Unified Classification of Bacterial Colonies on Different Agar Media Based on Hyperspectral Imaging and Machine Learning
Abstract
1. Introduction
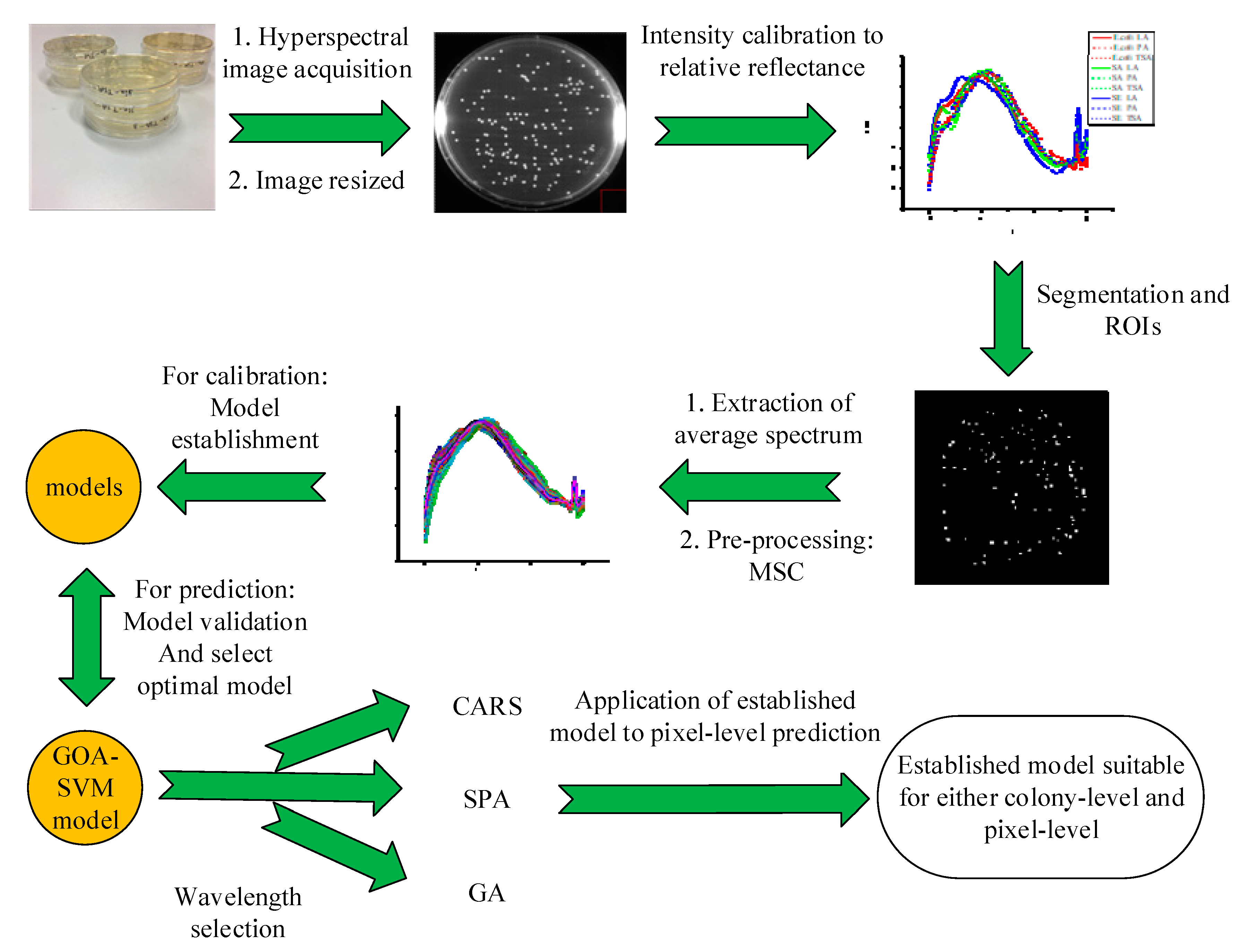
2. Results and Discussion
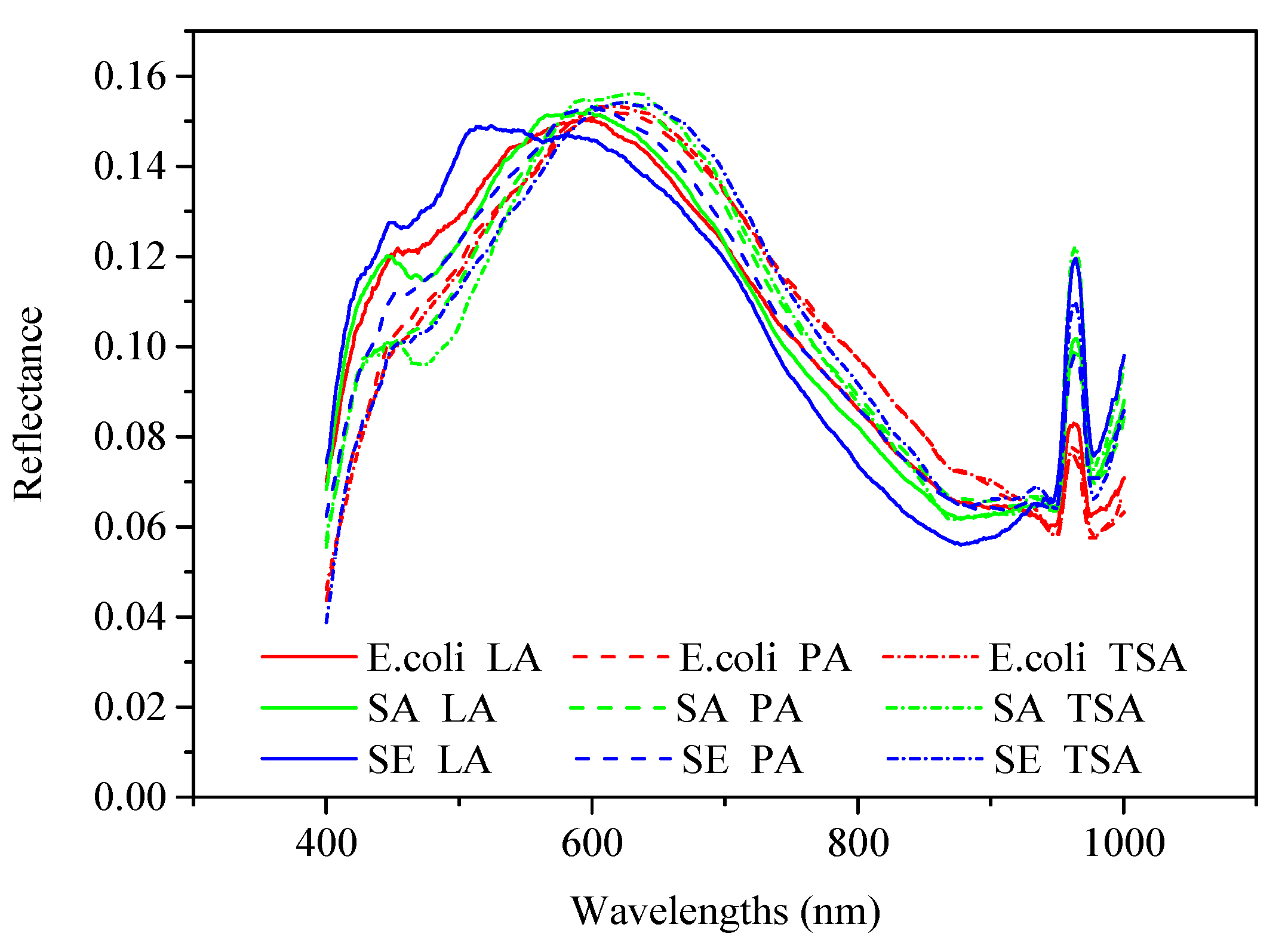
2.1. Spectral Analysis of Bacterial Colonies
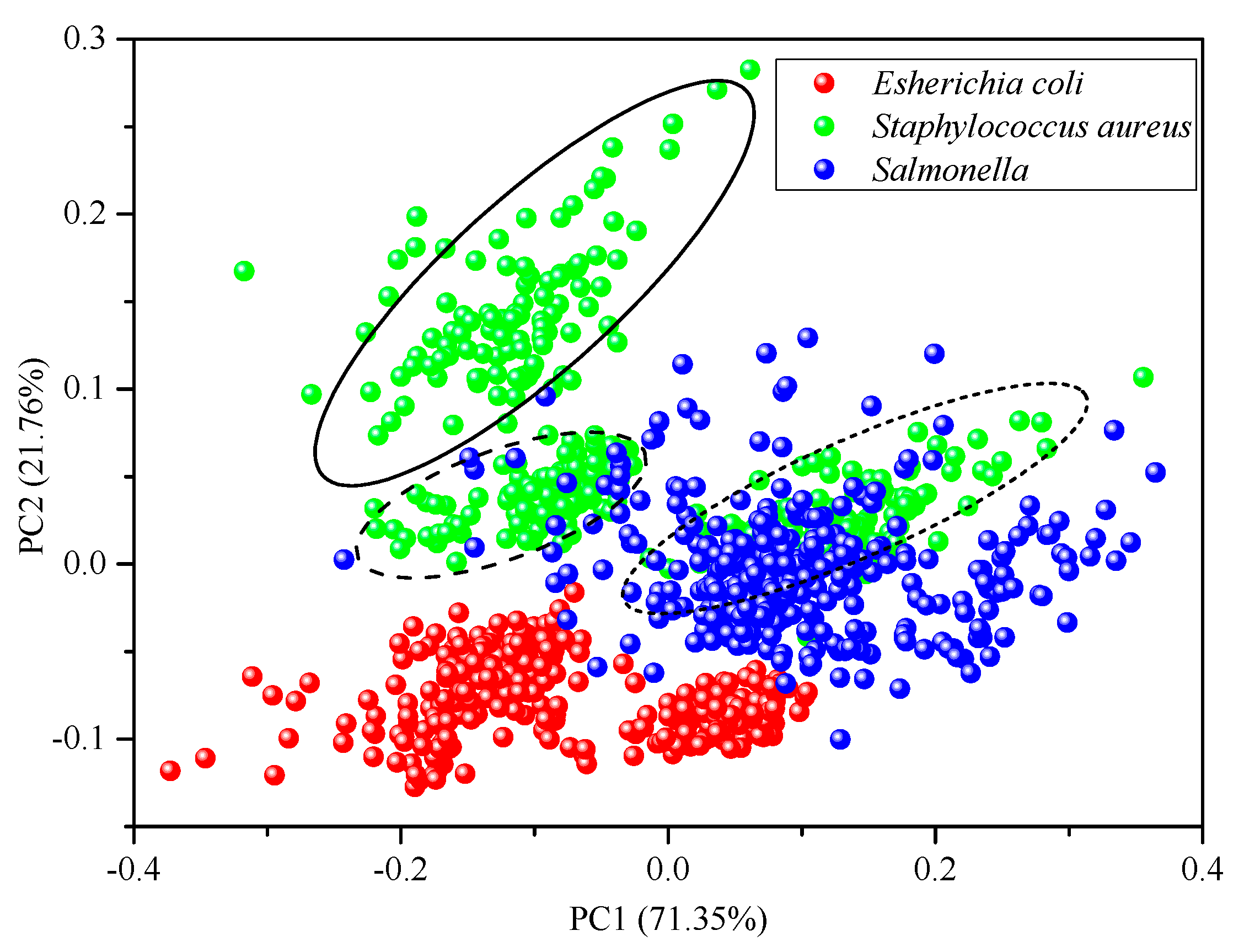
2.2. Principal Component Analysis
2.3. Full Wavelength Models
2.3.1. PLS-DA Classification Models
2.3.2. SVM Model Optimization
2.4. Simplified Classification Models
2.5. Pixel Analysis
3. Materials and Methods
3.1. Bacterial Culture and Sample Preparation
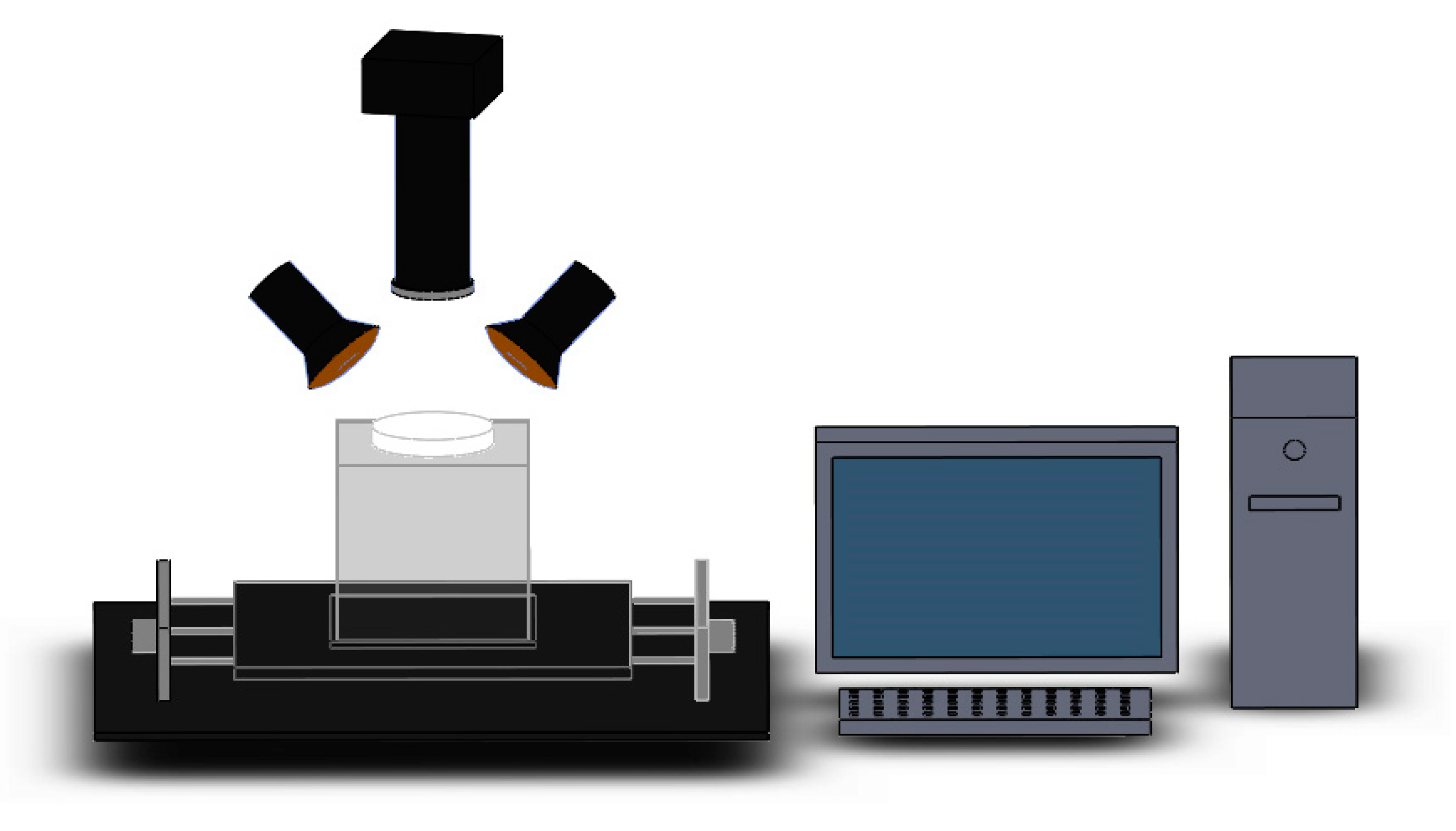
3.2. Hyperspectral Image Acquisition
3.3. Hyperspectral Image Processing and Data Acquisition
3.4. Sample Set Establishment
3.5. Model Establishment and Optimization
3.6. Grasshopper Optimization Algorithm (GOA)
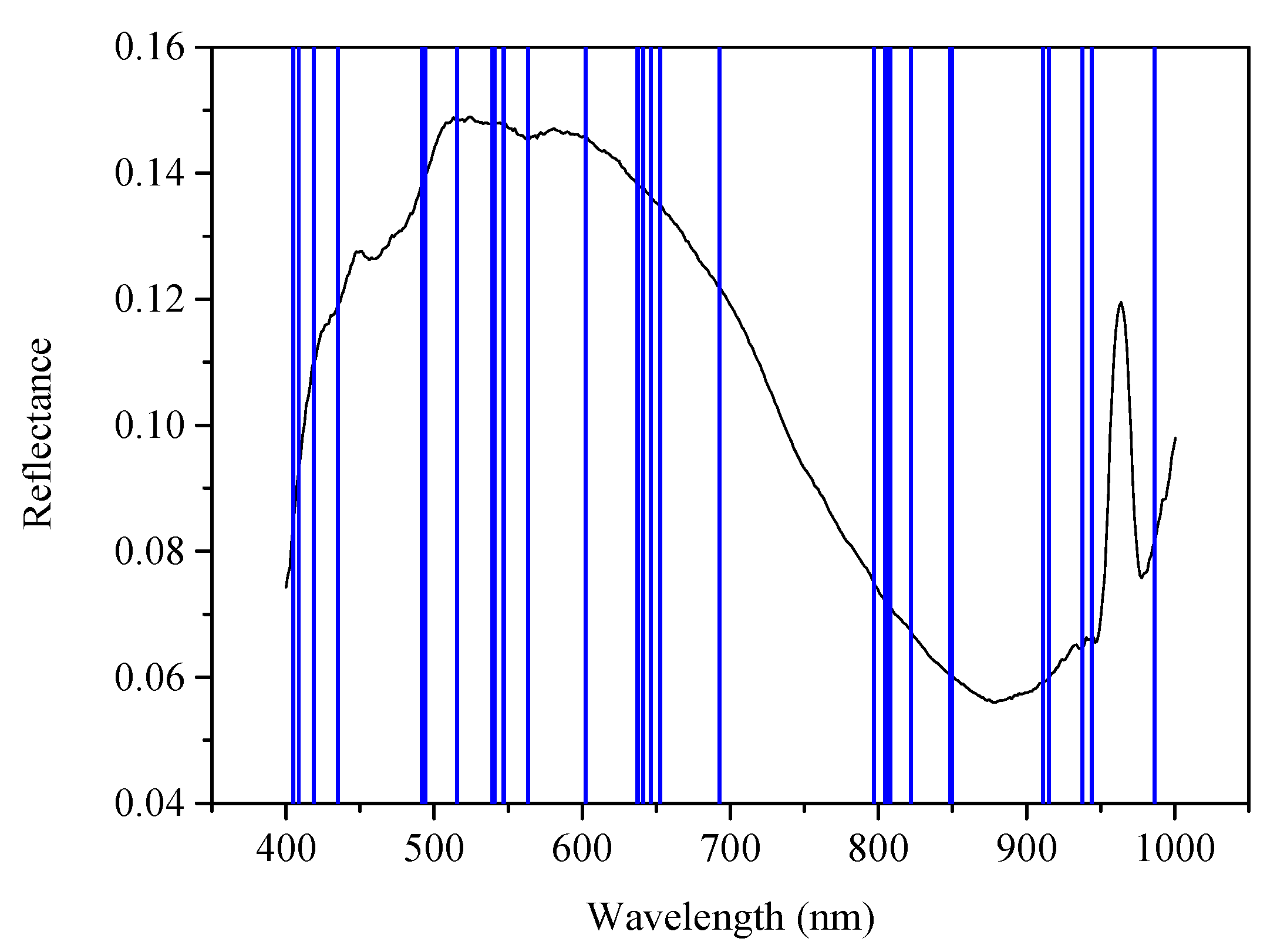
3.7. Wavelength Selection
3.8. Model Assessment
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Yan, W.-X.; Li, J.-G.; Wu, Y.-N.; Zhou, Y.-J. Burden of self-reported acute gastrointestinal illness in China: A population-based survey. BMC Public Health 2013, 13, 456. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, S.; Ping, F.; Liu, J.; Han, H.; Li, B.; Pei, X.; Ning, L.; Liu, X.; Guo, Y. National molecular tracing network for foodborne disease surveillance in China. Food Control 2017, 88, 28–32. [Google Scholar] [CrossRef]
