivity. The addition of suitable dopant substances, however, has been shown to overcome these limitations [53,54]. A nonlinear concentration range was previously described for IMS, requiring the careful monitoring of sample concentration to avoid sample saturation. Furthermore, the separation, which is based on CCS, often provides limited information regarding specific qualities concerning size and shape of analytes. However, the drawback of interference caused by spectral complexity and nonlinearities can be overcome by using computer-based analysis tools [55].
The complexity of biological samples results from the presence of a variety of compounds, which provide in their entirety a characteristic HS-GC-IMS spectrum, often referred to as the VOC profile or ‘fingerprint’ [56,57]. HS-GC-IMS has been demonstrated to be an effective technique for the evaluation of VOC profiles of biological samples due to its simple system setup, robustness, and price [44,58,59,60,61]. The chemical fingerprinting of food and beverages in combination with chemometric analysis is widely used for food authentication and ultimately to identify food adulteration and fraud [62]. Furthermore, the VOC profile is influenced by production processes as well as storage conditions. Consequently, process control and quality assurance, such as the control of food freshness or food safety, are topics of interest for NTS using HS-GC-IMS [63,64] techniques.
2. Motivation for Non-Targeted Screening Using HS-GC-IMS
Labelling fraud, e.g., of organic certifications or geographic origin, is the most common type of fraud in agricultural and food markets [65,66]. According to the European Commission, honey and olive oil are particularly affected by mislabeled botanical origin, as well as dilution with inferior or less expensive products [2,67]. Moreover, food adulteration and food fraud have led to cases of economic loss and may pose health risks [2]. The detection of food fraud or adulteration often involves the identification of compounds of unknown molecular composition. Since no identified chemical markers or sets of markers are commonly accessible for a target-based analysis, an analytical approach covering a multitude of parameters in parallel paired with strong discrimination power is required. The currently used methods to determine quality and authenticity, such as sensory analysis and physicochemical analysis [68], are time- and resource-consuming, while lacking sensitivity as well as prediction accuracy, not at least due to univariate analysis. To overcome the limitations of traditional, wet-chemistry-based assays, targeted and non-targeted approaches using chromatographic methods [69,70], often in combination with mass spectrometry [71,72,73], as well as infrared (IR)-based spectroscopy [74,75], and proton nuclear magnetic resonance (1H NMR) spectroscopy [76,77] have been discussed for various applications. However, to obtain the required reproducibility needed for chemometric analysis, time-consuming sample preparation, including precise adjustments of pH, water content or particle size, have been reported in combination with the mentioned methods. Furthermore, the high costs of ownership and maintenance, as well as the requirement for expert knowledge, may limit applications. Finally, high-end instrumentation also requires suitable laboratory infrastructures, which are usually not available at the point of care. Thus, robust, inexpensive, and fast analytical methods, such as HS-GC-MS, are needed, which require little or no sample preparation but deliver high selectivity.
Application examples for HS-GC-IMS with NTS:
A plethora of studies have shown the potential of HS-GC-IMS in combination with NTS for monitoring food quality or confirmation of geographical or botanical origin, despite the complexity of the samples. For example, HS-GC-IMS with NTS has been widely applied for the classification of olive oil between high-priced type 1 extra-virgin olive oil (EVOO), medium-priced type 2 virgin olive oil (VOO or OO), and non-edible type 3 olive oil, also known as pomace olive oil (POO) or lampante (virgin) olive oil (L(V)OO) [32,33,78,79]. Furthermore, HS-GC-IMS with NTS was successfully used for reliable classification of geographical origins for both olive oil (EVOO) [34,80,81] and wine [30]. Moreover, HS-GC-IMS with NTS was applied for the classification of honey according to botanical origin [52,81,82], as well as for the detection and quantification of honey adulterated with sugar cane or corn syrups [83,84]. Recently, HS-GC-IMS with NTS has been applied to assess the freshness of food [85] and for the detection of mold formation on milled rice [86], peanut kernels [87], and wheat kernels [88]. Further examples of recent studies using HS-GC-IMS with NTS are provided in Table 1.

Table 1.
Recent studies of HS-GC-IMS with NTS.
3. NTS-Workflow
NTS aims to identify the compounds of unknown molecular composition. The workflow for NTS generally consists of sample preparation, instrumental analysis, and post-acquisition data processing [93]. Since little or no a priori knowledge of the chemical structures and behavior of compounds is required, NTS approaches benefit from gentile sample preparation, robust instrumental analysis, and standardized data processing. The workflow for NTS using GC-IMS is shown in Figure 2. The first step, data acquisition, involves sample preparation and subsequent extraction and separation of VOCs. The collected data are then preprocessed and analyzed in the data-processing step. Since no pre-existing knowledge is used, the entire spectral fingerprint obtained by HS-GC-IMS analysis is subject to data analysis and classification or quantification models being built using machine learning tools. In the third step, model interpretation, key compounds are identified, which are extracted through back projection of loadings. Complementary analyses, such as sensory analysis of GC-MS measurements, provide further insight into sample composition and can be used to improve model accuracy. The model coherency is evaluated and finally applied for benchtop profiling.
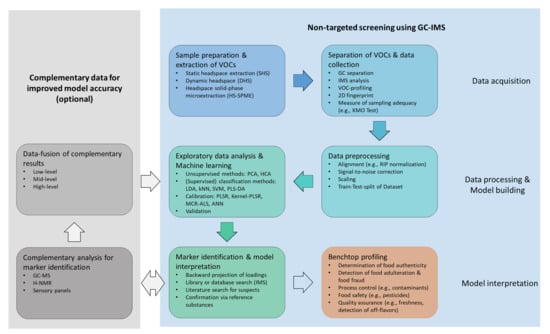
Figure 2.
Workflow for non-targeted screening using HS-GC-IMS.
3.1. Data Acquisition
In untargeted approaches, sample preparation strategies need to be suitable for a variety of matrices and enable the extraction of a wide range of compounds [49]. In order to minimize the extraction-related formation of artifacts, static headspace extraction (SHS) is commonly used for the analysis of volatile compounds in samples of different origin [94]. The limited sensitivity and the bias related to the extraction of low-volatile compounds are generally considered as the main limitations of SHS methods, which can be overcome through dynamic headspace extraction (DHS) [95]. If the preconcentration of analytes is needed for analysis, high-concentration-capacity HS techniques, such as SPME can be used, where the selective isolation of compounds of interest from samples with minimal matrix contamination is crucial [95,96,97]. However, SPME is less commonly applied to IMS with NTS, possibly due to drawbacks which have been associated with commercially available extraction phases, such as the low recovery of polar compounds, insufficient matrix-compatibility, and the reduced life-time and robustness of the coating against matrix macromolecules [98].
VOC profiling based on IMS in combination with NTS has been shown to be an effective tool for various classification tasks, such as the differentiation between various green teas with chestnut-like flavor [5]. Using PLS-DA, the authors were able to obtain an accuracy of 95.6% for the classification of tender, pure, and roasted green tea aromas. However, due to the complexity of many biogenic samples, IMS with direct sample introduction is often not sufficient for NTS approaches, requiring extraction and/or column separation techniques. Shuai and coworkers analyzed adulterated flaxseed oils after n-hexane extraction using a handheld IMS and subsequent data analysis. A recursive support vector machine (R-SVM) led to a model with an accuracy of 93.1% for the identification of adulterated flaxseed oil [44].
To address the complexity of biogenic samples, pre-IMS separation techniques are commonly applied. To avoid the clustering and formation of heteromers in the ionization or drift region, IMS devices are commonly coupled to GC. The spectra obtained by HS-GC-IMS are 2D, with the GC retention time as the first dimension and the IMS drift time as the second dimension. The complexity of biological samples results from a plethora of compounds, which provide in their entirety a characteristic GC-IMS spectrum that is often referred to as VOC profile or ‘fingerprint’ [56,57]. For GC separation in combination with IMS and NTS, both the capillary column (CC) and multicapillary column (MCC) have been coupled to IMS. Among others, nonpolar CCs, such as fused silica SE-54-CB (94% methyl, 5% phenyl, 1% vinyl silicone) [32,33,86,89,92], as well as polar CCs, such as DB-225 (25% phenyl, 25% cyanopropyl methyl siloxane) [79,81], were used for HS-GC-IMS with NTS. For MCCs, which are composed of a large number of parallel glass capillaries (~1000), nonpolar OV-5 (5% diphenyl, 95% dimethylpolysiloxane) was used [78,99]. Compared to the ordinary CC, MCCs allow the separation to be carried out at an elevated speed and can be operated at increased carrier gas flows, which may pose an advantage for IMS analysis [100]. However, Garrido-Delgado and coworkers obtained the better predictive accuracy for the classification (kNN classifier with k = 3) between different qualities of olive oil (EVOO, OO, and LVOO), when CC-IMS (83%) versus MCC-IMS (79%) was employed for the separation of the VOCs [32].
The data acquisition is finalized with the evaluation of the suitability of the collected data for factorial analysis. This sampling adequacy is commonly determined using the Kaiser–Meyer–Olkin (KMO) test, which is a measure of the proportion of variance among variables [101].
