Abstract
Because high-molecular-weight glutenin subunits (HMW-GS) are important contributors to wheat end-use quality, there is a need for high-throughput identification of HMW-GS in wheat genetic resources and breeding lines. We developed an optimized method using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) to distinguish individual HMW-GS by considering the effects of the alkylating reagent in protein extraction, solvent components, dissolving volume, and matrix II components. Using the optimized method, 18 of 22 HMW-GS were successfully identified in standard wheat cultivars by differences in molecular weights or by their associations with other tightly linked subunits. Interestingly, 1Bx7 subunits were divided into 1Bx7 group 1 and 1Bx7 group 2 proteins with molecular weights of about 82,400 and 83,000 Da, respectively. Cultivars containing the 1Bx7 group 2 proteins were distinguished from those containing 1Bx7OE using well-known DNA markers. HMW-GS 1Ax2* and 1Bx6 and 1By8 and 1By8*, which are difficult to distinguish due to very similar molecular weights, were easily identified using RP-HPLC. To validate the method, HMW-GS from 38 Korean wheat varieties previously evaluated by SDS-PAGE combined with RP-HPLC were analyzed by MALDI-TOF-MS. The optimized MALDI-TOF-MS method will be a rapid, high-throughput tool for selecting lines containing desirable HMW-GS for breeding efforts.
1. Introduction
The glutenins are abundant storage proteins accumulated in developing wheat endosperm that consist of high-molecular-glutenin subunits (HMW-GS) and low-molecular-weight subunits (LMW-GS) linked by disulfide bonds into large protein polymers. These polymers contribute to the viscoelastic properties of wheat flour dough in bread-making.
The HMW-GS are encoded at the Glu-A1, Glu-B1, and Glu-D1 loci on the long arms of chromosomes 1A, 1B, and 1D, respectively [1]. Each locus contains tightly-linked genes encoding two different types of HMW-GS, referred to as x- and y-type subunits [2,3]. The x-type subunits generally have higher molecular weights (MWs) than the y-type subunits. Most common wheat varieties express only three to five subunits due to the silencing of some genes, particularly those encoding the y subunits at the Glu-A1 locus [4]. Each subunit has been assigned a unique number and been given different quality scores related to bread-making properties [1,5,6,7]. For instance, the allelic pair 1Dx5 + 1Dy10 is associated with superior bread-making quality, especially dough strength. 1Ax2* and 1Dx17 + 1Dy18 also have positive effects on bread-making quality [8,9], while the allelic pair 1Dx2 + 1Dy12 is associated with poor bread-making quality [1,4,6]. In addition, overexpression of 1Bx7 subunit (1Bx7OE) in wheat with two functional copies of the 1Bx7 gene correlates with good dough strength [7,10,11,12,13]. Wheat varieties with different HMW-GS compositions are critical for providing the strength and extensibility of flour dough for different end uses [14].
In the past decades, a variety of techniques have been developed to characterize the allelic variation of HMW-GS. These include sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), two-dimensional electrophoresis (2-DE), polymerase chain reaction (PCR), high-performance capillary electrophoresis (HPCE), reversed-phase high-performance liquid chromatography (RP-HPLC), and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) [15,16,17,18,19,20,21,22,23]. SDS-PAGE is the method that is most frequently used in breeding programs because it is simple and easy to perform [20,24]. However, SDS-PAGE has poor reproducibility compared to other methods, and HMW-GS with similar electrophoretic mobilities are difficult to identify. 2-DE and tandem mass spectrometry techniques were also developed but their applications are limited because of the technical expertise required and high cost. PCR is a simpler technique and currently used in many breeding programs [25,26,27,28,29]. However, primers are not available for all HMW-GS alleles, particularly those identified recently. HPCE satisfied the requirements of efficient, fast, and automated separation [30,31,32]. However, it is difficult to achieve both high resolution and reproducibility and some HMW-GS yield multiple peaks [20,22]. RP-HPLC has been used generally to select specific subunits associated with superior quality [16,17,19,20,21,33]. In RP-HPLC, subunits are separated according to their surface hydrophobicities [34] and subunits with higher hydrophobicities elute faster than subunits with lower hydrophobicities. Compared with other methods, RP-HPLC is convenient, extremely precise, fully automated, and easy to use for identifying many HMW-GS. However, HMW-GS having similar hydrophobicities cannot be differentiated by this method. RP-HPLC also requires long separation times and consumes an abundance of chemicals. As a result, RP-HPLC is not available to many breeding programs. MALDI-TOF-MS overcomes many of the limitations of other methods and has become a powerful tool for characterizing wheat storage proteins because of its accuracy and sensitivity [35,36,37,38,39,40]. It also requires relatively small samples of protein and only a few minutes per sample for analysis [37].