- Pignata, C.; D’Angelo, D.; Fea, E.; Gilli, G. A review on microbiological decontamination of fresh produce with nonthermal plasma. J. Appl. Microbiol. 2017, 122, 1438–1455. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.N.; Chen, J.S. Food safety monitoring and surveillance in China: Past, present and future. Food Control 2018, 90, S0956713518301063. [Google Scholar] [CrossRef]
- Insalata, N.F.; Dunlap, W.G.; Mahnke, C.W. A comparison of cultural methods used with microcolony and direct fluorescent-antibody techniques to detect salmonellae. J. Milk Food Technol. 1975, 38, 201–203. [Google Scholar] [CrossRef]
- Park, C.H.; Vandel, N.M.; Hixon, D.L. Rapid immunoassay for detection of escherichia coli O157 directly from stool specimens. J. Clin. Microbiol. 1996, 34, 988–990. [Google Scholar] [CrossRef]
- Kong, R.; So, C.; Law, W.; Wu, R.; Kong, Y.C.R. A sensitive and versatile multiplex PCR system for the rapid detection of enterotoxigenic (ETEC), enterohaemorrhagic (EHEC) and enteropathogenic (EPEC) strains of Escherichia coli. Mar. Pollut. Bull. 1999, 38, 1207–1215. [Google Scholar] [CrossRef]
