Abstract
Citrus aurantium is a widespread tree in the Mediterranean area, and it is mainly used as rootstock for other citrus. In the present study, a vacuum infiltration centrifugation procedure, followed by solid phase extraction matrix-assisted laser desorption ionization tandem mass spectrometry (SPE MALDI MS/MS) analysis, was adopted to isolate proteins from leaves. The results of mass spectrometry (MS) profiling, combined with the top-down proteomics approach, allowed the identification of 78 proteins. The bioinformatic databases TargetP, SignalP, ChloroP, WallProtDB, and mGOASVM-Loc were used to predict the subcellular localization of the identified proteins. Among 78 identified proteins, 20 were targeted as secretory pathway proteins and 36 were predicted to be in cellular compartments including cytoplasm, nucleus, and cell membrane. The largest subcellular fraction was the secretory pathway, accounting for 25% of total proteins. Gene Ontology (GO) of Citrus sinensis was used to simplify the functional annotation of the proteins that were identified in the leaves. The Kyoto Encyclopedia of Genes and Genomes (KEGG) showed the enrichment of metabolic pathways including glutathione metabolism and biosynthesis of secondary metabolites, suggesting that the response to a range of environmental factors is the key processes in citrus leaves. Finally, the Lipase GDSL domain-containing protein GDSL esterase/lipase, which is involved in plant development and defense response, was for the first time identified and characterized in Citrus aurantium.
1. Introduction
Citrus species constitute one of the major tree fruit crops with great economic impact and is currently facing biotic and abiotic stresses. Rootstocks play a pivotal role in the success of the global commercial production of citrus fruits. The choice of rootstocks is of critical importance because their possibly unsatisfactory characteristics can cause serious failure in the citrus industry. In many instances, citrus rootstocks are the sole determining element that allows citrus to be grown in particular circumstances; they adapt trees to the effects of biotic and abiotic stresses and lead to excellent yields of high-quality fruits. Rootstocks are responsible for the ground anchoring and the proper development of trees, including water and nutrient absorption; they act as an energy source, provide carbohydrate storage, control the harvest time and fruit quality, ensure protection against soil borne diseases, and allow the adaptation of plants to soil and atmospheric conditions. Notwithstanding, every rootstock has one or more undesirable traits that preclude its universal use [1,2]. Those limitations have generally been highlighted from experimental data; alternatively, they have been described only after developing commercial experiences. Today, research projects are principally focused on the selection and preparation of new suitable citrus rootstocks; the improvement of their productivity and resistance characteristics remains the main objectives of the investigation. As a consequence, the study of plant proteome is fundamental in understanding protein differential expression and biological functions. Evidence led to the design of effective sample preparation methods for numerous plant species, tissues [3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21], and citrus leaves among others [22,23,24,25,26,27]. The rapid and significant upgrading of sensitivity, throughput and mass accuracy of modern mass spectrometers drastically improved gel-free proteomic approaches [28,29,30]. Mass spectrometry methodologies are rapid and sensitive tools for the identification [31] and quantitation [32,33] of metabolites, amino acids, proteins and their post-translational modifications [34,35]. Matrix-assisted laser desorption ionization mass spectrometry (MALDI MS), and tandem mass spectrometry (MS/MS) techniques are used as an alternative to Liquid Chromatography Electrospray Ionization (LC-ESI), for the highly sensitive analysis of low and high molecular weight compounds in complex matrices [36,37,38,39,40]. MALDI MS technique offers great advantages, such as short analysis times, high sensitivity, tolerance to contaminants, the ability to detect different components in highly complex mixtures, and the possibility to be combined with a rapid and simple preparation of the sample, preventing any possible analyte loss [41].
In this study, a simple procedure based on the vacuum infiltration and centrifugation with salt solutions, fractionation and trypsin digestion, followed by MALDI time of flight (TOF)/TOF mass spectrometry is applied to leaves of Citrus aurantium, an economically important fruit tree in the Mediterranean area, widely used as a rootstock for citrus. Vacuum infiltration with an extraction solution was adopted because it is applicable to small samples and allows the extraction of proteins reducing the contamination by non-protein components. The results of mass spectrometry (MS) profiling, combined with the top-down proteomics approach, allowed the identification of 78 proteins with a significant match. MS data were processed by the amino-acid sequence-based predictors TargetP, SignalP, ChloroP, WallProtDB and mGOASVM-Loc in order to establish the subcellular locations of the extracted proteome. Gene Ontology annotation of the Citrus sinensis genome was used to facilitate the functional annotation of the proteins that were identified in citrus. The main metabolic pathways including glutathione metabolism and biosynthesis of secondary metabolites were enriched suggesting that the response to a range of environmental factors is one of the key processes in citrus leaves. GDSL esterase/lipase variants (A0A067EBP6, A0A067EBA9, A0A067EF15, A0A067ENI5, A0A067EMQ7, and V4TXR3) and hydrolytic enzymes with multifunctional properties previously undescribed for citrus species, were characterized.
2. Results
2.1. Protein Identification
Figure 1 displays the strategy adopted in this study. The experimental procedure was divided in three stages: protein extraction, separation, and identification of proteins by MS analysis combined with bioinformatics.
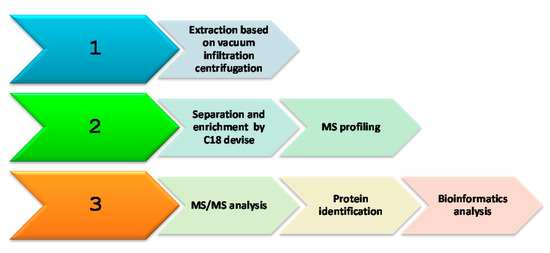
Figure 1.
Schematic overview of the workflow.
Many methods which are generally used to extract proteins from plant tissues have been based on a vacuum infiltration centrifugation together with an extraction solution process, followed by centrifugation [21,42]. We used a classical vacuum infiltration centrifugation method, slightly modified according to the description provided in the Materials and Methods section. It is known that proteins can be selectively solubilized depending on the chemical used for the extraction. The use of salt solutions is a commonly accepted tool [43,44], likewise NaCl has proven to be more effective in releasing the greatest number of proteins [43,44]. We applied a single washing step to obtain a protein fraction that was chromatographically fractionated by adopting a C18 cartridge. All chromatographic fractions were monitored by linear MALDI MS, in order to evaluate the intact protein mass information. Representative MALDI MS spectra are reported in Figure 2 (fractions 39 and 47; Figure S1). Peak overlapping and charge state ambiguity occur to some extent in a top-down analysis of intact proteins using MALDI TOF-TOF platforms. In fact, mono and multicharged protein ions (+1, +2, +3, +4 and +6) were detected in several fractions. The protein precursor ions and the dissociation method employed affects the structural information that can be produced in a MS/MS experiment. The dissociation of intact proteins is a more difficult process than the peptide fragmentation. Top-down protein identification by database search based on peptide sequence tags from the MS/MS spectrum has been reported only for platforms displaying high resolving power [45,46,47,48,49,50,51,52,53]. Several approaches have been applied to obtain primary structure information from entire protein ions for proteins with molecular weights as large as several tens of kilodaltons [45,46,47,48,49,50,51,52,53]. The ions observed for the unknown proteins from fraction 47 were 41 kDa, 40 kDa (which are the calculated average mass from +3 and +6 protein ions) and 31kDa (from +2 and +4 protein ions, Figure 2). The electrophoretic profile resulting from fraction 47 highlighted the presence of two protein bands within 30–44 kDa (Figure S2). Therefore, the protein profile displayed by linear MALDI experiments agreed with that obtained by SDS-PAGE. Sequence information for the unknown proteins were obtained by digesting all fractions and performing MS/MS experiments on the digestion products. MS data obtained from a typical digested fraction, e.g., fraction 47, were directly subjected to the National Center for Biotechnology Information (NCBI) database for protein identification against other green plants.