In this study, we established optimal conditions for the separation of HMW-GS by MALDI-TOF-MS. Twenty-two HMW-GS of bread wheat were distinguished successfully in standard wheat cultivars. The method was then applied to 38 varieties of Korean wheat to determine HMW-GS compositions.
2. Results and Discussion
2.1. Sample Optimization for MALDI-TOF-MS Analysis
To develop a reliable method for the resolution of HMW-GS by MALDI-TOF-MS, four different experimental parameters were optimized using flour from the wheat cultivar “Chinese Spring”. These included the use of an alkylating reagent during protein extraction, four different solvent components, three different dissolving volumes, and four different matrix II components (Figure 1, Table 1). In total, 96 experiments with different combinations were performed. Conditions that yielded the best spectra are shown in Figure 1.
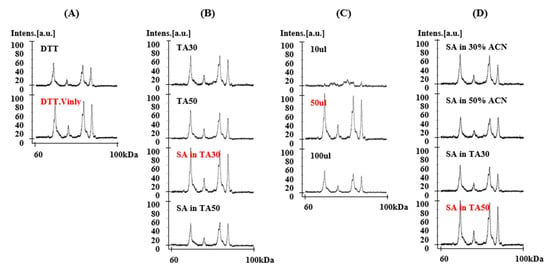
Figure 1.
Effects of four major factors on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) resolution of Chinese Spring using MALDI-TOF-MS. (A) treatment with alkylation reagent in glutenin extraction, (B) solvent components, (C) dilution volume, and (D) matrix II components. Parameters shown in red were found to be optimal and were incorporated into the new method.

Table 1.
Major factors influencing resolution of high-molecular-weight glutenin subunits (HMW-GS) by MALDI-TOF-MS.
2.1.1. Treatment of 4-Vinylpyridine as Alkylating Reagent in Protein Extraction
In the glutenin extraction step, 4-vinylpyridine (4-vp) was added to alkylate the cysteine residues and stop the formation of disulfide bonds [41]. 4-vp-treated samples showed increased resolution of mass spectra when compared to non-treated samples (Figure 1A). The molecular weights determined by MALDI-TOF-MS for 4-vp-treated or non-treated HMW-GS are shown in Table S1. The increase in molecular mass of individual HMW-GS 1Dy12, 1By8, 1Bx7, and 1Dx2 in 4-vp treated samples was 737.2, 714.9, 401.6, and 425.7 Da, respectively, which is in good agreement with previous knowledge that y-type HMW-GS have 7 cysteine residues, and most x-type HMW-GS have 4 cysteine residues (with the exception of 1Dx5 and 1Bx20 that have 5 and 2 cysteines, respectively). Although the measured increase in molecular mass differs somewhat from the 4-vp molecular mass of 105.14 Da, it is within the instrument’s common error range of a 90–115 Da increase for one cysteine [42]. Taking together the results of Wang et al. [42] and our results in this study, treatment with 4-vp showed enhanced resolution of mass spectra and could give useful information about the numbers of cysteine residues in the HMW-GS.
2.1.2. Solvent Components
HMW-GS are difficult to ionize because of their large molecular masses. Four different solvents for HMW-GS were evaluated, including TA30 (30% acetonitrile (ACN), 0.1% trifluoroacetic acid (TFA)), TA50 (50% ACN, 0.1% TFA), SA in TA30 (10 μg/μL sinapinic acid (SA) in TA30), and SA in TA50 (10 μg/μL SA in TA50) (Figure 1B). SA was added to the solvent to increase the ionization and is well known to be useful for the analysis of higher mass proteins [43]. The best resolution of the alkylated protein sample was obtained with SA in TA30 (Figure 1B).