- Stokes, D.L.; Allain, L.R.; Griffin, G.D.; Stratis-Cullum, D.N.; Wintenberg, A.L.; Maples, R.A.; Vo-Dinh, T.; Mobley, J. Biochip using a biofluidic system for detection of E. coli and other pathogens. In Proceedings of the SPIE—The International Society for Optical Engineering, San Jose, CA, USA, 22 July 2003; pp. 13–21. [Google Scholar]
- Jiang-Bo, L.I.; Rao, X.Q. Advance on application of hyperspectral imaging to nondestructive detection of agricultural products external quality. Spectrosc. Spectr. Anal. 2011, 31, 2021. [Google Scholar]
- Gowen, A.A.; Feng, Y.; Gaston, E.; Valdramidis, V. Recent applications of hyperspectral imaging in microbiology. Talanta 2015, 137, 43–54. [Google Scholar] [CrossRef]
- Turra, G.; Conti, N.; Signoroni, A. Hyperspectral image acquisition and analysis of cultured bacteria for the discrimination of urinary tract infections. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 759–762. [Google Scholar]
- Windham, W.R.; Yoon, S.C.; Ladely, S.R.; Haley, J.A.; Heitschmidt, J.W.; Lawrence, K.C.; Park, B.; Narrang, N.; Cray, W.C. Detection by hyperspectral imaging of shiga toxin-producing Escherichia coli serogroups O26, O45, O103, O111, O121, and O145 on rainbow agar. J. Food Prot. 2013, 76, 1129–1136. [Google Scholar] [CrossRef]