3.2. Data Processing and Model Building
The ion velocity and thus the signal position are highly dependent on temperature and ambient pressure; therefore, the drift-time alignment is a crucial preprocessing step [36,102]. For baseline correction, drift times are often normalized to the RIP position, which is, however, not sufficient for the determination of absolute mobilities [103]. Alternatively, a reference peak/substance can be used for alignment, which is recommended when GC-IMS measurements are performed with long separation columns. In a second preprocessing step, multiple measurements are averaged, and the background noise is subtracted. Scaling methods, such as unit variance (UnVa) scaling, Pareto (Par) scaling, and mean centering (Ctr) scaling, can be alternatively used for data set normalization [5]. In UnVa scaling, the variance of each variable is unified using the standard deviation, while in Par scaling, the square root of the standard deviation is used for normalization, whereas Ctr scaling emphasizes the analysis of data fluctuations and the large-fold change in the data [82]. For further noise reduction, smoothing algorithms, such as Savitzky-Golay or Gaussian smoothing, are applied to the spectra [52]. Interfering peaks and nonrelevant areas can be removed by the careful selection of relevant GC retention time and drift-time ranges. Prior to data analysis, the 2D spectra (GC retention time × drift time) are unfolded into arrays, which are then concatenated to the final data set (samples × measurements). For supervised data analyses, the data set is finally split into training and test sets, where the training set is used for model building and the test set for model validation.
Nonlinear behavior has been described for the ratio of the RIP and the distribution between the protonated monomer and the proton-bound dimer [36,37,104]. By increasing the molecular concentration of the analyte, the monomer peak decreases, while the dimer peak increases. These typical nonlinear monomer-dimer distributions in IMS are often accompanied by tailing effects of the monomer or dimer peak [105], requiring careful consideration for quantification. In univariate regression (UR), the peak area or intensity of a single peak is correlated against concentration [46]. Due to the nonlinear behavior between the monomer and dimer peak, linearity can only be approximated for narrow concentration ranges when using the regression analysis based on single-peak analysis [36]. Occasionally, the sum of the volume of monomer and dimer is correlated against the concentration [106]; however, in complex mixtures, quantification is further complicated by overlapping peaks, competitive ionization situations between coeluting analytes and the occurrence of multicomponent cluster ions (heterodimeric ions), which are composed of coeluting analytes [37].
3.2.1. Exploratory Data Analysis and Machine Learning Techniques
Due to the complexity of food matrices, abstract terms such as ‘food quality’ and ‘authenticity’ are the sum of multiple characteristics, which implies that the correlation to a single analyte or analytical technique is often not sufficient or even impossible; hence, multivariate variate data analysis (MVA) techniques are required [81]. MVA approaches can be divided into exploratory, classification, and quantitative regression methods. Exploratory methods, such as PCA or HCA, are unsupervised and typically used for pattern recognition, whereas classification methods such as PCA-LDA, kNN, or PLS-DA are supervised methods. In this context, PLS-DA is a special case of classification, as it basically uses a regression approach with class boundaries instead of single values, as in quantitative regressions. For sample quantification, the latter methods, such as PLS regression, Kernel-PLSR, MCR-ALS, or ANN are commonly applied.
Principal component analysis (PCA) is a powerful technique for unsupervised discovery of patterns in data, which is further used for dimension reduction [107]. The information extracted from a data matrix is explained by principal components (PCs), which are orthogonal (mathematically independent) to each other. Since PCA models are predicted without labels or validation by test data, they are generally considered unsupervised. Unsupervised statistical methods are exploratory methods that can be used to study data structures and search for clusters of samples [108]. Hierarchical cluster analysis (HCA) of PCA models in a tree-like diagram (dendrogram) is, e.g., used for the visualization of multivariate association and sample similarities [109]. An extension of PCA for processing three-dimensional (3D) data is provided by multiway principal component analysis (MPCA) [110], which has been applied for the feature extraction of GC-IMS matrices, without prior transformation of the 2D data [99]. Further alternatives to PCA and PLS are models based on PARAFAC [111] or Tucker3 [112], which may also be used with 3D data.
Compared to unsupervised techniques, which provide predictions without labels or target variables, supervised techniques aim to build models able to predict target variables. In supervised learning, several data points or samples are described using predictor variables or features and target variables. For classification tasks, the scores obtained by the unsupervised exploratory analysis are combined with subsequent supervised pattern recognition techniques to distinguish samples according to defined categories. Among PCA-based qualitative methods are linear discriminant analysis (LDA) and k-nearest neighbors (kNN). Whereas PCA-LDA maximizes the interclass variance, kNN assigns the category most common among the k-nearest neighbors. The downside of using PCA-based methods is that the correlation between dependent and independent variables are not considered during PCA analysis, which can result in the loss of information included in higher PCs [107]. An alternative is provided by partial least squares (PLS), where the scores are calculated by considering the relationship between the independent and dependent variables.
Other supervised methods used in NTS with HS-GC-IMS are gradient boosting (e.g., XGBoost) [31], decision tree classification (Tree) [91], logistic regression (Regressor) [91], orthogonal partial least-squares discriminant analysis (OPLS-DA), quadratic discriminant analysis (QDA) [30], or soft independent modeling of class analogy (SIMCA) [82]. Furthermore, nonlinear classifications are often performed using support vector machines (SVMs). By using the kernel trick, which transforms the input data into high-dimensional feature spaces, SVMs can perform nonlinear classifications, in addition to performing linear classification [113]. This is of particular importance when no linear hyperplanes are separating the respective classes.
3.2.2. Model Performance and Validation
The performance of a model is usually measured as ‘accuracy’, which is the fraction of correctly classified samples. The classification accuracy determines the fraction of correctly classified samples for a given sample set. The classification accuracy, however, is susceptible to overfitting and thus should only be used as reference. To prevent overfitting, the data set is split into training and test data. The ratio between training and test data, which is commonly referred to as ‘train–test split’, is usually between 2:1 [88] and 4:1 [33], and sometimes as low as 6:1 [31]. The test set is used to determine the prediction accuracy, which is usually lower than the classification accuracy but more meaningful [88]. For small and inhomogeneous data sets, a single split of the data into training and test set may give misleading results [114]. An alternative model validation is therefore provided by resample methods, such as cross validation (CV), bootstrapping, or permutation testing, where multiple random subsets are generated. For the CV of samples, a subset of the data is held out for use as a validation set, and a model is fit to the remaining data (training set) and used to predict the validation set. The process of generating a subset of data, model fitting, and evaluation is performed repeatedly, and an overall prediction accuracy is determined by averaging the quality of the predictions across the validation sets. Leave-one-out CV leaves out a single observation at a time, while k-fold CV splits the data into k subsets, which are one by one held out as the validation set [107]. For NTS using HS-GC-IMS, 10-fold CV is commonly performed [31,52], alongside with leave-one-out CV [80,85]. Bootstrapping is a resampling method that can be used as an alternative to CV to estimate the prediction performance of a model with a low number of training samples. Due to drawing with replacement, a bootstrapped data set contains the same number of cases as the original data set, and it can contain multiple instances of the same original cases [114]. Another resampling method is permutation testing, where labels are switched on data points when performing the test statistics. Both bootstrapping [32,78] and permutation tests [82,92] have been applied with HS-GC-IMS and NTS.
The success rate of different chemometric models for classification depends on many factors, such as botanical origin of the samples, number of samples or the selection of PCs, PLS, and k values. Gerhardt and coworkers compared different chemometric methods for the classification of the botanical origins of honey (acacia, canola, and honeydew) using a resolution-optimized HS-GC-IMS. They found a 98.6% accuracy with PCA-LDA, 86.1% with kNN (k = 5), and 97.0% with PLS-DA after employing 10-fold CV [52]. Quality assessment of olive oils based on temperature-ramped HS-GC-IMS and sensory evaluation conducted by Gerhardt and coworkers reported a 83.3% accuracy with PCA-LDA, 73.8% with kNN (k = 5) and 88.1% with SVM models, after employing 10-fold CV [79].
Artificial neural networks (ANNs) are a powerful modeling approach, which is vaguely inspired by the biological neural networks in the brain [115]. Due to their hidden layers, ANNs have the ability to capture complex interactions present in biological samples. Furthermore, ANNs can be applied for pattern recognition, classification tasks, and quantification problems, as well as data preprocessing. ANN has recently the gained attention for applications in food science and technology, while often being limited by the requirement of having sufficiently large data sets [116,117]. Zhu and coworkers have shown the superior prediction performance of ANN (89.5%) over PCA-LDA (65.7%), PLS-DA (58.7%), kNN (k = 5, 60.8%) and SVM (51.8%), and XGBoost (81.8%), for the classification of Sauvignon Blanc via SHS-GC-IMS [31]. Vega-Márquez and coworkers evaluated a deep learning network and five different benchmark methods for the classification of olive oil samples into EVOO, OO, and LVOO, based on HS-GC-IMS spectra obtained for 701 olive oil sample from two different harvests [91]. Among the five benchtop models used for comparison, XGBoost offers the best accuracy of 85.7%, compared to SVM (83.1%), kNN (84.5%), Tree (78.3%), and Regressor (85.5%). However, even better results were achieved with the deep learning approach, obtaining an accuracy of 88.8%, which underlines the potential of ANNs.