2.1.3. Dissolving Volume
To test dilution volumes, 1 μL sample was diluted to 10 μL, 50 μL, and 100 μL of the resolving solvents. Figure 1C shows the results when the sample was diluted with SA in TA30. The 10 μL sample did not perform properly because the protein concentration was too high. The best results were obtained with a dilution volume of 50 μL.
2.1.4. Matrix II Components
The matrix transfers the energy required for ionization from the laser to the sample molecules. Two matrixes were prepared according to the double layer method using SA recommended by manufacturer. SA is considered best matrix to analyze high molecular mass proteins over 5 kDa. Matrix I was SA in EtOH at a concentration of 10 mg/500 μL as recommended by the manufacturer’s manual. Four different matrix II compositions—SA in 30% ACN, SA in 50% ACN, SA in TA30, and SA in TA50—were tested and compared in Figure 1D. The addition of TFA to the matrix solution provides high intense peaks [43]. SA in TA50 was the most suitable as matrix II (Figure 1D).
As a result, in the optimized method alkylated proteins were dissolved in 50 μL SA in TA30. SA in TA50 was used for Matrix II.
2.2. Identification of HMW-GS Compositions in Standard Wheat Varieties
The HMW-GS compositions of 24 standard wheat varieties that have been well studied in various references were characterized by MALDI-TOF-MS using the optimized method [20,33,44,45,46,47,48,49]. Allelic variation of standard wheat varieties at Glu-A1, Glu-B1, and Glu-D1 includes 22 subunits: 1Ax1 and 1Ax2* at Glu-A1, 1Bx6, 1Bx7, 1Bx7OE, 1Bx13, 1Bx14, 1Bx17, 1Bx20, 1By8, 1By8*, 1By9, 1By15, 1By16, 1By18, 1By20 at Glu-B1 and 1Dx2, 1Dx2.2, 1Dx4, 1Dx5, 1Dy10, and 1Dy12 at Glu-D1 (Table 2). The MALDI-TOF-MS analysis of each variety was performed five times. The mass spectra of the HMW-GS for standard wheat cultivars showed 3–5 distinct peaks with good resolution which allowed accurate molecular masses to be obtained (Figure 2). Average molecular weights (MW) and relative standard deviations (RSD) are shown in Table 2. RSDs for all subunits were less than 0.092, indicating that the molecular masses for all subunits were highly reproducible within the experiment.

Table 2.
Reproducibility of molecular weights for HMW-GS determined by MALDI-TOF-MS in standard wheat cultivars.
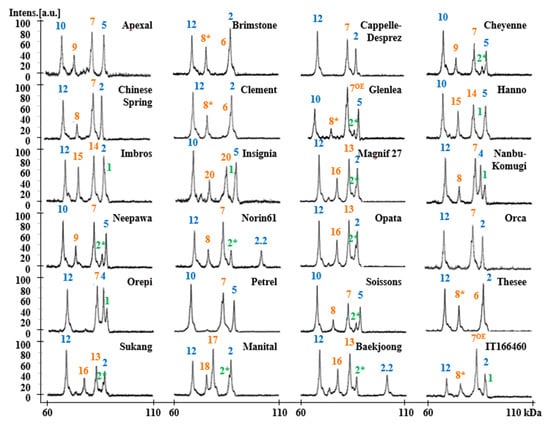
Figure 2.
HMW-GS profiles of standard wheat cultivars determined by MALDI-TOF-MS. Subunits corresponding to chromosomes 1A, 1B, and 1D are displayed in green, orange, and blue, respectively.