- Windham, W.R.; Yoon, S.C.; Ladely, S.R.; Heitschmidt, J.W.; Lawrence, K.C.; Park, B.; Narrang, N.; Cray, W.C. The effect of regions of interest and spectral pre-processing on the detection of non-0157 Shiga-toxin producing Escherichia coli serogroups on agar media by hyperspectral imaging. J. Near Infrared Spectrosc. 2012, 20, 547. [Google Scholar] [CrossRef]
- Arrigoni, S.; Turra, G.; Signoroni, A. Hyperspectral image analysis for rapid and accurate discrimination of bacterial infections: A benchmark study. Comput. Biol. Med. 2017, 88, 60. [Google Scholar] [CrossRef] [PubMed]
- Kammies, T.L.; Manley, M.; Gouws, P.A.; Williams, P.J. Differentiation of foodborne bacteria using NIR hyperspectral imaging and multivariate data analysis. Appl. Microbiol. Biotechnol. 2016, 100, 9305–9320. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Duschek, F.; Fellner, L.; Grünewald, K.M.; Handke, J. Stand-Off Detection: Distinction of Bacteria by Hyperspectral Laser Induced Fluorescence. In Proceedings of the Chemical, Biological, Radiological, Nuclear, & Explosives, Baltimore, MD, USA, 17–21 April 2016; pp. 98240Y-1–98240Y-7. [Google Scholar]
- Signoroni, A.; Savardi, M.; Pezzoni, M.; Guerrini, F.; Turra, G. Combining the use of CNN classification and strength-driven compression for the robust identification of bacterial species on hyperspectral culture plate images. IET Comput. Vis. 2018, 12, 941–949. [Google Scholar] [CrossRef]