3.2.3. Quantification Tasks
For quantification tasks, partial least squares regression (PLSR) has become the standard method used in chemometrics, including the fields of sensorial analysis in food chemistry [107,110]. PLSR is used to describe the relationship between two data matrices, X (experimental data) and Y (actual concentrations), which are decomposed into X = TPT + E and Y = UQT + F, by finding the maximum covariance and linear relationship between the score matrices T and U. P and Q represent the loading matrices and E and F the matrices of residuals. After multiplying X with a nonlinear function, linear PLS can be applied as described above. After the successful implementation of multivariate models for the interpretation of the training data, the model performance is commonly tested using test data. The model quality can be determined by different figures of merit, such as the determination coefficient (R2) and the relative percentage error of prediction (RE), as well as root mean square error (RMSE), systematic error (Bias), or standard error of prediction (SEP).
One limitation to PLSR is the typically lower performance on nonlinear and heteroscedastic data, as is partially the case for IMS data. Several studies have analyzed nonlinear IMS data, as shown by the quantification of histamine in tuna stomach [7] or allergenic fragrance compounds, such as citral, in complex cosmetic products [38]. Nonlinear relationships between the matrices T and U can be described by kernel PLSR (also known as nonlinear PLSR), where the data are transformed into higher-dimensional spaces using the kernel trick [118].
An alternative to PLSR or kernel PLSR is provided by multivariate curve resolution alternating least squares (MCR-ALS). MCR decomposes the initial data matrix D with n sample spectra and m data points in the spectra to D = CST + E. The matrices C (n × l) and S (l × m) represent the concentration and spectra profiles of D for l components, while the matrix E (n × m) contains the residuals not explained by C and S [119]. MCR-ALS may deconvolute overlaying spectra from coelution and reconstitute the pure spectra for quantitation. A comparison of UR, PLSR, MCR-ALS, and Kernel-PLSR for the quantification of fragrances using GC-IMS analysis was provided by Brendel and coworkers. For both, a mixture of geraniol and citral as well as a mixture of the citral and cinnamal, Kernel-PLSR demonstrated the superior ability for the quantification of the nonlinear relationships of GC-IMS data since Kernel-PLSR was able to significantly reduce the RE of prediction and increase R2 of calibration.
3.3. Model Interpretation
Once a robust and reliable model is built, whose predictive abilities are sufficient for the chosen task, the model should be checked for plausibility. Therefore, the influencing variables, also referred to as markers, are extracted, identified, and subsequently used for interpreting and explaining the model. By using PCA for marker extraction, the loadings obtained by multivariate analysis are projected backwards into the original data space and are subsequently evaluated either manually or automatically. This allows one to identify the signals with the main influence for separation in the respective principal components with relation to the original data (Figure 3). Another example is the use of PLS-DA, where volcano plots and variable importance in projection (VIP) analysis have been performed to determine the final markers for each discrimination task [56]. Compounds with a VIP score > 1 are generally considered as suitable markers [120].
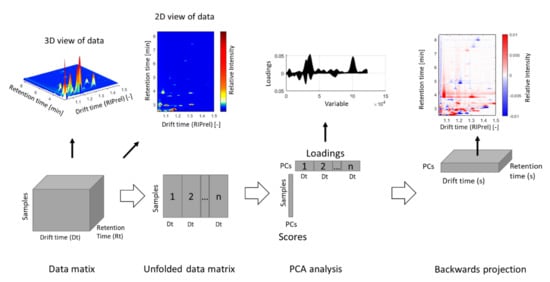
Figure 3.
Data analysis and backwards projection of loadings for identification of key substances.
Web-based platforms, such as MetaboAnalyst (http://www.metaboanalyst.ca, accessed on 1 July 2021) [121] and XCMS Online (https://xcmsonline.scripps.edu/, accessed on 1 July 2021) [122], are designed to handle comprehensive untargeted metabolomic data. Compared to GC-MS data, which include m/z information, GC-IMS data are intrinsically limited to (normalized) drift times and retention times. While MetaboAnalyst has also been used for the processing of GC-IMS data, including the preprocessing (normalization and scaling) of data [82], VIP analysis [83], and entire chemometric analysis [123], the lack of m/z information limits the use of these metabolomic data processing tools for compound identification. However, the combination of retention indices and normalized drift times is considered as a reliable alternative; as in particular, drift times are highly reproducible. By comparing markers obtained from the GC-IMS data to databases, which are often provided by IMS manufacturers, the substances of interest can be identified [124,125]. Furthermore, a search of the literature and subsequent confirmation by reference substances can be used for substance identification [126,127,128]. Since coelution and matrix effects can influence GC-IMS data, a common technique is to spike a complex sample with the pure substance. To increase the number of substances identified and to further increase the model’s accuracy, complementary data, such as GC-MS [123], can be used as described in Section 3.4. This procedure is optional, as reliable models for classification and quantification tasks can be built solely from HS-GC-IMS data [44,80]. Typically, these MS detectors are unit-resolved and, as such, again limited in their selectivity in comparison to high-resolution systems, such as TOF or Orbitrap systems. However, a full spectral interpretation is not always necessary in NTS approaches. Some studies solely detect markers without subsequent identification of substances [43,80,84].
NTS is a powerful approach for complex classification and quantification tasks. However, next to model overfitting, the transferability of a model to new data poses a major challenge. A dramatic example is given by Contreras and coworkers who analyzed 701 olive oil samples from the years 2014–15 and from 2015–16 for the classification into EVOO, OO, and LVOO, using HS-GC-IMS with NTS [33]. A model built with olive oil samples from 2014–15 obtained a prediction percentage of 67.8%. A better prediction success was achieved for a model built with olive oil samples from 2014–15 and 2015–16, obtaining 79.4% accuracy. However, when applying the NTS model built with samples from the year 2014–15 to predict the years 2015–16, a prediction success of only 36.0% was achieved, revealing the low transferability of the model. This is however not an effect solely to be attributed to GC-IMS but rather is a general issue of profiling techniques so far and is mainly driven by typically high analytical variance together with a high biological variance of the samples.
To achieve good predictive abilities and transferability of a model to new data, large sample sets containing independent and diverse samples are necessary; however, studies are often limited by sample availability. To overcome this issue and to reveal influencing factors, a comprehensive evaluation of the model should be performed. The identification of influencing substances, for instance, may reveal the characteristic compounds or classes of substances in which the classes differ. A subsequent comprehensive evaluation can determine which factors may be caused by systematic errors and which factors allow for true differentiation between classes. High predictive accuracies for classification and quantification tasks can be achieved without substance identification, yet a comprehensive evaluation of the model helps to detect strategic errors, such as a narrow sample distribution, and thus should always be part of NTS approaches. Due to its robustness and comparably low prices, HS-GC-IMS is suitable for benchtop profiling. In combination with NTS approaches, HS-GC-IMS data can be used for the implementation of models, which are suitable for classification and quantification task in various fields.
3.4. Complementary Data (Optional)
Due to the complexity of biological matrices, the substance identification with GC-IMS data alone can be challenging, which is why stand-alone IMS are rarely used to investigate the sample composition. Complementary techniques, such as GC-MS [129] or 1H NMR [130], are often used to identify decisive marker substances [131].
Next to marker identification, complementary data can be combined with GC-IMS data to build a multimodal model. The process of integrating multiple data sources, which is commonly known as ‘data fusion’, has the potential to increase model accuracy and reliability, while reducing interferences and error rates [132]. The process of data fusion is categorized as low-level, mid-level (or intermediate-level), and high-level data fusion, depending on the fusion strategies used [133]. In low-level (or data-level) fusion approaches, data from all sources are preprocessed, concatenated into a common data matrix, and subsequently analyzed using classical multivariate methods, such as PCA or PLS (see Section 3.2). Mid-level data fusion approaches, which are also referred to as feature-level data fusion, are based on the extraction of relevant features from each data source separately [87]. Several latent variables, such as the score values from PCA or PLS, are selected for this feature-based data fusion, concatenated, and subsequently analyzed using classical multivariate methods. The third fusion approach is high-level or decision-level data fusion, where completely independent models are calculated from each data set; this level is the most challenging due to the determination of the ideal parameters for each separate multivariate model. Careful considerations of parameters, together with the implementation of a voting or scoring scheme, which prioritizes results from different data sources, can provide a combined model, which outperforms the individual models [134].
Schwolow and coworkers showed that the data fusion approach significantly increased both the predictive power and the robustness of the resulting classification model for the determination of geographical origin of olive oil [81]. The prediction accuracy obtained for Fourier-transform infrared (FT-IR) data alone was 67% and for GC-IMS 78%, while resulting a perfect score for a low-level data fusion approach using the complementary chemical information provided by GC-IMS and FT-IR analysis. The extra effort needed for data fusion approaches, however, is not always rewarded. Gu and coworkers classified fungal growth on peanut kernels into potentially aflatoxigenic fungi and non-aflatoxin producing fungal species using HS-GS-IMS and fluorescence measurements [87]. The predictive accuracy for HS-GC-IMS measurements alone was 93.3% with an OPLS-DA model, while low-level data fusion reduced the accuracy to 90%. The predictive accuracy of the OPLA-DA after mid-level data fusion using VIP > 1.3 further decreased to 86.7%. Only a mid-level data fusion using 10 PCs increased the model accuracy to 96.7%.