Molecular weights of HMW-GS determined by MALDI-TOF-MS were compared with those determined from mature protein sequences deduced from HMW-GS genes with the exception of 1By8* and 1Dx4 for which gene sequences were not available in public databases (Table 3). N-termini of proteins were as determined by Shewry et al. [52] and the molecular weight of 4-vp (105.14 Da for each cysteine residue) was deducted from molecular weights determined by MALDI-TOF-MS. Predicted and measured molecular weights differed by less than 230 Da with errors less than −0.30% for all HMW-GS except 1Bx7, 1Bx17, and 1Dx2.2. Considering the error rate of our mass spectrometer, this indicates that our optimized method for sample extraction and spectral analysis is reliable. In comparison, 32 of 94 HMW-GS measured by Gao et al. [20] differed by more than 230 Da from their predicted molecular weights. It is more difficult to compare the accuracy of measurements in the Liu et al. [50] study since the measured values were presented as an average for the protein type and rounded to the nearest hundred. Nonetheless, values obtained for Bx14, By15, and Dy12 differed by more than 352 from molecular weights predicted from gene sequences. As more HMW-GS gene sequences have become available, small differences in the sequences of genes from different cultivars have become apparent, resulting in minor changes in the molecular weights of the encoded proteins. Nonetheless, in the current study the same subunit generally displayed similar molecular masses in different genotypes. However, two distinct molecular weights with a difference of ~600 Da were detected in genotypes containing the 1Bx7 HMW-GS, 82,400 Da (group 1) and 83,000 Da (group 2) (Table 3). Interestingly, the molecular masses detected in genotypes containing 1Bx7OE were also 83,000 Da. In previous studies, it was not possible to discriminate 1Bx7 and 1Bx7OE by either mobility in SDS-PAGE or retention time in RP-HPLC [33]. A molecular weight difference of about 600 Da suggests that the group 2 proteins may contain an additional six amino acid residues. Butow et al. [10] revealed that compared to 1Bx7, 1Bx7OE has an 18 bp insertion in the repetitive domain of its coding sequence, resulting in an additional six amino acids near the C-terminal end. To further investigate the observed differences in the 1Bx7 subunits among cultivars, we performed PCR using DNA primers from Ma et al. [53] that span the region of the 18 bp insertion in 1Bx7OE (Figure 3). Apexal, Cheyenne, Neepawa, Norin 61, Orca and Soissons each yielded two fragments of 650 bp and 750 bp, while the amplification products in Cappelle-Despre, Chinese Spring, Nanbu-Komugi, Orepi, and Petrel were slightly larger: ~670 bp and 770 bp (Figure 3A). In addition, we used primers developed by Ragupathy et al. [54] that detect the gene duplication found in cultivars containing 1Bx7OE (Figure 3B,C). These primers amplified only Glenlea and IT166460 that are known to contain the duplicated gene. Our results suggest that there are two groups of similar 1Bx7 proteins that differ in molecular weight by about 600 Da. The group with the higher molecular weight is divided further into 1Bx7 group 2 and 1Bx7OE that differ by the presence of a gene duplication.

Table 3.
Comparison of molecular weights of HMW-GS predicted from mature proteins deduced from gene sequences with actual molecular weights determined by MALDI-TOF-MS in standard cultivars.
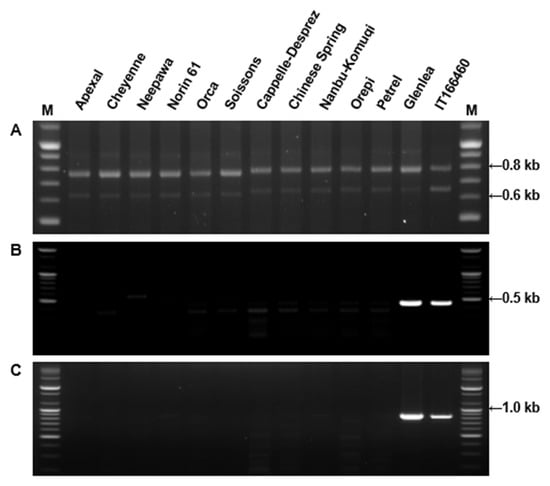
Figure 3.
Amplification of DNA from standard cultivars containing 1Bx7 or 1Bx7OE genes. (A) PCR primers span the region of the 18 bp insertion in 1Bx7OE; (B) PCR primers from the left junction of the retroelement and duplicated region of Bx7OE; (C) PCR primers from right junction of the retroelement and duplicated region of Bx7OE. Glenlea and IT166460 were used as positive controls for Bx7OE. The 100 bp Plus DNA Ladder is shown in lanes (M).