- Feng, Y.Z.; Yu, W.; Chen, W.; Peng, K.K.; Jia, G.F. Invasive weed optimization for optimizing one-agar-for-all classification of bacterial colonies based on hyperspectral imaging. Sens. Actuators B Chem. 2018, 269, 264–270. [Google Scholar] [CrossRef]
- Yoon, S.K.; Lawrence, K.C.; Siragusa, G.R.; Line, J.E.; Park, B.; Feldner, P.W. Hyperspectral reflectance imaging for detecting a foodborne pathogen: Campylobacter. Trans. ASABE 2009, 52, 651–662. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Grefenstette, J.J. Genetic Algorithms and Machine Learning; Springer: New York, NJ, USA, 1993. [Google Scholar]
- Eberhart, R.C.; Kennedy, J. New Optimizer Using Particle Swarm Theory. In Proceedings of the Mhs95 Sixth International Symposium on Micro Machine & Human Science, Nagoya, Japan, 4–6 October 1995. [Google Scholar]
- Aljarah, I.; Al-Zoubi, A.M.; Faris, H.; Hassonah, M.A.; Mirjalili, S.; Saadeh, H. Simultaneous feature selection and support vector machine optimization using the grasshopper optimization algorithm. Cogn. Comput. 2018, 10, 478–495. [Google Scholar] [CrossRef]
- Saremi, S.; Mirjalili, S.; Lewis, A. Grasshopper optimisation algorithm: Theory and application. Adv. Eng. Softw. 2017, 105, 30–47. [Google Scholar] [CrossRef]