Another approach to increase the discrimination power of a classification model is the parallel analysis of IMS and MS, which has recently gained attention for classification and quantification tasks [90,106,127]. Although IMS and MS cannot be seen as fully complementary methods, in particular due to the identical volatile fraction being monitored by both techniques [90], it was shown that the soft ionization and drift-time-based ion separation on the one hand and a hard ionization and m/z-based separation on the other hand improved substance identification in the case of coelution in hops analysis [127]. While complementary data in general can increase the interpretability and accuracy of a model developed with HS-GC-IMS, they are optional for NTS via GC-IMS.
4. Comparison of NTS and Targeted Strategies
An alternative approach to NTS is targeted screening. While NTS of HS-GC-IMS data uses no pre-existing knowledge and the entire spectral fingerprint is subject to data analysis, for targeted analysis, specific markers are chosen prior to the data analysis. The markers used for targeted screening can be either handpicked or mathematically determined [33,135]. One-way analysis of variance (ANOVA), using for example a Tukey’s test, is often applied to identify volatile compounds which exhibit significant differences, commonly quantified at a 5% significance level (p ≤ 0.05) [59]. Various other methods, such as Gabor filters, local binary pattern, Haar, and histograms of oriented gradients (HOG), have been proposed for feature extraction [136,137]. Chen and coworkers applied MPCA and HOG for feature extraction and data reduction of MCC-IMS data, with subsequent canonical discriminant analysis for the generation of nonlinear boundaries, for the successful quantification of the adulteration degree of canola oil. A predictive accuracy of more than 95.2% was reported for a PLS model, which was obtained using a train–test split of 70–30 [99]. Using targeted approaches and applying PCA-kNN, the same authors reported a successful classification of rapeseed oils according to their quality (grade 1–4) and a successful determination of vegetable oil according to its botanical origin (sesame oil, rapeseed oil, and camellia oil). For the classification of the rapeseed oil quality, the colorized differences method was applied to CC-IMS data, resulting in 34 peaks of interest and a predictive accuracy of 100% [138]. Furthermore, Otsu’s method and colorized differences method was used for automatic peak detection, resulting in 88 peaks of interest and a predictive accuracy of 98.3% for the classification of vegetable oil using MCC-IMS data [139]. The advantage of preselecting markers with significant differences is the simultaneous reduction of noise in the data, which, however, includes the risk of overlooking valuable information.
NTS approaches (spectral fingerprinting) have also been directly compared to targeted approaches (extraction of specific markers) for the analysis of IMS data. Garrido-Delgado and coworkers compared targeted and NTS approaches for the classification of olive oil into EVOO, OO, and LVOO, using data obtained by MCC coupled to IMS [78]. A PCA-LDA model was used for data reduction and data clustering, followed by kNN (k = 3) for classification, obtaining a prediction percentage of 79% for the targeted strategy and 85% for NTS strategy. For the classification of olive oil harvested in 2014–15, Contreras and coworkers obtained a prediction percentage of 56.9% for the targeted strategy and 67.8% for the NTS strategy [33]. An improved prediction success was achieved for models built with olive oil samples from 2014–15 and 2015–16, obtaining 74.3% for the targeted strategy and 79.4% for the NTS, hence suggesting superior abilities of the NTS approach versus the selection of specific markers. By contrast, the authors also reported that a targeted model built with samples from the years 2014–15 (prediction success of 51.6%) was superior to the NTS approach (prediction success of 36.0%) when applied to the years 2015–16. Both models built with the data from the years 2014–15 show weak prediction abilities for the prediction of samples from the following year, revealing some fundamental challenges in data science: the predictive ability of a model is highly dependent on the number of samples as well as on the sample diversity. Both approaches include the risk of overfitting to a specific problem [33].
Arroyo-Manzanares and coworkers likewise obtained superior classification accuracy using HS-GC-IMS for a model based on a targeted marker selection (100%) compared to a model based on the whole spectral fingerprint (90%) for the distinction between dry-cured Iberian ham from pigs fattened on acorns and pasture or on feed [89]. However, the model based on marker selection was built using OPLS-DA, while kNN (k = 3) and PCA-LDA were used for the model based on spectral fingerprints; hence, the differences in the predictability of the models may result from the use of different mathematical tools and do not provide inferences about targeted and non-targeted approaches. Gu and coworkers by contrast obtained better classification results with the NTS versus a targeted approach for distinguishing between fungal infections of wheat kernels, as well as for the quantification of fungal colony counts [88]. In conclusion, NTS and targeted screening approaches are both effective tools for data analysis, with different challenges and application areas.
5. Conclusions
HS-GC-IMS in combination with an NTS approach provides an effective tool for classification and quantification tasks not at least due to low maintenance and easy handling of the instruments [9]. In combination with HS-GC and machine learning, IMS has been demonstrated to be an effective technique for various classification and quantification tasks, such as for the determination of food authenticity as well as for the detection of food adulteration and food fraud. Furthermore, HS-GC-IMS has been applied in the field of process control, for example for the early detection of microbial contaminants or in the field of food safety for the detection of pesticides. HS-GC-IMS is also found in the field of quality assurance for the detection of food freshness or the detection of off-flavors. Recently, HS-GC-IMS in combination with deep learning approaches has shown promising results for further improvement of model accuracies. HS-GC-IMS is suitable for benchtop profiling, due to its robustness and comparably low purchase and running costs. For benchtop analysis, MDV-based methods for HS-GC-IMS devices can be implemented in the laboratory and subsequently applied for food analysis in various fields. A common approach for further market acceptance is the combination of IMS instrumentation with HS-GC-MS, which alone has already been applied to a wide variety of applications.
Author Contributions
Writing—original draft preparation, C.C.; funding acquisition, P.W.; writing—review and editing, P.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Zentrales Innovationsprogramm Mittelstand (ZIM) of the Federal Ministry for Economic Affairs and Energy, grant number FZ4014707AW9.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds, etc., are available from the authors of the respective publications.
Glossary
| Abbreviations | Description |
| 1H NMR | proton nuclear magnetic resonance |
| 2D | two-dimensional |
| 3D | three-dimensional |
| Am-241 | americium-241 |
| ANN | artificial neural networks |
| ANOVA | one-way analysis of variance |
| APPI | atmospheric pressure photo ionization |
| Bias | systematic error |
| CC | capillary column |
| CCS | collision cross section |
| CD | corona discharge |
| Ctr | mean centering |
| CV | cross validation |
| D(I)MS | differential ion mobility spectrometry |
| DHS | dynamic headspace extraction |
| DTIMS | drift-tube ion mobility spectrometry |
| E-nose | electric nose |
| EVOO | extra-virgin olive oil |
| FAIMS | high-field asymmetric waveform ion mobility spectrometry |
| ft | film thickness |
| FT-IR | Fourier-transform infrared spectroscopy |
| GC | gas chromatography |
| GC–IMS | gas chromatography-ion mobility spectrometry |
| H+[H2O]n | proton-water clusters with n number of water molecules |
| H-3 | tritium |
| HCA | hierarchical cluster analysis |