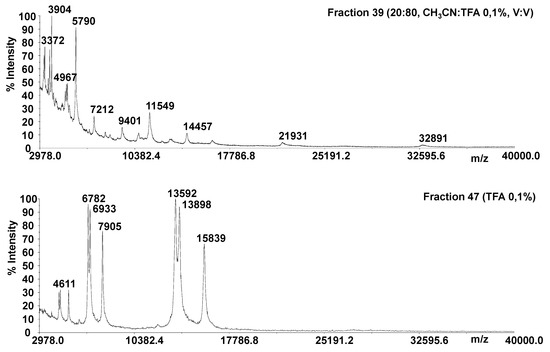
Figure 2.
Linear matrix-assisted laser desorption ionization mass spectrometry (MALDI MS) of the chromatographic fractions 39 and 47.
The database output allowed to identify a lipase-GDSL, by using 11 masses corresponding to six possible peptide sequences (gi|641833486, gi|641833485, gi|641833487, gi|568850564, gi|567901604, gi|641833488), characterized by significant protein scores (>60).
Therefore, spectral data collected from MS/MS experiments performed on all digested chromatographic fractions were subjected to a database search (Protein Pilot software) for the identification of proteins. A total of 78 proteins, belonging to Citrus aurantium and Citrus sinensis species, were identified with a significant match (Table 1). MS and MS/MS searches were performed against Citrus aurantium [43165] and Citrus sinensis [2711] protein sequence database, including sequences derived from SwissProt and TrEMBL (Translated EMBL Nucleotide Sequence Data Library). Alanine-tRNA ligase (A0A067FLL5, Table 1, row 1), Glutathione S-transferase (Q3HM93, Table 1, row 4), non-specific serine/threonine protein kinase (A0A067F884, Table 1, row 5), and RING-type E3 ubiquitin transferase (A0A067GDZ1, Table 1, row 7) represent the only named, however unreviewed MS/MS identified sequences.

Table 1.
Identified proteins by matrix-assisted laser desorption ionization tandem mass spectrometry (MALDI MS/MS) and Protein Pilot Paragon Method. The tandem mass spectrometry (MS/MS) data were processed using a mass tolerance of 10 ppm and 0.2 Da for the precursor and fragment ions, respectively.
2.2. Bioinformatic Analysis
2.2.1. Prediction of Biological Processes and Protein Class
The proteomics generated dataset (Table 1) was sorted into 12 different functional classes, cell adhesion molecule, cytoskeletal protein, enzyme modulator, hydrolase, ligase, nucleic acid binding, oxidoreductase, signaling molecule, storage protein, transcription factor, transferase and transporter. The dataset was analyzed using the genome of Citrus sinensis as the reference. PANTHER classification resulted in 45 gene entries which were analyzed for functional classification (Figure 3).
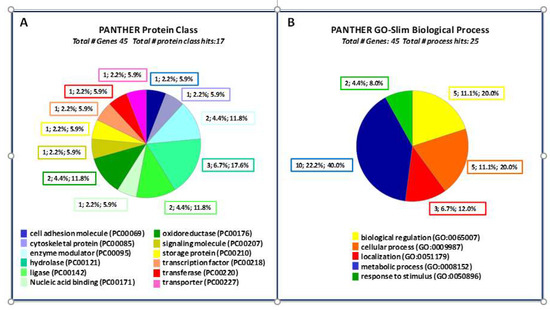
Figure 3.
PANTHER functional classification viewed in pie chart. (A) Protein Class; (B) Biological Process.
2.2.2. Subcellular Localization Prediction
The subcellular localization of a protein is an important step in understanding its function. In this study TargetP, SignalP, ChloroP, WallProtDB and mGOASVM-Loc were used to predict the subcellular localization of the 78 identified proteins. The FASTA format of all proteins identified using Citrus aurantium [43165] and Citrus sinensis [2711] database was used for bioinformatic analysis.
The following strategy was adopted to distinguish the subcellular localization: (a) proteins having a signal peptide predicted by TargetP and SignalP were gathered in “secretory pathways”, (b) TargetP was used to predict the mitochondrial and chloroplast localization, (c) mGOASVM-Loc (Multi-Label Protein Subcellular Localization) was used for multi-location proteins, and (d) WallProtDB was used as a database resource for plant cell wall proteomics. The identified proteins (Table 1) were classified for their subcellular localization as deduced by TargetP1.1 [54]. The location assignment is based on the predicted presence of any of the N-terminal pre-sequences: chloroplast transit peptide (cTP), mitochondrial targeting peptide (mTP), or secretory pathway signal peptide (SP). TargetP output revealed 4 proteins containing a chloroplast transit peptide (cTP), 16 proteins containing secretory pathway signal peptides (SP), and 56 proteins were labeled as “other”, choosing specificity > 0.95 (cut-off restrictions were set as follows: 0.730 (cTP), 0.860 (mTP), 0.430 (SP) and 0.840 (other), respectively). The presence of a SP in a protein is considered as the signature of a secretory protein because much of the secretory protein is translocated across the endoplasmic reticulum (ER). Secretory proteins are directed to the ER membrane by an N-terminal signal peptide and are translocated by the same mechanism. Since TargetP also predicted proteins containing a chloroplast transit peptide (cTP), the FASTA formats of all proteins were submitted to ChloroP for a detailed report of the cTP scores along the sequences (Table S1).
The ChloroP output highlighted seven proteins (A0A067GUC9, A0A067GUN6, V4TXR3, A0A067EZE8, A0A067ECH7, A0A067EJ84, and A0A067FBM6) containing a cTP (Table S1). The predicted TargetP results were selected, and since this database can deal with multi-location proteins, the localization generated by mGOASVM-Loc was accepted. The proteins did not univocally assign to a compartment and so they were categorized as uncertain due to the discrepancies among the five programs. Finally, SignalP, TargetP, mGOASVM-Loc and WallProtDB tools for proteomics-generated data sets revealed the presence of eight subcellular fractions, including secretory pathway (20), cytoplasm (16), nucleus (8), cell membrane (6), chloroplast (4), endoplasmic-reticulum (3) peroxisome (1) and uncertain (29) (Table S1). According to the annotation protein function in Prosite (https://prosite.expasy.org), 20 proteins localized in the secretory pathway were sorted into nine groups: peptidase family A1, membrane lipoprotein, aspartyl proteases, sugar transport, soluble glutathione S-transferase N-terminal, soluble glutathione S-transferase C-terminal, sugar transport proteins, specific tyrosine protein kinases, and protein kinases (Table S2).
2.2.3. Pathways Enrichment Analysis
The domains are the structural and functional units of proteins and can be used to assign an undescribed sequenced protein to a specific family of proteins, and to further formulate hypotheses about its function (Figure 4).
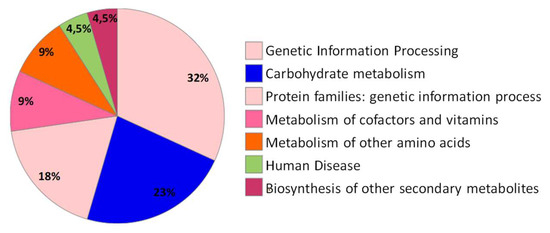
Figure 4.
The protein sequences of citrus-specific genes were functionally annotated with metabolic information from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database using KEGG Orthology And Links Annotation (BlastKOALA) program.