- Arora, S.; Anand, P. Chaotic grasshopper optimization algorithm for global optimization. Neural Comput. Appl. 2018, 31, 4385–4405. [Google Scholar] [CrossRef]
- Mu, K.X.; Feng, Y.Z.; Wei, C.; Wei, Y. Near infrared spectroscopy for classification of bacterial pathogen strains based on spectral transforms and machine learning. Chemom. Intell. Lab. Syst. 2018, 179, 46–53. [Google Scholar] [CrossRef]
- Vieira, S.M.; Kaymak, U.; Sousa, J.M.C. Cohen’s kappa coefficient as a performance measure for feature selection. In Proceedings of the IEEE International Conference on Fuzzy Systems, Barcelona, Spain, 18–23 July 2010. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |
| Model | Optimization Methods | LVs | cp | g | OCCR (%) | Kappa | |
|---|---|---|---|---|---|---|---|
| Calibration | Prediction | ||||||
| PLS-DA | - | 5 | - | - | 75 | 74.03 | 0.71 |
| SVM | GA | - | 41.74 | 51.53 | 100 | 97.33 | 0.96 |
| PSO | - | 13.69 | 16.56 | 99.45 | 98.78 | 0.98 | |
| GOA | - | 70.61 | 5.49 | 99.45 | 98.82 | 0.98 | |
| Predicted Label | ||||||
|---|---|---|---|---|---|---|
| Bacteria | E. coli | SA | SE | Unclassified | ||
| PLS-DA | True Label | E. coli | 80.39% | 7% | 0 | 4.30% |
| SA | 9.51% | 75.59% | 14.71% | |||
| SE | 0 | 38.24% | 61.13% | |||
| GOA-SVM | True Label | E. coli | 97.33% | 0 | 2.67% | 0 |