In this study, HMW-GS 1Dx 2.2 with a predicted molecular weight over 100 kD was identified in the Japanese wheat Norin 61 and the Korean wheat Baekjoong. Although the mass range of the instrument used in this study was set to 60,000–110,000 Da, the difference between the measured molecular weight (100,390 Da for Norin 61 and 100,388 Da for Baekjoong) and the calculated molecular weight (100,866 Da) was −0.49%. It is estimated that the measurement error was due to the high molecular mass of the proteins. 1Dx2.2 was either not identified in other MALDI-TOF-MS studies [23,50] or the measured molecular weights had much higher errors [20]. For example, in the study of Gao et al. [20], the measured molecular weight of 1Dx2.2 was only 86,340 Da.
The precision of MALDI-TOF-MS measurements in the Gao et al. [20] study and the current study can be compared in cases where HMW-GS gene sequences are available from the same cultivar that was analyzed in both studies. The molecular weights of the mature Bx7, By8, Dx2, and Dy12 proteins deduced from gene sequences from Chinese Spring are 83,123, 75,131, 87,105, and 68,528, respectively (Huo et al., in preparation). The measured molecular weights of these proteins in the Gao et al. [20] study were 82,749, 75,488, 87,120, and 68,935, respectively, differing by more than 357 Da for three of the four subunits. The molecular weights measured for these HMW-GS in the current study were 83,067, 75,150, 87,072, and 68,674, differing by only 19 to 146 Da. Gene sequences for Bx14 (KF733216) and By15 (KF733215) from the cultivar Hanno have also been reported. The predicted molecular weights determined from the gene sequences were 82,343 for Bx14 and 74,739 for By15. In the Gao et al. [20] study, the measured molecular weights of Bx14 and By15 from Hanno were 82,505 and 75,282, differing by 162 and 543 Da, respectively, while those obtained in the current study were 82,365 and 74,790, differing by less than 51 Da.
Figure 4 summarizes the average MWs and the margins of error for each subunit. 1Ax1, 1Bx14, 1Bx17, 1Bx20, 1By9, 1By16, 1Dx2, 1Dx2.2, 1Dx4, 1Dx5, 1Dy10, and 1Dy12 differ by more than 500 Da and can be distinguished easily. Subunits that differ in MWs by less than 500 Da—1Ax2*, 1Bx6 and 1Bx7, 1Bx7 OE, 1Bx13 and 1By8, 1By8*, 1By15, 1By18, and 1By20—cannot not be differentiated. However, 1Bx13, 1By15, 1By18, and 1By20 could be identified by considering the sizes of their tightly linked pairs, 1By16, 1Bx14, 1Bx17, and 1Bx20, respectively. Identification of other subunits is difficult because they have very similar molecular weights. 1Ax2 * and 1Bx6 have average molecular weights of 86,594 Da and 86,843 Da, respectively, which differ by only 249 Da (Table 2; Figure 4). 1By8 and 1By8* with average MWs of 75,874 Da and 75,966 Da, respectively, also are difficult to distinguish because they differ by only 92 Da (Table 2; Figure 4). However, both 1Ax2* and 1Bx6 and 1By8 and 1By8* can be distinguished easily by RP-HPLC as shown in Figure 5 and our previous study [33].
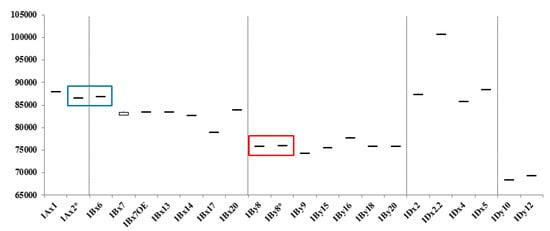
Figure 4.
Box and whisker plot of molecular weights of individual subunits in standard wheat cultivars. Red and blue boxes indicate subunits differing by less than 300 Da that were not distinguished by MALDI-TOF-MS, but could be differentiated by RP-HPLC.