A critical step in understanding protein functions is the identification of relevant protein–protein interactions, such as the direct physical binding, indirect interaction and participation in the same metabolic pathways or cellular processes. Protein association network analysis is usually performed by the STRING database (Search Tool for the Retrieval of Interacting Genes/Proteins) [55]. This database includes interactions which have been described in the literature on the basis of experimentally studied relationships, as well as those obtained from the genome analysis performed using several methods that establish domain fusion, phylogenetic profiling and gene neighborhood concepts. Accordingly, a confidence score for every protein–protein association is assigned to the network. Higher scores designate an association supported by several types of evidence. In the present investigation, STRING analysis was exploited on the organism Citrus sinensis, using the list of 78 protein annotations (accession number from UniProtKB) reported in Table 1. STRING associated only 37 proteins matching the input list and failed the analysis, since the data set probably is a random collection of proteins that are not very well connected. This does not necessarily mean that it is not a biologically meaningful selection of proteins, but it could simply be that these proteins have not been studied and that their interactions might not yet be known to STRING.
BlastKOALA (Basic Local Alignment Search Tool-KEGG Orthology and Links Annotation, http://www.kegg.jp/blastkoala/) is automatic annotation server for genome and metagenome sequences, which perform KO (KEGG Orthology) assignments to characterize individual gene functions and reconstruct KEGG pathways. The 78 unique protein identifiers (Table 1, ID) were subjected to BlastKOALA [56] to obtain their corresponding K-numbers to further investigate the biological function of the proteins in the citrus leaves. Of the 78 entries, 22 entries (28.2%) were annotated. This provided a list of 22 unique K numbers that was then used for Kyoto Encyclopedia of Genes and Genomes (KEGG) mapping [57]. The K number assigned sequences were categorized according to the KEGG Orthology system (ko00001). The highlighted functional categories of annotated genes according to the KO system were genetic information processing (7 entries), carbohydrate metabolism (5 entries), protein families: genetic information processing (4 entries), metabolism of other amino acids (2 entries), metabolism of cofactors and vitamins (2 entries), human diseases (1 entries) and biosynthesis of other secondary metabolites (1 entries) (Figure 4). These 22 K numbers were mapped to 18 KEGG pathways (Tables S3 and S4) and 5 modules. The main pathways were “Metabolic pathways—Citrus sinensis (Valencia orange)” (score 81), “glutathione metabolism—Citrus sinensis (Valencia orange)” (score 58) and “biosynthesis of secondary metabolites—Citrus sinensis (Valencia orange)”(score 23).
2.3. GDSL Esterase-Lipase Characterization
The data reported in Table 1 highlight that several proteins belong to the GDSL lipase family (Table 1, lanes 61–66, gray region). GDSL lipases have been found in various plant species, including Arabidopsis thaliana, rice and maize, and their roles in plant development, morphogenesis and the defense response have been described [58,59]. Therefore, to improve the sequence coverage and characterization of proteins, all tandem mass spectra recorded for the single fraction 47 were evaluated by the MASCOT database searching. The oxidation of methionine and acetylation of protein N-term were also taken in account as the variable modifications. The results were carefully validated by a manual check of the corresponding MS/MS spectra. Six isoforms were recognized by direct submission of MALDI MS/MS data for protein identification. Table 2 shows collectively the peptide sequences produced by trypsin digestion and useful in identifying GDSL family. The alignment of the six identified GDSL variants with the GDSL sequence of A. thaliana (GDL79_ARATH) is reported in Figure 5. The MS/MS identified regions are colored, red used for the catalytic sites, and yellow for the GDSL motif. GDSL lipases represent a subfamily of lipolytic enzymes and possess a conserved catalytic triad (Ser, Asp, and His) [60]. However, unlike lipases that commonly contain a GxSxG motif, GDSL lipases exhibit a GxSxxxxG motif, in which the active site Ser is located near the N–terminus [60]. The alignment of peptides identified by MS/MS allowed to validate the expressed protein sequence (Figure 6, Table S5). The main protein microheterogeneity region suggested four isoforms to be present in the sample. Catalytic sites are not included in the identified peptides, and their positions are deduced only from the alignment with the validated sequence of A. thaliana. The prediction of protein functions and/or functional domains by bioinformatics tools is commonly used to classify an unknown. In these cases, the assumption is that proteins sharing functional domains have the same activity. The results obtained by comparing amino acid sequences (A0A067EBP6, A0A067EBA9, A0A067EF15, A0A067ENI5, A0A067EMQ7, V4TXR3) were combined with searches for functional domains (http://www.ebi.ac.uk/InterProScan/).

Table 2.
MS/MS identified peptides from GDSL esterase family by trypsin digestion.
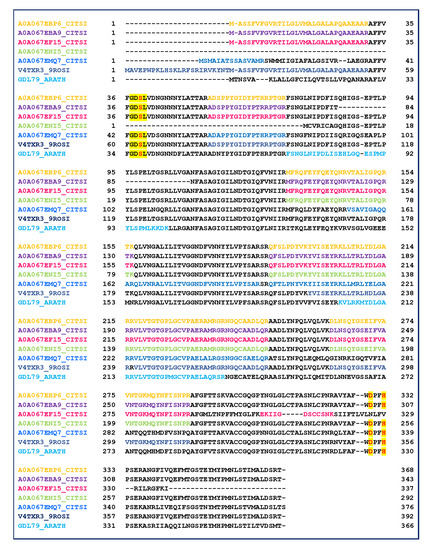
Figure 5.
Alignment of the six identified GDSL variants with the sequence of A. Thaliana (GDL79_ARATH). MS/MS validated sequences are colored, while the yellow highlight the GDSL motif and the active sites (red amino acids).

Figure 6.
Sequence of GDSL from citrus by alignment of MS/MS validated peptides, using as reference sequence A0A067EBP6_CITSI (blue string). In bold is reported the validated sequences, in red is indicated the punctual modifications and in yellow is highlighted the catalytic triad, while the gray highlights the region with the major microheterogeneity of the protein.
The predicted functional class was GDSL lipase/esterase-like (IPR035669), a plant specific subfamily of the SGNH-family of hydrolases, acting on ester bonds. The SGNH hydrolase superfamily represents a subgroup of the GDSL family, based on the presence of four residues Ser, Gly, Asn, and His which are present in four conserved regions (blocks I, II, III, and V, respectively). This subgroup of enzymes has been found to be secreted and involved in the response to stimuli [61]. The signal peptide indicated in UniProt as 1–28 for the sequences A0A067EBP6_CITSI, A0A067EBA9_CITSI, A0A067EF15_CITSI, confirmed the secreted nature of those proteins.
3. Discussion
In the present study, a MS-based proteomic analysis was used for the analysis of leaves from Citrus aurantium, growing under normal conditions. A total of 78 proteins belonging to citrus species were identified through proteomics-generated data sets. MS and MS/MS searches were performed against the Citrus aurantium [43165] and Citrus sinensis [2711] protein sequence database, including sequences derived from SwissProt and TrEMBL (Translated EMBL Nucleotide Sequence Data Library). Although, the Citrus aurantium database is the most appropriate for identifying species-specific gene products, it suffers from the inherent limitation due to reviewed sequences (only 101 entries). To overcome this limitation and to expand the dataset of the identified proteins, the TrEMBL [43064 entries] database research was performed.