| SA | 0 | 99.71% | 0.29% | |||
| SE | 0.63% | 0.21% | 99.16% | |||
| Wavelength Selection Methods | Number of Wavelengths | PLS-DA | GOA-SVM | ||||
|---|---|---|---|---|---|---|---|
| OCCRC% | OCCRP% | Kappa | OCCRC% | OCCRP% | Kappa | ||
| CARS | 30 | 79.15 | 71.72 | 0.63 | 99.45 | 98.73 | 0.98 |
| SPA | 24 | 75.66 | 71.72 | 0.62 | 99.24 | 98.69 | 0.98 |
| GA | 69 | 78.50 | 73.53 | 0.64 | 99.45 | 98.60 | 0.98 |
| Bacteria | Predicted Label | Unclassified | |||
|---|---|---|---|---|---|
| E. coli | SA | SE | |||
| True Label | E. coli | 97.48% | 0 | 2.52% | 0 |
| SA | 0 | 99.8% | 0.20% | ||
| SE | 1.47% | 0.21% | 98.32% | ||
| Predicted Label | ||||||
|---|---|---|---|---|---|---|
| Bacteria | E. coli | SA | SE | OCCRP (%) | ||
| GOA-SVM | True Label | E. coli | 94.99% | 0.12% | 4.89% | 91.86 |
| SA | 6.09% | 90.77% | 3.14% | |||
| SE | 21.06% | 0.88% | 78.06% | |||
| CARS-GOA-SVM | True Label | E. coli | 95.35% | 0.12% | 4.53% | 93.32 |
| SA | 1.18% | 95.51% | 3.31% | |||
| SE | 19.27% | 0.64% | 80.09% | |||
| Calibration Set | Prediction Set | |||||||
|---|---|---|---|---|---|---|---|---|
| LA | PA | TSA | Total | LA | PA | TSA | Total | |
| E. coli | 103 | 83 | 94 | 280 | 325 | 167 | 222 | 714 |
| SA | 110 | 114 | 101 | 325 | 347 | 289 | 384 | 1020 |