| HOG | histograms of oriented gradients |
| HS | headspace |
| ID | inner diameter |
| IMS | ion mobility spectrometry |
| K0 | reduced ion mobility |
| KMO | Kaiser–Meyer–Olkin |
| kNN | k-nearest neighbors |
| L(V)OO | lampante (virgin) olive oil |
| LC | liquid chromatography |
| LDA | linear discriminant analysis |
| LDI | laser desorption/ionization technique |
| LOO | leave-one-out |
| LS | library search |
| M2H+ | proton-bound dimers |
| MCC | multicapillary column |
| MCR-ALS | multivariate curve resolution alternating least squares |
| MH+ | protonated monomers |
| MPCA | multiway principal component analysis |
| MVA | multivariate variate data analysis |
| Ni-63 | nickel-63 |
| NTS | non-targeted screening |
| OPLS-DA | orthogonal partial least-squares discriminant analysis |
| Par | Pareto |
| PC(A) | principal component (analysis) |
| PLS(R) | partial least squares (regression) |
| POO | pomace olive oil |
| (Q)DA | (quadratic) discriminant analysis |
| R² | determination coefficient |
| RE | relative error of prediction |
| Regressor | logistic regression |
| RIP | reactant ion peak |
| RMSE | root mean square error |
| RS | reference substances |
| (R)-SVM | (recursive) support vector machine |
| SEP | standard error of prediction |
| SHS | static headspace extraction |
| SIMCA | soft independent modeling of class analogy |
| SPME | solid-phase microextraction |
| TIMS | trapped ion mobility spectrometry |
| Tree | decision tree classification |
| TWIMS | travelling tube ion mobility spectrometry |
| UnVa | unit variance |
| UR | univariate regression |
| UV | ultraviolet light |
| VIP | variable importance in projection |
| VOC | volatile organic compound |
| (V)OO | (virgin) olive oil |
References
- Jackson, L.S. Chemical food safety issues in the United States: Past, present, and future. J. Agric. Food Chem. 2009, 57, 8161–8170. [Google Scholar] [CrossRef]
- Moore, J.C.; Spink, J.; Lipp, M. Development and application of a database of food ingredient fraud and economically motivated adulteration from 1980 to 2010. J. Food Sci. 2012, 77, R118–R126. [Google Scholar] [CrossRef]
- Van de Brug, F.J.; Lucas Luijckx, N.B.; Cnossen, H.J.; Houben, G.F. Early signals for emerging food safety risks: From past cases to future identification. Food Control 2014, 39, 75–86. [Google Scholar] [CrossRef]
- Vuckovic, D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2012, 403, 1523–1548. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, H.; Yao, Y.; Hua, J.; Yang, Y.; Dong, C.; Deng, Y.; Wang, J.; Li, H.; Jiang, Y.; et al. Rapid volatiles fingerprinting by dopant-assisted positive photoionization ion mobility spectrometry for discrimination and characterization of Green Tea aromas. Talanta 2019, 191, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Maecker, R.; Vyhmeister, E.; Meisen, S.; Rosales Martinez, A.; Kuklya, A.; Telgheder, U. Identification of terpenes and essential oils by means of static headspace gas chromatography-ion mobility spectrometry. Anal. Bioanal. Chem. 2017, 409, 6595–6603. [Google Scholar] [CrossRef]
- Cohen, G.; Rudnik, D.D.; Laloush, M.; Yakir, D.; Karpas, Z. A Novel Method for Determination of Histamine in Tuna Fish by Ion Mobility Spectrometry. Food Anal. Methods 2015, 8, 2376–2382. [Google Scholar] [CrossRef]
- Borsdorf, H.; Eiceman, G.A. Ion Mobility Spectrometry: Principles and Applications. Appl. Spectrosc. Rev. 2006, 41, 323–375. [Google Scholar] [CrossRef]
- Cumeras, R.; Figueras, E.; Davis, C.E.; Baumbach, J.I.; Gràcia, I. Review on ion mobility spectrometry. Part 2: Hyphenated methods and effects of experimental parameters. Analyst 2015, 140, 1391–1410. [Google Scholar] [CrossRef]
- Schmidt, C.; Jaros, D.; Rohm, H. Ion Mobility Spectrometry as a Potential Tool for Flavor Control in Chocolate Manufacture. Foods 2019, 8, 460. [Google Scholar] [CrossRef]
- Karpas, Z.; Guamán, A.V.; Calvo, D.; Pardo, A.; Marco, S. The potential of ion mobility spectrometry (IMS) for detection of 2,4,6-trichloroanisole (2,4,6-TCA) in wine. Talanta 2012, 93, 200–205. [Google Scholar] [CrossRef]
- Tzschoppe, M.; Haase, H.; Höhnisch, M.; Jaros, D.; Rohm, H. Using ion mobility spectrometry for screening the autoxidation of peanuts. Food Control 2016, 64, 17–21. [Google Scholar] [CrossRef]
- Rearden, P.; Harrington, P.B. Rapid screening of precursor and degradation products of chemical warfare agents in soil by solid-phase microextraction ion mobility spectrometry (SPME–IMS). Anal. Chim. Acta 2005, 545, 13–20. [Google Scholar] [CrossRef]
- Cook, G.W.; LaPuma, P.T.; Hook, G.L.; Eckenrode, B.A. Using gas chromatography with ion mobility spectrometry to resolve explosive compounds in the presence of interferents. J. Forensic Sci. 2010, 55, 1582–1591. [Google Scholar] [CrossRef] [PubMed]
- Zalewska, A.; Pawłowski, W.; Tomaszewski, W. Limits of detection of explosives as determined with IMS and field asymmetric IMS vapour detectors. Forensic Sci. Int. 2013, 226, 168–172. [Google Scholar] [CrossRef]
- Eiceman, G.A.; Karpas, Z.; Hill, H.H. Ion Mobility Spectrometry, 3rd ed.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2016. [Google Scholar]
- Browne, C.A.; Forbes, T.P.; Sisco, E. Detection and identification of sugar alcohol sweeteners by ion mobility spectrometry. Anal. Methods 2016, 8, 5611–5618. [Google Scholar] [CrossRef]
- Reyes-Garcés, N.; Gómez-Ríos, G.A.; Souza Silva, E.A.; Pawliszyn, J. Coupling needle trap devices with gas chromatography-ion mobility spectrometry detection as a simple approach for on-site quantitative analysis. J. Chromatogr. A 2013, 1300, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Siems, W.F.; Hill, J.H.H.; Hannan, R.M. Analytical determination of nicotine in tobacco by supercritical fluid chromatography–ion mobility detection. J. Chromatogr. A 1998, 811, 157–161. [Google Scholar] [CrossRef]
- Raatikainen, O.; Reinikainen, V.; Minkkinen, P.; Ritvanen, T.; Muje, P.; Pursiainen, J.; Hiltunen, T.; Hyvönen, P.; Wright, A.v.; Reinikainen, S.-P. Multivariate modelling of fish freshness index based on ion mobility spectrometry measurements. Anal. Chim. Acta 2005, 544, 128–134. [Google Scholar] [CrossRef]
- Pfeifer, K.B.; Rumpf, A.N. Measurement of ion swarm distribution functions in miniature low-temperature co-fired ceramic ion mobility spectrometer drift tubes. Anal. Chem. 2005, 77, 5215–5220. [Google Scholar] [CrossRef]
- Laakia, J.; Adamov, A.; Jussila, M.; Pedersen, C.S.; Sysoev, A.A.; Kotiaho, T. Separation of different ion structures in atmospheric pressure photoionization-ion mobility spectrometry-mass spectrometry (APPI-IMS-MS). J. Am. Soc. Mass Spectrom. 2010, 21, 1565–1572. [Google Scholar] [CrossRef][Green Version]
- Vautz, W.; Baumbach, J.I.; Jung, J. Beer Fermentation Control Using Ion Mobility Spectrometry—Results of a Pilot Study. J. Inst. Brew. 2006, 112, 157–164. [Google Scholar] [CrossRef]
- Sielemann, S.; Baumbach, J.I.; Schmidt, H.; Pilzecker, P. Quantitative analysis of benzene, toluene, and m-xylene with the use of a UV-ion mobility spectrometer. Field Anal. Chem. Technol. 2000, 4, 157–169. [Google Scholar] [CrossRef]
- Parchami, R.; Kamalabadi, M.; Alizadeh, N. Determination of biogenic amines in canned fish samples using head-space solid phase microextraction based on nanostructured polypyrrole fiber coupled to modified ionization region ion mobility spectrometry. J. Chromatogr. A 2017, 1481, 37–43. [Google Scholar] [CrossRef]
- Sheibani, A.; Tabrizchi, M.; Ghaziaskar, H.S. Determination of aflatoxins B1 and B2 using ion mobility spectrometry. Talanta 2008, 75, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Seyed Khademi, S.M.; Telgheder, U.; Valadbeigi, Y.; Ilbeigi, V.; Tabrizchi, M. Direct detection of glyphosate in drinking water using corona-discharge ion mobility spectrometry: A theoretical and experimental study. Int. J. Mass Spectrom. 2019, 442, 29–34. [Google Scholar] [CrossRef]
- Illenseer, C.; Löhmannsröben, H.-G. Investigation of ion–molecule collisions with laser-based ion mobility spectrometry. Phys. Chem. Chem. Phys. 2001, 3, 2388–2393. [Google Scholar] [CrossRef]
- Council Directive 2013/59/Euratom of 5 December 2013 Laying Down Basic Safety Standards for Protection against the Dangers Arising from Exposure to Ionising Radiation. Off. J. Eur. Union 2013. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:32013L0059&rid=6 (accessed on 14 May 2021).