Among the 78 identified only four proteins are “named” but “unreviewed”: Alanine-tRNA ligase (A0A067FLL5, Table 1, row 1), Glutathione S-transferase (Q3HM93, Table 1, row 4), non-specific serine/threonine protein kinase (A0A067F884, Table 1, row 5), and RING-type E3 ubiquitin transferase (A0A067GDZ1, Table 1, row 7). Alanine-tRNA ligase (A0A067FLL5, Table 1, row 1) catalyzes the attachment of alanine to tRNA. Literature data reported RNA ligases to be active in vitro on a variety of substrates, and capable of inter- and intra-molecular RNA joining. Their role in vivo might comprise yet unknown essential functions aside from their involvement in pre-tRNA splicing [62]. Glutathione S-transferase (GST, Q3HM93, Table 1, row 4) is involved in the metabolic process of transport and/or accumulation of both anthocyanins and proanthocyanidins in the vacuole, that are well known plant pigments sharing common flavonoid intermediates until the formation of anthocyanidins. Literature data on A. thaliana report that the GST binding activity is affected by a single amino acid substitution. GST overexpression has been found to enhance the growth of transgenic tobacco seedlings during stress [63,64]. The non-specific serine/threonine protein kinase (A0A067F884, Table 1, row 5) plays an important role in the plant defense response in A. thaliana [65,66]. The RING-type E3 ubiquitin transferase (A0A067GDZ1, Table 1, row 7) regulates the defense response of a plant to pathogenic agents. E3 ubiquitin ligase activity is correlated to the cell death and defense in Solanaceae and Brassicaceae, as reported in the literature [67].
The list of MS/MS identified proteins (Table 1) does not represent the whole predicted proteome of citrus leaves. There are several reasons that can limit the coverage of proteome observed for C. aurantium leaves. It must be underlined that the mild extraction procedures employed did not allow extracting highly hydrophobic proteins, which may be considered a limitation of this approach. It could be thought that some proteins were probably present at concentrations that might be undetectable by the currently employed separation and sequencing techniques, although the highly sensitive MALDI TOF/TOF platform used in this investigation can generally enable the detection of very low amounts of analytes (10 pmol/μL).
According to SignalP, TargetP, mGOASVM-Loc, and WallProtDB the proteomics-generated data set (Table 1) was sorted into eight subcellular fractions, recognized as secretory pathway, cytoplasm, nucleus, cell membrane, chloroplast, endoplasmic-reticulum, peroxisome and “uncertain”. The largest subcellular fraction was the secretory pathway, accounting for 25% of total proteins. However, only 11 of the 20 secretory pathway proteins were indicated as cell wall proteins, according to CellWallDB. Recently, it has been reported that proteins present in the cell wall, lacking a signal peptide, may be excluded via more than a single non-classical secreted mechanism, such as secretory exosomes, lysosomes membrane, transporting and unknown [44,68,69,70]. In fact, proteins A0A067DDE4 and A0A067EPP0 (Table 1, row 13 and 30) were also retrieved in WallCellDB although they did not hold a signal peptide. A0A067EPP0 (LRR receptor-like serine threonine-protein kinase) and A0A067DDE4 (protein kinase) are two protein kinase domain-containing proteins (Table S2) found in grapevine [71] and thought to be involved in the development and stress responses. According to the annotate protein function in Prosite (https://prosite.expasy.org), 20 proteins localized in the secretory pathway were sorted into nine groups: peptidase family A1, membrane lipoprotein, aspartyl proteases, sugar transport, soluble glutathione S-transferase N-terminal, soluble glutathione S-transferase C-terminal, sugar transport proteins, specific tyrosine protein kinases, and protein kinases (Table S2). The role and the biological functions of the proteins belonging to peptidase family A1 and aspartyl proteases are still hypothetical. These proteases are involved in protein processing and/or degradation in different plant organs, as well as in plant senescence, stress responses, programmed cell death and reproduction.
The MS-based approach was also successfully used for the identification of six isoforms of GDSL (A0A067EBP6, A0A067EBA9, A0A067EF15, A0A067ENI5, A0A067EMQ7, V4TXR3), displaying point mutations in the region 186–214, as is well established by MS/MS experiments. The characterization of these stress responsive hydrolytic enzymes in C. aurantium is here reported for the first time. A GDSL-lipase family protein, called GLIP, has previously been identified as stress responsive secreted proteins in Arabidopsis thaliana [72]. This subclass of lipolytic enzymes has been related to seed development, lipid metabolism [73], and cutin formation [74]. Studies focusing on the secretion of GDSL-lipase family proteins have suggested the multiple functions that these enzymes exert in plants under normal growth and stress conditions [43,44].
GO enrichment analysis highlight some features of leaves proteome. Firstly, the main metabolic pathways including glutathione metabolism and biosynthesis of secondary metabolites were enriched suggesting that the response to a range of environmental factors is the key processes in citrus leaves. Plants deploy secondary metabolites to assist the interactions with the biotic and abiotic environment, including the essential role of chemical defense against herbivores and pathogens. The deployment of secondary metabolites, i.e., molecules that have no direct role in the primary functions, depends on genetic variability and can also be modified in response to environmental factors [75]. Glutathione metabolism is also correlated to the plant defense system and is directly linked to sulfur metabolism. Holler reported a link between the activation of cysteine and glutathione metabolism with sulfur-induced resistance in tobacco plants [76]. In particular, glutathione is known to be involved in plant defense reactions as a signaling molecule, and it has also been reported to cross-communicate with other established signaling molecules [76]. The key enzyme of pathways is Glutathione S-transferase (GST, EC. 1.1.18). GSTs represent a multifunctional family of enzymes may be involved in the conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. Evidence suggests that GSTs play an important role in the detoxification of both endogenous and xenobiotic compounds, and they are also involved in intracellular transport, bio-synthesis of hormones, and protection against oxidative stress [77,78,79,80,81]. The analysis of the gene expressions in orange leaf indicated that the isoforms GSTU1, and GSTU2 are distinctly expressed in the leaf [82] It was also showed that the expression of U1 gene was remarkably induced in response to stress while the U2 isoform was constitutively expressed playing some sort of“ default scavenging” activity in vivo. GSTs provide a tool to control weeds in agronomic crops [83,84]. The overexpression of heterologous GST genes is widely related to enhance the crop qualitative and quantitative features. The antioxidant activity of GST limits the damages and the extent of programmed cell death during the hypersensitive response. In particular, the GST expression is up-regulated during the resistance process against pathogenic attack and represents a positive regulator. Therefore, proteomic data reported here highlight that Citrus aurantium might be a rootstock with good features for the correct and optimal growth of citrus fruit trees.
4. Materials and Methods
4.1. Chemicals
Trifluoroacetic acid (TFA), methanol (MeOH), acetonitrile (ACN), H2O, acetone (CH3COCH3) ammonium bicarbonate (NH4HCO3, 99.5%), trypsin, α-cyano-4-hydroxy-trans-cynnamic acid (α-CHCA, 99.0%) and sodium chloride (NaCl, ≥ 99%) were purchased from Sigma-Aldrich (Italy).
4.2. Plant Materials
Experiments were carried out on leaves from three Citrus aurantium var. amara plants. To minimize errors, three biological repeats were conducted for proteome analysis. For each biological repeat sample, ten leaves from 3 Citrus aurantium plants were pooled. All experiments were repeated in three independent times, resulting in three technical and three biological replicates. Plants were grown in a botanical garden (Orto Botanico, 964H + QJ Arcavacata, Rende CS) under natural conditions. Standard cultural practices included drip irrigation. Drip irrigation frequency was modified to seasons and ranged from once weekly (winter) to five days/week (summer), with 40 L tree-1 per irrigation. Leaves were harvested during October 2018 (wet season). The age of the plants was 3 years.