| SE | 96 | 108 | 107 | 311 | 114 | 211 | 151 | 476 |
| Total | 309 | 305 | 302 | 916 | 786 | 667 | 757 | 2210 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, P.; Feng, Y.-Z.; Zhu, L.; Kong, L.-Q.; Zhang, X.-l.; Zhang, S.; Li, S.-W.; Jia, G.-F. Unified Classification of Bacterial Colonies on Different Agar Media Based on Hyperspectral Imaging and Machine Learning. Molecules 2020, 25, 1797. https://doi.org/10.3390/molecules25081797
Gu P, Feng Y-Z, Zhu L, Kong L-Q, Zhang X-l, Zhang S, Li S-W, Jia G-F. Unified Classification of Bacterial Colonies on Different Agar Media Based on Hyperspectral Imaging and Machine Learning. Molecules. 2020; 25(8):1797. https://doi.org/10.3390/molecules25081797
Chicago/Turabian StyleGu, Peng, Yao-Ze Feng, Le Zhu, Li-Qin Kong, Xiu-ling Zhang, Sheng Zhang, Shao-Wen Li, and Gui-Feng Jia. 2020. "Unified Classification of Bacterial Colonies on Different Agar Media Based on Hyperspectral Imaging and Machine Learning" Molecules 25, no. 8: 1797. https://doi.org/10.3390/molecules25081797
APA StyleGu, P., Feng, Y.-Z., Zhu, L., Kong, L.-Q., Zhang, X.-l., Zhang, S., Li, S.-W., & Jia, G.-F. (2020). Unified Classification of Bacterial Colonies on Different Agar Media Based on Hyperspectral Imaging and Machine Learning. Molecules, 25(8), 1797. https://doi.org/10.3390/molecules25081797