- Chen, T.; Qi, X.; Chen, M.; Lu, D.; Chen, B. Discrimination of Chinese yellow wine from different origins based on flavor fingerprint. Acta Chromatogr. 2020, 32, 139–144. [Google Scholar] [CrossRef]
- Zhu, W.; Benkwitz, F.; Kilmartin, P. Assessment of Sauvignon Blanc Aroma and Quality Gradings Based on Static Headspace-Gas Chromatography-Ion. Mobility Spectrometry (SHS-GC-IMS): Merging Analytical Chemistry with Machine Learning; Cambridge University Press & Assessment: Cambridge, UK, 2020. [Google Scholar] [CrossRef]
- Garrido-Delgado, R.; Del Dobao-Prieto, M.M.; Arce, L.; Valcárcel, M. Determination of volatile compounds by GC-IMS to assign the quality of virgin olive oil. Food Chem. 2015, 187, 572–579. [Google Scholar] [CrossRef]
- Del Contreras, M.M.; Jurado-Campos, N.; Arce, L.; Arroyo-Manzanares, N. A robustness study of calibration models for olive oil classification: Targeted and non-targeted fingerprint approaches based on GC-IMS. Food Chem. 2019, 288, 315–324. [Google Scholar] [CrossRef]
- Gerhardt, N.; Birkenmeier, M.; Sanders, D.; Rohn, S.; Weller, P. Resolution-optimized headspace gas chromatography-ion mobility spectrometry (HS-GC-IMS) for non-targeted olive oil profiling. Anal. Bioanal. Chem. 2017, 409, 3933–3942. [Google Scholar] [CrossRef]
- Griffin, G.W.; Dzidic, I.; Carroll, D.I.; Stillwell, R.N.; Horning, E.C. Ion mass assignments based on mobility measuremets. Validity of plasma chromatographic mass mobility correlations. Anal. Chem. 1973, 45, 1204–1209. [Google Scholar] [CrossRef]
- Pomareda, V.; Guamán, A.V.; Mohammadnejad, M.; Calvo, D.; Pardo, A.; Marco, S. Multivariate curve resolution of nonlinear ion mobility spectra followed by multivariate nonlinear calibration for quantitative prediction. Chemom. Intell. Lab. Syst. 2012, 118, 219–229. [Google Scholar] [CrossRef]
- Ewing, R.G.; Eiceman, G.A.; Stone, J. Proton-bound cluster ions in ion mobility spectrometry. Int. J. Mass Spectrom. 1999, 193, 57–68. [Google Scholar] [CrossRef]
- Brendel, R.; Schwolow, S.; Rohn, S.; Weller, P. Comparison of PLSR, MCR-ALS and Kernel-PLSR for the quantification of allergenic fragrance compounds in complex cosmetic products based on nonlinear 2D GC-IMS data. Chemom. Intell. Lab. Syst. 2020, 205, 104128. [Google Scholar] [CrossRef]
- Borsdorf, H.; Rudolph, M. Gas-phase ion mobility studies of constitutional isomeric hydrocarbons using different ionization techniques. Int. J. Mass Spectrom. 2001, 208, 67–72. [Google Scholar] [CrossRef]
- Vautz, W.; Bödeker, B.; Baumbach, J.I.; Bader, S.; Westhoff, M.; Perl, T. An implementable approach to obtain reproducible reduced ion mobility. Int. J. Ion Mobil. Spectrom. 2009, 12, 47–57. [Google Scholar] [CrossRef]
- May, J.C.; McLean, J.A. Ion mobility-mass spectrometry: Time-dispersive instrumentation. Anal. Chem. 2015, 87, 1422–1436. [Google Scholar] [CrossRef]
- Ewing, M.A.; Glover, M.S.; Clemmer, D.E. Hybrid ion mobility and mass spectrometry as a separation tool. J. Chromatogr. A 2016, 1439, 3–25. [Google Scholar] [CrossRef]
- Schneider, B.B.; Nazarov, E.G.; Londry, F.; Vouros, P.; Covey, T.R. Differential mobility spectrometry/mass spectrometry history, theory, design optimization, simulations, and applications. Mass Spectrom. Rev. 2016, 35, 687–737. [Google Scholar] [CrossRef]
- Shuai, Q.; Zhang, L.; Li, P.; Zhang, Q.; Wang, X.; Ding, X.; Zhang, W. Rapid adulteration detection for flaxseed oil using ion mobility spectrometry and chemometric methods. Anal. Methods 2014, 6, 9575–9580. [Google Scholar] [CrossRef]
- Spietelun, A.; Pilarczyk, M.; Kloskowski, A.; Namieśnik, J. Current trends in solid-phase microextraction (SPME) fibre coatings. Chem. Soc. Rev. 2010, 39, 4524–4537. [Google Scholar] [CrossRef]
- Ruzsanyi, V.; Mochalski, P.; Schmid, A.; Wiesenhofer, H.; Klieber, M.; Hinterhuber, H.; Amann, A. Ion mobility spectrometry for detection of skin volatiles. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 911, 84–92. [Google Scholar] [CrossRef]
- Thomas, C.F.; Zeh, E.; Dörfel, S.; Zhang, Y.; Hinrichs, J. Studying dynamic aroma release by headspace-solid phase microextraction-gas chromatography-ion mobility spectrometry (HS-SPME-GC-IMS): Method optimization, validation, and application. Anal. Bioanal. Chem. 2021, 413, 2577–2586. [Google Scholar] [CrossRef]
- Gómez-Ramos, M.M.; Ferrer, C.; Malato, O.; Agüera, A.; Fernández-Alba, A.R. Liquid chromatography-high-resolution mass spectrometry for pesticide residue analysis in fruit and vegetables: Screening and quantitative studies. J. Chromatogr. A 2013, 1287, 24–37. [Google Scholar] [CrossRef]
- Baduel, C.; Mueller, J.F.; Tsai, H.; Gomez Ramos, M.J. Development of sample extraction and clean-up strategies for target and non-target analysis of environmental contaminants in biological matrices. J. Chromatogr. A 2015, 1426, 33–47. [Google Scholar] [CrossRef]
- Krauss, M.; Singer, H.; Hollender, J. LC-high resolution MS in environmental analysis: From target screening to the identification of unknowns. Anal. Bioanal. Chem. 2010, 397, 943–951. [Google Scholar] [CrossRef]
- Masiá, A.; Campo, J.; Blasco, C.; Picó, Y. Ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry to identify contaminants in water: An insight on environmental forensics. J. Chromatogr. A 2014, 1345, 86–97. [Google Scholar] [CrossRef]
- Gerhardt, N.; Birkenmeier, M.; Schwolow, S.; Rohn, S.; Weller, P. Volatile-Compound Fingerprinting by Headspace-Gas-Chromatography Ion-Mobility Spectrometry (HS-GC-IMS) as a Benchtop Alternative to 1H NMR Profiling for Assessment of the Authenticity of Honey. Anal. Chem. 2018, 90, 1777–1785. [Google Scholar] [CrossRef]
- Brendel, R.; Rohn, S.; Weller, P. Nitrogen monoxide as dopant for enhanced selectivity of isomeric monoterpenes in drift tube ion mobility spectrometry with 3H ionization. Anal. Bioanal. Chem. 2021, 413, 3551–3560. [Google Scholar] [CrossRef]
- Puton, J.; Nousiainen, M.; Sillanpää, M. Ion mobility spectrometers with doped gases. Talanta 2008, 76, 978–987. [Google Scholar] [CrossRef]
- Gabelica, V.; Marklund, E. Fundamentals of ion mobility spectrometry. Curr. Opin. Chem. Biol. 2018, 42, 51–59. [Google Scholar] [CrossRef]
- Wang, S.; Chen, H.; Sun, B. Recent progress in food flavor analysis using gas chromatography-ion mobility spectrometry (GC-IMS). Food Chem. 2020, 315, 126158. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Mesa, M.; Ropartz, D.; García-Campaña, A.M.; Rogniaux, H.; Dervilly-Pinel, G.; Le Bizec, B. Ion Mobility Spectrometry in Food Analysis: Principles, Current Applications and Future Trends. Molecules 2019, 24. [Google Scholar] [CrossRef]
- Márquez-Sillero, I.; Cárdenas, S.; Sielemann, S.; Valcárcel, M. On-line headspace-multicapillary column-ion mobility spectrometry hyphenation as a tool for the determination of off-flavours in foods. J. Chromatogr. A 2014, 1333, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Delgado, R.; Dobao-Prieto, M.M.; Arce, L.; Aguilar, J.; Cumplido, J.L.; Valcárcel, M. Ion mobility spectrometry versus classical physico-chemical analysis for assessing the shelf life of extra virgin olive oil according to container type and storage conditions. J. Agric. Food Chem. 2015, 63, 2179–2188. [Google Scholar] [CrossRef] [PubMed]
- Denawaka, C.J.; Fowlis, I.A.; Dean, J.R. Evaluation and application of static headspace-multicapillary column-gas chromatography-ion mobility spectrometry for complex sample analysis. J. Chromatogr. A 2014, 1338, 136–148. [Google Scholar] [CrossRef]
- Krisilova, E.V.; Levina, A.M.; Makarenko, V.A. Determination of the volatile compounds of vegetable oils using an ion-mobility spectrometer. J. Anal. Chem 2014, 69, 371–376. [Google Scholar] [CrossRef]
- Danezis, G.P.; Tsagkaris, A.S.; Camin, F.; Brusic, V.; Georgiou, C.A. Food authentication: Techniques, trends & emerging approaches. TrAC Trends Anal. Chem. 2016, 85, 123–132. [Google Scholar] [CrossRef]
- Karpas, Z. Applications of ion mobility spectrometry (IMS) in the field of foodomics. Food Res. Int. 2013, 54, 1146–1151. [Google Scholar] [CrossRef]
- Hernández-Mesa, M.; Escourrou, A.; Monteau, F.; Le Bizec, B.; Dervilly-Pinel, G. Current applications and perspectives of ion mobility spectrometry to answer chemical food safety issues. TrAC Trends Anal. Chem. 2017, 94, 39–53. [Google Scholar] [CrossRef]
- Charlebois, S.; Schwab, A.; Henn, R.; Huck, C.W. Food fraud: An exploratory study for measuring consumer perception towards mislabeled food products and influence on self-authentication intentions. Trends Food Sci. Technol. 2016, 50, 211–218. [Google Scholar] [CrossRef]
- Ehmke, M.D.; Bonanno, A.; Boys, K.; Smith, T.G. Food fraud: Economic insights into the dark side of incentives. Aust. J. Agric. Resour. Econ. 2019, 63, 685–700. [Google Scholar] [CrossRef]
- Aries, E.; Burton, J.; Carrasco, L.; de Rudder, O.; Maquet, A. Scientific Support to the Implementation of a Coordinated Control Plan with a View to Establishing the Prevalence of Fraudulent Practices in the Marketing of Honey: N° SANTE/2015/E3/JRC/SI2.706828. JRC Technical Report 2016, JRC104749. 2016, p. 38. Available online: https://ec.europa.eu/food/sites/food/files/safety/docs/oc_control-progs_honey_jrc-tech-report_2016.pdf (accessed on 12 May 2021).