4.3. Protein Extraction
Citrus aurantium leaves (5 g) were washed with deionized water and then cut into segments. Leaf segments (5 cm) were placed in a centrifuge tube and added with 15 mL of NaCl 50 mM to extract proteins by constant horizontal shaking (200 rpm) for 1 h on ice, followed by vacuum-infiltration and centrifugation at 1500× g for 10 min at 4 °C. Thereafter, the supernatant was added with 5 mL of CH3COCH3 vortexed for 10 min. After centrifugation (1500× g, 5 min), the organic solvent was removed under nitrogen flow, the aqueous proteins solution was reduced to 4 mL in a vacuum centrifuge (Speed-Vac, Cryo Rivoire) and stored at −20 °C until analysis. Protein concentration (100 μg/mL) was determined using the UV-160 spectrophotometer (Shimadzu, Kyoto, Japan) by the Bradford method. Bovine Serum Albumin (BSA) was used as the standard.
4.4. Solid Phase Extraction (SPE) Procedures
Protein extract was pre-purified by SPE (55 um, 70 A, Phenomenex, USA) equilibrated with acidified water (0.1% trifluoroacetic acid, TFA). Four milliliters of protein extract (100 μg/mL) were added with 2 µL of TFA 2% and loaded and washed with 1ml water. Elutions were performed with 20%, 40%, 60% and 80% acetonitrile in acidified water (4 mL for each step) [85]. All fractions were freeze-dried in a vacuum centrifuge (Speed-Vac, Cryo Rivoire) and subsequently reconstituted with NH4HCO3 (50 mM) at 1/10 of the initial volume. An aliquot (1 μL) of each fraction was analyzed by MALDI MS in linear mode. Fractions showing the same MALDI protein profile were unified.
4.5. SDS PAGE
A series of SPE fractions were separated by one-dimensional SDS-PAGE (Electronic Supplementary Material, Figure S1 ESM). Each SPE fraction (≈ 8 μg) was mixed with 5× gel loading buffer, containing 2-mercaptoethanol and bromophenol blue, denatured at 95 °C for 10 min before electrophoresis analysis in 12.5% sodium dodecyl sulphate-polyacrylamide gel electrophoresis. A homemade protein molecular weight marker (Lactoferrin 87 kDa, L9507; Bovine Serum Albumin 66 kDa, A2153; Albumin from chicken 44 kDa, A5503; Mioglobin from equine skeletal muscle 17 kDa, M0630; Cytocrome C 12 kDa, C2506) was loaded in the molecular weight marker lane. Proteins were stained with Comassie Brillant Blu R-250 for 4 h and destained overnight with a solution containing 40% MeOH, 10% CH3COOH and 50% H2O.
4.6. In-Solution Digestion
In-solution protein digestion was performed by adding 2 µL of trypsin (4 pmol/µL) to each chromatographic fraction. The complete microwave-assisted digestion was obtained after three treatments in the microwave (MWD 246 SL, Whirlpool Europe, Italy) at 250 W irradiation power each lasting for 3 min.
4.7. Mass Spectrometry Analysis
A 1 μL amount of each protein chromatographic fraction was mixed with 10 μL of α-CHCA (5 mg/mL). A 1 μL portion of sample−matrix solution was spotted on a MALDI matrix target, dried at room temperature, and directly analyzed by MALDI mass spectrometry. MS analyses were performed using a 5800 MALDI TOF/TOF analyzer (AB SCIEX, Germany) equipped with a neodymium: yttrium-aluminium-garnet laser (349 nm). Linear MALDI MS spectra were acquired averaging 4000 laser shots with a mass accuracy of 500 ppm in default calibration mode that was performed using the following set of standards: aldolase (rabbit, [M+H]+avg = 39905), BSA (bovin serum albumin [M+H]+avg = 66431) and IgG1 (murine myeloma [M+H]+avg = 148500).
Tryptic peptide solution (1 μL) was mixed with 10 μL of α-CHCA. A 1 μL portion of sample−matrix solution was spotted on a MALDI matrix target, dried at room temperature, and directly analyzed by MALDI mass spectrometry in reflectron positive mode with a mass accuracy of 5 ppm. Typically, 4000 laser shots were accumulated with a laser pulse rate of 400 Hz in the MS mode, whereas in the MS/MS mode spectra up to 5000 laser shots were acquired and averaged with a pulse rate of 1000 Hz. MS/MS experiments were performed at a collision energy of 1 kV, and ambient air was used as the collision gas with a medium pressure of 10−6 Torr. Protein identification was performed by the Protein Pilot 4.0 software program (AB Sciex) using the Paragon (AB Sciex) protein database search algorithm. The data analysis parameters were as follows: Sample Type: Identification; Cys Alkylation: None; Digestion: Trypsin; Instrument: 5800; Special factors: None; Species: None ID; Focus: Biological modifications—Amino acid substitution; Database: uniprot-taxonomy_Citrus + aurantium + (Bitter+orange) + (Citrus+vulgaris) + [43165]_.fasta and uniprot-taxonomy_ Citrus + sinensis + (Sweet + orange) + (Citrus+aurantium + var + sinensis) + [2711]_.fasta; Search Effort: Thorough ID; FDR analysis: Yes; Used Modified Parameter Files: No; Detected Protein Threshold [Unused ProtScore (Conf)]:1.5 (95.0%). Spectra were also handled using Data Explorer version 4.11 (AB Sciex). The MS/MS data were processed to assign candidate peptides in the NCBI and UniProt database using the MASCOT search program (http://www.matrixscience.com). The mass tolerance of the parent and fragments for MS/MS data search was set at 10 ppm and 0.20 Da, respectively. The query was made for “Other Green Plants” taxonomy allowing 2 missed cleavage. A Peak-list of 50 fragment ions of intensity higher than 10% above the noise level was generically used for the database search. All spectra were manually checked to verify the validity of the MASCOT results.
4.8. Database Proteomics, Targeting Predictions and Functional Classification
The presence and location of signal peptide cleavage sites are predicted by the SignalP 3.0 server, which contains two prediction programs (SignalP-HMM and Signal-NN; http://www.cbs.dtu.dk/services/SignalP) [54].
SignalP software (http://www.cbs.dtu.dk/services/SignalP), which searches for signal sequences and their cleavage sites, is generally used to determine whether identified secreted proteins contain signal peptides. TargetP [86] is applied to predict the mitochondrial localization and not to consider them as secreted proteins. Additionally, Multi-Label Protein Subcellular Localization Prediction (mGOASVM (V1), http://bioinfo.eie.polyu.edu.hk/mGoaSvmServer/mGOASVM_v1.html) [87] was adopted for general localization prediction purpose. Identification of conserved domains in identified proteins is performed using the Prosite (https://prosite.expasy.org).
Network analysis was performed submitting the orthologous Arabidopsis ID to the STRING (Search Tool for the Retrieval of Interacting Genes) software (v.11) (http://stringdb.org/) [55] Functional and Gene Ontology (GO) analysis was performed by PANTHER program (http://www.pantherdb.org/) [88] and BlastKOALA (Query dataset: 78 entries; Taxonomy group: Eukaryotes, Plants; KEGG database searched: family_eukaryotes.pep,genus_prokaryotes.pep; 22 entries (28.2%) annotated) [56,57].
Supplementary Materials
The following are available online at https://www.mdpi.com/1420-3049/25/7/1485/s1, Figure S1: Linear MALDI spectra of the chromatographic fractions. Figure S2: SDS-PAGE of fraction 47. Table S1. Predicted subcellular localization of the 78 identified proteins. Table S2: PROSITE output for the 20 proteins localized in the secretory pathway. Table S3: K numbers (KO) by BlastKOALA (https://www.kegg.jp/blastkoala/). Table S4: KEGG Mapper Search Result. Table S5. MS/MS identified peptides of GDSLs.
Author Contributions
Conceptualization, A.N.; methodology, C.S., F.M., D.A.; software, D.A.; investigation, A.N.; D.A.; writing—original draft preparation, A.N.; writing—review and editing, L.D.D., and R.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Castle, W.S. A Career Perspective on Citrus Rootstocks, Their Development, and Commercialization. HortScience 2010, 45, 11–15. [Google Scholar] [CrossRef]
- Nimbolkar, P.K.; Awachare, C.; Reddy, Y.T.N.; Chander, S.; Hussain, F. Role of Rootstocks in Fruit Production—A Review. J. Agric. Eng. Food Technol. 2016, 3, 183–188. [Google Scholar]
- Vensel, W.H.; Tanaka, C.K.; Cai, N.; Wong, J.H.; Buchanan, B.B.; Hurkman, W.J. Developmental changes in the metabolic protein profiles of wheat endosperm. Proteomics 2005, 5, 1594–1611. [Google Scholar] [CrossRef]
- Balmer, Y.; Vensel, W.H.; Dupont, F.M.; Buchanan, B.B.; Hurk-Man, W.J. Proteome of amyloplasts isolated from developing wheat endosperm presents evidence of broad metabolic capability. J. Exp. Bot. 2006, 57, 1591–1602. [Google Scholar] [CrossRef]
- Song, X.; Ni, Z.; Yao, Y.; Xie, C.; Li, Z.; Wu, H.; Zhang, Y.; Professo, Q.S. Wheat (Triticumaestivum L.) root proteome and differentially expressed root proteins between hybridand parents. Proteomics 2007, 7, 3538–3557. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, B.E.; Madden, R.D.; Ayoubi, P.; Porter, R.; Dillwith, J.W. The wheat (Triticum aestivum L.) leaf proteome. Proteomics 2005, 5, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, A.M.; Echevarria-Zomeno, S.; Jean-Baptiste, S.; Hernandez, M.; Jorrin-Novo, J.V. Evaluation of three different protocols of protein extraction for Arabidopsis thaliana leaf proteome analysis by two-dimensional electrophoresis. J. Proteom. 2008, 71, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.P.; Ventura, J.A.; Zingali, R.B.; Fernandes, R.M.B. Evaluation of sample preparation methods for the analysis of papaya leaf proteins through 2wo-dimensional gel electrophoresis. Phytochem. Anal. 2009, 20, 456–464. [Google Scholar] [CrossRef]
- Da Silva, M.A.O.; Garcia, J.S.; de Souza, G.; Eberlin, M.N.; Gozzo, F.C.; Arruda, M.A.Z. Evaluation of sample preparation protocols for proteomic analysis of sunflower leaves. Talanta 2010, 80, 1545–1551. [Google Scholar] [CrossRef]
- Xie, C.J.; Wang, D.; Yang, X.Y. Protein extraction methods compatible with proteomic analysis for the cotton seedling. Crop Sci. 2009, 49, 395–402. [Google Scholar] [CrossRef]
- Zheng, Q.; Song, J.; Doncaster, K.; Rowland, E.; Byers, D.M. Qualitative and quantitative evaluation of protein extraction protocols for apple and strawberry fruit suitable for two-dimensional electrophoresis and mass spectrometry analysis. J. Agric. Food Chem. 2007, 55, 1663–1673. [Google Scholar] [CrossRef] [PubMed]
- Napoli, A.; Aiello, D.; Di Donna, D.; Moschidis, P.; Sindona, G. Vegetable proteomics: The detection of Ole e 1 isoallergens by peptide matching of MALDI MS/MS spectra of underivatized and dansylated glycopeptides. J. Prot. Res. 2008, 7, 2723–2732. [Google Scholar] [CrossRef] [PubMed]
- Delaplace, P.; van der Wal, F.; Dierick, J.F.; Cordewener, J.H.G.; Fauconnier, M.L.; du Jardin, P.; America, A.H.P. Potato tuber proteomics: Comparison of two complementary extraction methods designed for 2-DE of acidic proteins. Proteomics 2006, 6, 6494–6497. [Google Scholar] [CrossRef] [PubMed]
- Jellouli, N.; Ben Salem, A.; Ghorbel, A.; Ben Jouira, H. Evaluation of protein extraction methods for Vitis vinifera leaf and root proteome analysis by two-dimensional electrophoresis. J. Integr. Plant Biol. 2010, 52, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Negri, A.S.; Prinsi, B.; Scienza, A.; Morgutti, S.; Cocucci, M.; Espen, L. Analysis of grape berry cell wall proteome: A comparative evaluation of extraction methods. J. Plant Physiol. 2008, 165, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Shi, M.J.; Lu, X.L.; Ma, R.F.; Wu, C.G.; Guo, A.P.; Peng, M.; Tian, W. A method for protein extraction from different subcellular fractions of laticifer latex in Hevea brasiliensis compatible with 2-DE and MS. Proteome Sci. 2010, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Yang, Y.W.; Liu, J.Y. An efficient protein preparation for proteomic analysis of developing cotton fibers by 2-DE. Electrophoresis 2006, 27, 4559–4569. [Google Scholar] [CrossRef]
- Carpentier, S.C.; Witters, E.; Laukens, K.; Deckers, P.; Swennen, R.; Panis, B. Preparation of protein extracts from recalcitrant plant tissues: An evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 2005, 5, 2497–2507. [Google Scholar] [CrossRef]
- Napoli, A.; Aiello, D.; Di Donna, L.; Sajjad, A.; Perri, E.; Sindona, G. Profiling of hydrophilic proteins from Olea europaea olive pollen by MALDI TOF mass spectrometry. Anal. Chem. 2006, 78, 3434–3443. [Google Scholar] [CrossRef]
- Fan, P.X.; Wang, X.C.; Kuang, T.Y.; Li, Y.X. An efficient method for the extraction of chloroplast proteins compatible for 2-DE and MS analysis. Electrophoresis 2009, 30, 3024–3033. [Google Scholar] [CrossRef]
- Witzel, K.; Shahzad, M.; Matros, A.; Mock, H.P.; Mühling, K.H. Comparative evaluation of extraction methods for apoplastic proteins from maize leaves. Plant Methods 2011, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Zukas, A.A.; Breksa, A.P., III. Extraction methods for analysis of Citrus leaf proteins by two-dimensional gel electrophoresis. J. Chromatogr. A 2005, 1078, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Maserti, B.E.; Della Croce, C.M.; Luro, F.; Morillon, R.; Cini, M.; Caltavuturo, L. A general method for the extraction of citrus leaf proteins and separation by 2D electrophoresis: A follow up. J. Chromatogr. B 2007, 849, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Maserti, B.E.; Del Carratore, R.; Della Croce, C.M.; Podda, A.; Migheli, Q.; Froelicher, Y.; Luro, F.; Morillon, R.; Ollitrault, P.; Talon, M.; et al. Comparative analysis of proteome changes induced by the two spotted spider mite Tetranychus urticae and methyl jasmonate in citrus leaves. J. Plant Physiol. 2011, 168, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Lliso, I.; Tadeo, F.R.; Phinney, B.S.; Wilkerson, C.G.; Talon, M. Protein changes in the albedo of citrus fruits on post-harvesting storage. J. Agric. Food Chem. 2007, 55, 9047–9053. [Google Scholar] [CrossRef] [PubMed]
- Cantú, M.D.; Mariano, A.G.; Palma, M.S.; Carrilho, E.; Wulff, N.A. Proteomic analysis reveals suppression of bark chitinases and proteinase inhibitors in Citrus plants affected by the Citrus Sudden Death Disease. Phytopathology 2008, 98, 1084–1092. [Google Scholar] [CrossRef]