- Louveaux, J.; Maurizio, A.; Vorwohl, G. Methods of Melissopalynology. BEE World 1978, 59, 139–157. [Google Scholar] [CrossRef]
- Cavazza, A.; Corradini, C.; Musci, M.; Salvadeo, P. High-performance liquid chromatographic phenolic compound fingerprint for authenticity assessment of honey. J. Sci. Food Agric. 2013, 93, 1169–1175. [Google Scholar] [CrossRef]
- Zhou, J.; Yao, L.; Li, Y.; Chen, L.; Wu, L.; Zhao, J. Floral classification of honey using liquid chromatography-diode array detection-tandem mass spectrometry and chemometric analysis. Food Chem. 2014, 145, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Karabagias, I.K.; Badeka, A.; Kontakos, S.; Karabournioti, S.; Kontominas, M.G. Characterisation and classification of Greek pine honeys according to their geographical origin based on volatiles, physicochemical parameters and chemometrics. Food Chem. 2014, 146, 548–557. [Google Scholar] [CrossRef]
- Robotti, E.; Campo, F.; Riviello, M.; Bobba, M.; Manfredi, M.; Mazzucco, E.; Gosetti, F.; Calabrese, G.; Sangiorgi, E.; Marengo, E. Optimization of the Extraction of the Volatile Fraction from Honey Samples by SPME-GC-MS, Experimental Design, and Multivariate Target Functions. J. Chem. 2017, 2017, 6437857. [Google Scholar] [CrossRef]
- Jandrić, Z.; Haughey, S.A.; Frew, R.D.; McComb, K.; Galvin-King, P.; Elliott, C.T.; Cannavan, A. Discrimination of honey of different floral origins by a combination of various chemical parameters. Food Chem. 2015, 189, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, T.; Fagan, C.C.; O’Donnell, C.P.; Downey, G. Application of Near and Mid-Infrared Spectroscopy to Determine Cheese Quality and Authenticity. Food Bioprocess Technol. 2008, 1, 117–129. [Google Scholar] [CrossRef]
- Gok, S.; Severcan, M.; Goormaghtigh, E.; Kandemir, I.; Severcan, F. Differentiation of Anatolian honey samples from different botanical origins by ATR-FTIR spectroscopy using multivariate analysis. Food Chem. 2015, 170, 234–240. [Google Scholar] [CrossRef]
- Boffo, E.F.; Tavares, L.A.; Tobias, A.C.; Ferreira, M.M.; Ferreira, A.G. Identification of components of Brazilian honey by 1H NMR and classification of its botanical origin by chemometric methods. LWT 2012, 49, 55–63. [Google Scholar] [CrossRef]
- Schievano, E.; Finotello, C.; Uddin, J.; Mammi, S.; Piana, L. Objective Definition of Monofloral and Polyfloral Honeys Based on NMR Metabolomic Profiling. J. Agric. Food Chem. 2016, 64, 3645–3652. [Google Scholar] [CrossRef]
- Garrido-Delgado, R.; Arce, L.; Valcárcel, M. Multi-capillary column-ion mobility spectrometry: A potential screening system to differentiate virgin olive oils. Anal. Bioanal. Chem. 2012, 402, 489–498. [Google Scholar] [CrossRef]
- Gerhardt, N.; Schwolow, S.; Rohn, S.; Pérez-Cacho, P.R.; Galán-Soldevilla, H.; Arce, L.; Weller, P. Quality assessment of olive oils based on temperature-ramped HS-GC-IMS and sensory evaluation: Comparison of different processing approaches by LDA, kNN, and SVM. Food Chem. 2019, 278, 720–728. [Google Scholar] [CrossRef]
- Garrido-Delgado, R.; Mercader-Trejo, F.; Sielemann, S.; de Bruyn, W.; Arce, L.; Valcárcel, M. Direct classification of olive oils by using two types of ion mobility spectrometers. Anal. Chim. Acta 2011, 696, 108–115. [Google Scholar] [CrossRef]
- Schwolow, S.; Gerhardt, N.; Rohn, S.; Weller, P. Data fusion of GC-IMS data and FT-MIR spectra for the authentication of olive oils and honeys-is it worth to go the extra mile? Anal. Bioanal. Chem. 2019, 411, 6005–6019. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Rogers, K.M.; Li, Y.; Yang, S.; Chen, L.; Zhou, J. Untargeted and Targeted Discrimination of Honey Collected by Apis cerana and Apis mellifera Based on Volatiles Using HS-GC-IMS and HS-SPME-GC-MS. J. Agric. Food Chem. 2019, 67, 12144–12152. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, S.; He, J.; Chen, L.; Zhang, J.; Jin, Y.; Zhou, J.; Zhang, Y. A green triple-locked strategy based on volatile-compound imaging, chemometrics, and markers to discriminate winter honey and sapium honey using headspace gas chromatography-ion mobility spectrometry. Food Res. Int. 2019, 119, 960–967. [Google Scholar] [CrossRef]
- Arroyo-Manzanares, N.; García-Nicolás, M.; Castell, A.; Campillo, N.; Viñas, P.; López-García, I.; Hernández-Córdoba, M. Untargeted headspace gas chromatography–Ion mobility spectrometry analysis for detection of adulterated honey. Talanta 2019, 205, 120123. [Google Scholar] [CrossRef]
- Cavanna, D.; Zanardi, S.; Dall’Asta, C.; Suman, M. Ion mobility spectrometry coupled to gas chromatography: A rapid tool to assess eggs freshness. Food Chem. 2019, 271, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Chen, W.; Wang, Z.; Wang, J.; Huo, Y. Rapid detection of Aspergillus spp. infection levels on milled rice by headspace-gas chromatography ion-mobility spectrometry (HS-GC-IMS) and E-nose. LWT 2020, 132, 109758. [Google Scholar] [CrossRef]
- Gu, S.; Chen, W.; Wang, Z.; Wang, J. Rapid determination of potential aflatoxigenic fungi contamination on peanut kernels during storage by data fusion of HS-GC-IMS and fluorescence spectroscopy. Postharvest Biol. Technol. 2021, 171, 111361. [Google Scholar] [CrossRef]
- Gu, S.; Wang, Z.; Chen, W.; Wang, J. Targeted versus Nontargeted Green Strategies Based on Headspace-Gas Chromatography-Ion Mobility Spectrometry Combined with Chemometrics for Rapid Detection of Fungal Contamination on Wheat Kernels. J. Agric. Food Chem. 2020, 68, 12719–12728. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Manzanares, N.; Martín-Gómez, A.; Jurado-Campos, N.; Garrido-Delgado, R.; Arce, C.; Arce, L. Target vs spectral fingerprint data analysis of Iberian ham samples for avoiding labelling fraud using headspace–gas chromatography–ion mobility spectrometry. Food Chem. 2018, 246, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Brendel, R.; Schwolow, S.; Rohn, S.; Weller, P. Volatilomic Profiling of Citrus Juices by Dual-Detection HS-GC-MS-IMS and Machine Learning-An Alternative Authentication Approach. J. Agric. Food Chem. 2021, 69, 1727–1738. [Google Scholar] [CrossRef] [PubMed]
- Vega-Márquez, B.; Nepomuceno-Chamorro, I.; Jurado-Campos, N.; Rubio-Escudero, C. Deep Learning Techniques to Improve the Performance of Olive Oil Classification. Front. Chem. 2019, 7, 929. [Google Scholar] [CrossRef]
- Yuan, Z.-Y.; Li, J.; Zhou, X.-J.; Wu, M.-H.; Li, L.; Pei, G.; Chen, N.-H.; Liu, K.-L.; Xie, M.-Z.; Huang, H.-Y. HS-GC-IMS-Based metabonomics study of Baihe Jizihuang Tang in a rat model of chronic unpredictable mild stress. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1148, 122143. [Google Scholar] [CrossRef]
- Antignac, J.-P.; Courant, F.; Pinel, G.; Bichon, E.; Monteau, F.; Elliott, C.; Le Bizec, B. Mass spectrometry-based metabolomics applied to the chemical safety of food. TrAC Trends Anal. Chem. 2011, 30, 292–301. [Google Scholar] [CrossRef]
- Snow, N.H.; Slack, G.C. Head-space analysis in modern gas chromatography. TrAC Trends Anal. Chem. 2002, 21, 608–617. [Google Scholar] [CrossRef]
- Soria, A.C.; García-Sarrió, M.J.; Sanz, M.L. Volatile sampling by headspace techniques. TrAC Trends Anal. Chem. 2015, 71, 85–99. [Google Scholar] [CrossRef]
- Poole, C.F. (Ed.) Liquid-Phase Extraction; Handbooks in Separation Science; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Guo, Z.; Zhu, Z.; Huang, S.; Wang, J. Non-targeted screening of pesticides for food analysis using liquid chromatography high-resolution mass spectrometry-a review. Food Addit. Contam. Part A 2020, 37, 1180–1201. [Google Scholar] [CrossRef]
- Souza Silva, E.A.; Risticevic, S.; Pawliszyn, J. Recent trends in SPME concerning sorbent materials, configurations and in vivo applications. TrAC Trends Anal. Chem. 2013, 43, 24–36. [Google Scholar] [CrossRef]
- Chen, T.; Chen, X.; Lu, D.; Chen, B. Detection of Adulteration in Canola Oil by Using GC-IMS and Chemometric Analysis. Int. J. Anal. Chem. 2018, 2018, 3160265. [Google Scholar] [CrossRef]