- Shi, J.X.; Chen, S.; Gollop, N.; Goren RGoldschmidt, E.E.; Porat, R. Effects of anaerobic stress on the proteome of citrus fruit. Plant Sci. 2008, 175, 478–486. [Google Scholar] [CrossRef]
- Napoli, A.; Aiello, D.; Di Donna, L.; Prendushi, H.; Sindona, G. Exploitation of endogenous protease activity in raw mastitic milk by MALDI-TOF/TOF. Anal. Chem. 2007, 79, 5941–5948. [Google Scholar] [CrossRef]
- Aiello, D.; Materazzi, S.; Risoluti, R.; Thangavel, H.; Di Donna, L.; Mazzotti, F.; Casadonte, F.; Siciliano, C.; Sindona, G.; Napoli, A. Major allergen in rainbow trout (Oncorhynchus mykiss): Complete sequences of Parvalbumin by MALDI tandem mass spectrometry. Mol. BioSyst. 2015, 11, 2373–2381. [Google Scholar] [CrossRef]
- Aiello, D.; Casadonte, F.; Terracciano, R.; Damiano, R.; Savino, R.; Sindona, G.; Napoli, A. Targeted proteomics approach in prostatic tissue: A panel of potential biomarkers for cancer detection. Oncoscience 2016, 3, 220–241. [Google Scholar]
- Aiello, D.; Cardiano, P.; Cigala, R.M.; Gans, P.; Giacobello, F.; Giuffrè, O.; Napoli, A.; Sammartano, S. Sequestering ability of oligophosphate ligands toward Al3+ in Aqueous Solution. J. Chem. Eng. Data 2017, 62, 3981–3990. [Google Scholar] [CrossRef]
- Napoli, A.; Athanassopoulos, C.M.; Moschidis, P.; Aiello, D.; Di Donna, D.; Mazzotti, F.; Sindona, G. Solid phase isobaric mass tag reagent for simultaneous protein identification and assay. Anal. Chem. 2010, 82, 5552–5560. [Google Scholar] [CrossRef] [PubMed]
- Mazzotti, F.; Di Donna, L.; Napoli, A.; Aiello, D.; Siciliano, C.; Athanassopoulos, C.M.; Sindona, G. N-hydroxysuccinimidyl p-methoxybenzoate as suitable derivative reagent for isotopic dilution assay of biogenic amines in food. J. Mass Spectrom. 2014, 49, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Reddy, C.E.; Albanito, L.; De Marco, P.; Aiello, D.; Maggiolini, M.; Napoli, A.; Musti, A.M. Multisite phosphorylation of c-Jun at threonine 91/93/95 triggers the onset of c-Jun pro-apoptotic activity in cerebellar granule neurons. Cell Death Disease 2013, 4, e852. [Google Scholar] [CrossRef]
- Jahouh, F.; Saksena, R.; Aiello, D.; Napoli, A.; Sindona, G.; Kovàc, P.; Banoub, J.H. Glycation sites in neoglycoglycoconjugates from the terminal monosaccharide antigen of the O-PS of Vibrio cholerae O1; serotype Ogawa; and BSA revealed by matrix-assisted laser desorption–ionization tandem mass spectrometry. J. Mass. Spectrom. 2010, 45, 1148–1159. [Google Scholar] [CrossRef]
- Van Kampen, J.J.; Burgers, P.C.; de Groot, R.; Gruters, R.A.; Luider, T.M. Biomedical application of MALDI mass spectrometry for small-molecule analysis. Mass Spectrom. Rev. 2011, 30, 101–120. [Google Scholar] [CrossRef]
- Persike, M.; Karas, M. Rapid simultaneous quantitative determination of different small pharmaceutical drugs using a conventional matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system. Rapid Commun. Mass Spectrom. 2009, 23, 3555–3562. [Google Scholar] [CrossRef]
- Persike, M.; Zimmermann, M.; Klein, J.; Karas, M. Quantitative Determination of Acetylcholine and Choline in Microdialysis Samples by MALDI-TOF MS. Anal. Chem. 2010, 82, 922–929. [Google Scholar] [CrossRef]
- Meesters, R.J.; Hooff, G.P.; Gruters, R.; van Kampen, J.J.; Luider, T.M. Incurred sample reanalysis comparison of dried blood spots and plasma samples on the measurement of lopinavir in clinical samples. Bioanalysis 2012, 4, 237–240. [Google Scholar] [CrossRef]
- Van Kampen, J.J.; Burgers, P.C.; de Groot, R.; Luider, T.M. Qualitative and quantitative analysis of pharmaceutical compounds by MALDI-TOF mass spectrometry. Anal. Chem. 2006, 78, 5403–5411. [Google Scholar] [CrossRef][Green Version]
- Aiello, D.; Giambona, A.; Leto, F.; Passarello, C.; Damiani, G.; Maggio, A.; Siciliano, C.; Napoli, A. Human coelomic fluid investigation: A MS-based analytical approach to prenatal screening. Sci. Rep. 2018, 8, 10973. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, L.H.; Zhao, J.F.; Song, Y.; Zhang, C.J.; Guo, Y. Identification of an Apoplastic Protein Involved in the Initial Phase of Salt Stress Response in Rice Root by Two-Dimensional Electrophoresis. Plant Physiol. 2009, 149, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Ghahremani, M.; Stigter, K.A.; Plaxton, W. Extraction and characterization of extracellular proteins and their post-translational modifications from arabidopsis thaliana suspension cell cultures and seedlings: A critical review. Proteomes 2016, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Albenne, C.; Canut, H.; Jamet, E. Plant cell wall proteomics: The leadership of arabidopsis thaliana. Front. Plant Sci. 2013, 4, 111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Malmirchegini, G.R.; Clubb, R.T.; Loo, J.A. Native top-down mass spectrometry for the structural characterization of human hemoglobin. Eur. J. Mass Spectrom. 2015, 21, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Geer, L.Y.; Markey, S.P.; Kowalak, J.A.; Wagner, L.; Xu, M.; Maynard, D.M.; Yang, X.Y.; Shi, W.Y.; Bryant, S.H.J. Open mass spectrometry search algorithm. Proteome Res. 2004, 3, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Han, X.M.; Jin, M.; Breuker, K.; McLafferty, F.W. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science 2006, 314, 109–112. [Google Scholar] [CrossRef]
- Risoluti, R.; Piazzese, D.; Napoli, A.; Materazzi, S. Study of [2-(2′-pyridyl) imidazole] complexes to confirm two main characteristic thermoanalytical behaviors of transition metal complexes based on imidazole derivatives. J. Anal. Appl. Pyrolysis 2016, 117, 82–87. [Google Scholar] [CrossRef]
- Zubarev, R.A.; Kelleher, N.L.; McLafferty, F.W. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J. Am. Chem. Soc. 1998, 120, 3265–3266. [Google Scholar] [CrossRef]
- Syka, J.E.P.; Coon, J.J.; Schroeder, M.J.; Shabanowitz, J.; Hunt, D.F. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA 2004, 101, 9528–9533. [Google Scholar] [CrossRef]
- Pitteri, S.J.; Chrisman, P.A.; Hogan, J.M.; McLuckey, S.A. Electron-Transfer Ion/Ion Reactions of Doubly Protonated Peptides: Effect of Elevated Bath Gas Temperature. Anal. Chem. 2005, 77, 1831–1839. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Little, D.P.; Speir, J.P.; Senko, M.W.; Oconnor, P.B.; McLafferty, F.W. Infrared Multiphoton Dissociation of Large Multiply Charged Ions for Biomolecule Sequencing. Anal. Chem. 1994, 66, 2809–2815. [Google Scholar] [CrossRef] [PubMed]
- Laskin, J.; Futrell, J.H. Activation of large ions in FT-ICR mass spectrometry. Mass Spectrom. Rev. 2005, 24, 135–167. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2019, 29. [Google Scholar] [CrossRef]
- Brick, D.J.; Brumlik, M.J.; Buckley, J.T.; Cao, J.X.; Davies, P.C.; Misra, S.; Tranbarger, T.J.; Upton, C. A new family of lipolytic plant enzymes with members in rice; arabidopsis and maize. FEBS Lett. 1995, 377, 475–480. [Google Scholar]
- Lee, D.S.; Kim, B.K.; Kwon, S.J.; Jin, H.C.; Park, O.K. Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. Commun. 2009, 379, 1038–1042. [Google Scholar] [CrossRef]
- Akoh, C.C.; Lee, G.C.; Liaw, Y.C.; Huang, T.H.; Shaw, J.F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004, 43, 534–552. [Google Scholar] [CrossRef]
- Martinez-Esteso, M.J.; Selles-Marchart, S.; Vera-Urbina, J.C.; Pedreno, M.A.; Bru-Martinez, R. Changes of defense proteins in the extracellular proteome of grapevine (Vitis vinifera cv. Gamay) cell cultures in response to elicitors. J. Proteom. 2009, 73, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Englert, M.; Beier, H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005, 33, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gao, P.; Cui, D.; Wu, L.; Parkn, I.; Saberianfar, R.; Menassa, R.; Pan, H.; Westcottn, M.Y.; Gruber, M.Y. The Arabidopsis tt19-4 mutant differentially accumulates proanthocyanidin and anthocyanin through a 3’ amino acid substitution in glutathione S-transferase. Plant Cell Environ. 2011, 34, 374–388. [Google Scholar] [CrossRef] [PubMed]
- Roxas, V.P.; Smith, R.K., Jr.; Allen, E.R.; Allen, R.D. Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 1997, 15, 988–991. [Google Scholar] [CrossRef]
- Held, K.; Pascaud, F.; Eckert, C.; Gajdanowicz, P.; Hashimoto, K.; Corratgé-Faillie, C.; Offenborn, J.N.; Lacombe, B.; Dreyer, I.; Thibaud, J.B.; et al. Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 2011, 21, 1116–1130. [Google Scholar] [CrossRef]
- Romeis, T.; Ludwig, A.A.; Martin, R.; Jones, J.D. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 2001, 20, 5556–5567. [Google Scholar] [CrossRef]
- Yang, C.W.; Gonzàlez-Lamothe, R.; Ewan, R.A.; Rowland, O.; Yoshioka, H.; Shenton, M.; Ye, H.; O’Donnell, E.; Jones, J.D.G.; Sadanandoma, A. The E3 ubiquitin ligase activity of arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 2006, 18, 1084–1098. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Jwa, N.S.; Lebrun, M.H.; Job, D.; Rakwal, R. Plant secretome: Unlocking secrets of the secreted proteins. Proteomics 2010, 10, 799–827. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Lee, S.J. Straying off the highway: Trafficking of secreted plant proteins and complexity in the plant cell wall proteome. Plant Physiol. Biochem. 2010, 153, 433–436. [Google Scholar] [CrossRef]
- Zhu, J.M.; Chen, S.X.; Alvarez, S.; Asirvatham, V.S.; Schachtman, D.P.; Wu, Y.J.; Sharp, R.E. Cell wall proteome in the maize primary root elongation zone. I. Extraction and identification of water-soluble and lightly ionically bound proteins. Plant Physiol. 2006, 140, 311–325. [Google Scholar] [CrossRef]
- De Miccolis Angelini, R.M.; Rotolo, C.; Gerin, D.; Abate, D.; Pollastro, S.; Faretra, F. Global transcriptome analysis and differentially expressed genes in grapevine after application of the yeast-derived defense inducer cerevisane. Pest Manag. Sci. 2019, 75, 2020–2033. [Google Scholar] [CrossRef] [PubMed]
- Oh, I.S.; Park, A.R.; Bae, M.S.; Kwon, S.J.; Kim, Y.S.; Lee, J.E.; Kang, N.Y.; Lee, S.; Cheong, H.; Park, O.K. Secretome analysis reveals an Arabidopsis lipase involved in defence against Alternaria brassicicola. Plant Cell 2005, 17, 2832–2847. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.L.; Yan, S.Z.; Tan, R.K.; Zhang, Z.Y.; Wang, Z.; Chen, J. Characterization and expression of a GDSL-like lipase gene from Brassica napus in Nicotiana benthamiana. Protein J. 2014, 33, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.L.; Mounet, F.; Lemaire-Chamley, M.; Gaillard, C.; Elmorjani, K.; Vivancos, J.; Runavot, J.L.; Quemener, B.; Petit, J.; Germain, V.; et al. Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 2012, 24, 3119–3134. [Google Scholar] [CrossRef]
- Neilson, E.H.; Goodger, J.Q.D.; Woodrow, I.E.; Lindberg Møller, B. Plant chemical defense: At what cost? Trends Plant Sci. 2013, 18, 250–258. [Google Scholar] [CrossRef]
- Höller, K.; Király, L.; Künstler, A.; Müller, M.; Gullner, G.; Fattinger, M.; Zechmann, B. Enhanced glutathione metabolism is correlated with sulfur-induced resistance in tobacco mosaic virus-infected genetically susceptible Nicotiana tabacum plants. Mol. Plant Microbe Interact. 2010, 23, 1448–1459. [Google Scholar] [CrossRef]
- Lo Piero, A.R.; Puglisi, I.; Petrone, G. Gene isolation; analysis of expression and invitro synthesis of a glutathione S-transferase from orange fruit. [Citrus sinensis L. (Osbeck)]. J. Agric. Food Chem. 2006, 54, 9227–9233. [Google Scholar] [CrossRef]
- Mauch, F.; Dudler, R. Differential induction of distinct glutathione transferases of wheat by xenobiotics and by pathogen attack. Plant Physiol. 1993, 102, 1193–1201. [Google Scholar] [CrossRef]
- Moons, A. Osgstu3 and osgtu4; encoding tau class glutathione S-transferases; are heavy metal and hypoxic stress-induced and differentially salt stress-responsive in rice roots. FEBS Lett. 2003, 553, 427–432. [Google Scholar] [CrossRef]
- Bianchi, M.W.; Roux, C.; Vartanian, N. Drought regulation of GST8; encoding the Arabidopsis homologue of ParC/Nt107 glutathione transferase/peroxidase. Physiol. Plant 2002, 116, 96–105. [Google Scholar] [CrossRef]
- Edwards, R.; Dixon, D.P.; Walbot, V. Plant glutathione transferases: Enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000, 5, 193–198. [Google Scholar] [CrossRef]
- Lo Piero, A.R.; Mercurio, V.; Puglisi, I.; Petrone, G. Gene isolation and expression analysis of two distinct sweet orange [Citrus sinensis L. (Osbeck)] tau-type glutathione transferases. Gene 2009, 443, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Axarli, A.; Rigden, D.J.; Labrou, N.E. Characterization of the ligandin site of maize glutathione transferase I. Biochem. J. 2004, 382, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Kreuz, K.; Tommasini, R.; Martinoia, E. Herbicide detoxification in plants. Plant Physiol. 1996, 111, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Napoli, A.; Aiello, D.; Aiello, G.; Cappello, M.S.; Di Donna, L.; Mazzotti, F.; Materazzi, S.; Fiorillo, M.; Sindona, G. Mass spectrometry-based proteomic approach in Oenococcus oeni enological starter. J. Prot. Res. 2014, 13, 2856–2866. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP; SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Mak, M.-W.; Kun, S.-Y. mGOASVM: Multi-label protein subcellular localization based on gene ontology and support vector machines. BMC Bioinform. 2012, 13, 290. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes; a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).