- Baumbach, J.I.; Eiceman, G.A.; Klockow, D.; Sielemann, S.; Irmer, A.V. Exploration of a Multicapillary Column for Use in Elevated Speed Gas Chromatography. Int. J. Environ. Anal. Chem. 1997, 66, 225–239. [Google Scholar] [CrossRef]
- Cureton, E.E.; D’Agostino, R.B. Factor Analysis; Psychology Press: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Zheng, P.; de Harrington, P.B.; Davis, D.M. Quantitative analysis of volatile organic compounds using ion mobility spectrometry and cascade correlation neural networks. Chemom. Intell. Lab. Syst. 1996, 33, 121–132. [Google Scholar] [CrossRef]
- Kaur-Atwal, G.; O’Connor, G.; Aksenov, A.A.; Bocos-Bintintan, V.; Paul Thomas, C.L.; Creaser, C.S. Chemical standards for ion mobility spectrometry: A review. Int. J. Ion Mobil. Spectrom. 2009, 12, 1–14. [Google Scholar] [CrossRef]
- Liedtke, S.; Seifert, L.; Ahlmann, N.; Hariharan, C.; Franzke, J.; Vautz, W. Coupling laser desorption with gas chromatography and ion mobility spectrometry for improved olive oil characterisation. Food Chem. 2018, 255, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Borsdorf, H.; Neitsch, K. Ion mobility spectra of cyclic and aliphatic hydrocarbons with different substituents. Int. J. Ion Mobil. Spectrom. 2009, 12, 39–46. [Google Scholar] [CrossRef]
- Budzyńska, E.; Sielemann, S.; Puton, J.; Surminski, A.L.R.M. Analysis of e-liquids for electronic cigarettes using GC-IMS/MS with headspace sampling. Talanta 2020, 209, 120594. [Google Scholar] [CrossRef]
- Kessler, W. Multivariate Datenanalyse: Für die Pharma-, bio- und Prozessanalytik: Ien Lehrbuch; Wiley-VCH: Weinheim, Germany, 2007. [Google Scholar]
- Maimon, O.; Rokach, L. Data Mining and Knowledge Discovery Handbook; Springer: New York, NY, USA, 2005. [Google Scholar]
- Granato, D.; Santos, J.S.; Escher, G.B.; Ferreira, B.L.; Maggio, R.M. Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective. Trends Food Sci. Technol. 2018, 72, 83–90. [Google Scholar] [CrossRef]
- Wold, S.; Geladi, P.; Esbensen, K.; Öhman, J. Multi-way principal components-and PLS-analysis. J. Chemom. 1987, 1, 41–56. [Google Scholar] [CrossRef]
- Bro, R. PARAFAC. Tutorial and applications. Chemom. Intell. Lab. Syst. 1997, 38, 149–171. [Google Scholar] [CrossRef]
- Kroonenberg, P.M.; Ten Berge, J.M.F. The equivalence of Tucker3 and Parafac models with two components. Chemom. Intell. Lab. Syst. 2011, 106, 21–26. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Varmuza, K.; Filzmoser, P. Introduction to Multivariate Statistical Analysis in Chemometrics; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Chen, Y.-Y.; Lin, Y.-H.; Kung, C.-C.; Chung, M.-H.; Yen, I.-H. Design and Implementation of Cloud Analytics-Assisted Smart Power Meters Considering Advanced Artificial Intelligence as Edge Analytics in Demand-Side Management for Smart Homes. Sensors 2019, 19, 2047. [Google Scholar] [CrossRef]
- Torkashvand, A.M.; Ahmadi, A.; Nikravesh, N.L. Prediction of kiwifruit firmness using fruit mineral nutrient concentration by artificial neural network (ANN) and multiple linear regressions (MLR). J. Integr. Agric. 2017, 16, 1634–1644. [Google Scholar] [CrossRef]
- Scheinemann, A.; Sielemann, S.; Walter, J.; Doll, T. Evaluation Strategies for Coupled GC-IMS Measurement including the Systematic Use of Parametrized ANN. Open J. Appl. Sci. 2012, 2, 257–266. [Google Scholar] [CrossRef]
- Rosipal, R. Nonlinear Partial Least Squares An Overview. In Chemoinformatics and Advanced Machine Learning Perspectives; Lodhi, H., Yamanishi, Y., Eds.; IGI Global: Hershey, PA, USA, 2011; pp. 169–189. [Google Scholar] [CrossRef]
- De Juan, A.; Jaumot, J.; Tauler, R. Multivariate Curve Resolution (MCR). Solving the mixture analysis problem. Anal. Methods 2014, 6, 4964–4976. [Google Scholar] [CrossRef]
- Liu, P.; Duan, J.; Wang, P.; Qian, D.; Guo, J.; Shang, E.; Su, S.; Tang, Y.; Tang, Z. Biomarkers of primary dysmenorrhea and herbal formula intervention: An exploratory metabonomics study of blood plasma and urine. Mol. Biosyst. 2013, 9, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Zhang, X.; Dai, Z.; Fan, X.; Liu, M.; Ma, J.; Shang, W.; Liu, J.; Strappe, P.; Blanchard, C.; Zhou, Z. A study on volatile metabolites screening by HS-SPME-GC-MS and HS-GC-IMS for discrimination and characterization of white and yellowed rice. Cereal Chem. 2020, 97, 496–504. [Google Scholar] [CrossRef]
- Chen, T.; Liu, C.; Meng, L.; Lu, D.; Chen, B.; Cheng, Q. Early warning of rice mildew based on gas chromatography-ion mobility spectrometry technology and chemometrics. Food Meas. 2021, 15, 1939–1948. [Google Scholar] [CrossRef]
- Lv, W.; Lin, T.; Ren, Z.; Jiang, Y.; Zhang, J.; Bi, F.; Gu, L.; Hou, H.; He, J. Rapid discrimination of Citrus reticulata ‘Chachi’ by headspace-gas chromatography-ion mobility spectrometry fingerprints combined with principal component analysis. Food Res. Int. 2020, 131, 108985. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, R.; Zhang, H.; Wang, S.; Chen, D.; Lin, S. Development of a flavor fingerprint by HS-GC-IMS with PCA for volatile compounds of Tricholoma matsutake Singer. Food Chem. 2019, 290, 32–39. [Google Scholar] [CrossRef]
- Brendel, R.; Schwolow, S.; Rohn, S.; Weller, P. Gas-phase volatilomic approaches for quality control of brewing hops based on simultaneous GC-MS-IMS and machine learning. Anal. Bioanal. Chem. 2020, 412, 7085–7097. [Google Scholar] [CrossRef] [PubMed]
- Pu, D.; Zhang, H.; Zhang, Y.; Sun, B.; Ren, F.; Chen, H.; He, J. Characterization of the aroma release and perception of white bread during oral processing by gas chromatography-ion mobility spectrometry and temporal dominance of sensations analysis. Food Res. Int. 2019, 123, 612–622. [Google Scholar] [CrossRef]
- Tang, Z.-S.; Zeng, X.-A.; Brennan, M.A.; Han, Z.; Niu, D.; Huo, Y. Characterization of aroma profile and characteristic aromas during lychee wine fermentation. J. Food Process. Preserv. 2019, 43, e14003. [Google Scholar] [CrossRef]
- Gerhardt, N.; Birkenmeier, M.; Kuballa, T.; Ohmenhaeuser, M.; Rohn, S.; Weller, P. Differentiation of the botanical origin of honeys by fast, non-targeted 1H-NMR profiling and chemometric tools as alternative authenticity screening tool. In Proceedings of the XIII International Conference on the Applications of Magnetic Resonance in Food Science, Karlsruhe, Germany, 7–10 June 2016; IM Publications: Charlton, UK, 2016; p. 33. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, D.; Dong, Y.; Ju, H.; Wu, C.; Lin, S. Characteristic volatiles fingerprints and changes of volatile compounds in fresh and dried Tricholoma matsutake Singer by HS-GC-IMS and HS-SPME-GC-MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018, 1099, 46–55. [Google Scholar] [CrossRef]
- Klein, L.A. Sensor and Data Fusion: A Tool for Information Assessment and Decision Making; SPIE Press: Bellingham, WA, USA, 2010. [Google Scholar]
- Borràs, E.; Ferré, J.; Boqué, R.; Mestres, M.; Aceña, L.; Busto, O. Data fusion methodologies for food and beverage authentication and quality assessment—a review. Anal. Chim. Acta 2015, 891, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kuncheva, L.I. Combining Pattern Classifiers: Methods and Algorithms; John Wiley & Sons: New York, NY, USA, 2010. [Google Scholar]
- Martín-Gómez, A.; Arroyo-Manzanares, N.; Rodríguez-Estévez, V.; Arce, L. Use of a non-destructive sampling method for characterization of Iberian cured ham breed and feeding regime using GC-IMS. Meat Sci. 2019, 152, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Chatterji, B.N. Feature Extraction Methods for Character Recognition. IETE Tech. Rev. 1986, 3, 9–22. [Google Scholar] [CrossRef]
- Lowe, D.G. Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Chen, T.; Qi, X.; Chen, M.; Chen, B. Gas Chromatography-Ion Mobility Spectrometry Detection of Odor Fingerprint as Markers of Rapeseed Oil Refined Grade. J. Anal. Methods Chem. 2019, 2019, 3163204. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Qi, X.; Lu, D.; Chen, B. Gas chromatography-ion mobility spectrometric classification of vegetable oils based on digital image processing. Food Meas. 2019, 13, 1973–1979. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).