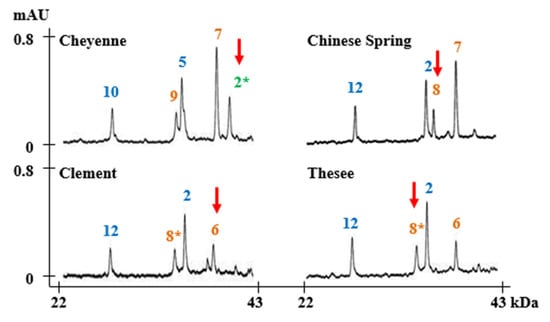
Figure 5.
RP-HPLC analysis of subunits that could not be distinguished by MALDI-TOF-MS. Subunits corresponding to chromosomes 1A, 1B, and 1D are displayed in green, orange, and blue, respectively. Unidentified subunits 1Ax2*, 1Bx6 and 1Bx8, and 1Bx8* are indicated with red arrows.
2.3. Identification of HMW-GS in 38 Korean Wheat Cultivars by MALDI-TOF-MS
To determine the feasibility of the methods developed in this study for use in breeding programs, the optimized MALDI-TOF-MS method was used to assess the HMW-GS compositions of 38 Korean wheat lines (Table 4, Table S2). MALDI-TOF analyses were performed five times for each variety and the average MWs and RSDs of HMW-GS from these analyses are shown in Table 4. The RSDs ranged from 0.011 to 0.096, indicating that the identification of HMW-GS by our MALDI-TOF-MS method is very reliable. In order to identify HMW-GS of each variety, first of all, the molecular weight measured by MALDI-TOF-MS was compared with those of the box and whisker plots of standard wheat varieties in Figure 4. Subunits not distinguished by apparent molecular weight differences were judged by tightly linked subunit pairs or by RP-HPLC as described above for 1Ax2* and 1Bx6 and 1By8 and 1By8*. The identified HMW-GS of each variety in this study was exactly in agreement with the results obtained using RP-HPLC combined with SDS-PAGE, a far more time-consuming method (Table 4; Figure 6) [33].

Table 4.
Reproducibility of molecular weights (MW) for HMW-GS in Korean wheat cultivars by MALDI-TOF-MS.
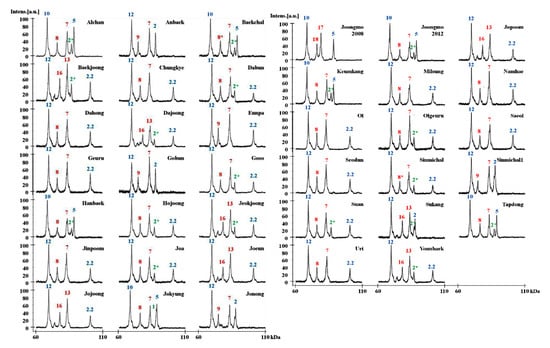
Figure 6.
HMW-GS profiles of 38 Korean wheat cultivars of individual subunits determined by MALDI-TOF-MS. Subunits corresponding to chromosomes 1A, 1B, and 1D are shown in green, red, and blue, respectively.
Interestingly, 29 out of 38 Korean wheat varieties (76.3%) contained 1Bx7. As shown in Supplementary Figure S1, after subtraction of the molecular weight of 4-vp, molecular weights of proteins determined by MALDI-TOF were approximately 82,400 in 16 varieties (1Bx7 group 1) and ~83,000 Da in 13 varieties (1Bx7 group 2). PCR analysis using primers of [53] revealed larger amplification products for the group 2 varieties, suggesting that these 1Bx7 group 2 genes contain the 18 bp insertion (Figure S1). Indeed, recent DNA sequence analysis confirmed that the 1Bx7 genes in the group 2 cultivars Keumkang and Olgeuru were identical to 1Bx7OE gene, EU157184 (unpublished observations). However, PCR analysis with the primers of [54] indicated that none of the Korean lines contain the gene duplication (Figure S1). Additionally, it is notable that a number of other HMW-GS were not found among the 38 Korean wheat varieties. These include 1Bx6, 1Bx14, 1Bx20, 1By15, 1By20, and 1Dx4.
3. Materials and Methods
3.1. Plant Materials
The standard wheat (Triticum aestivum L.) cultivars for HMW-GS analysis were kindly provided from the National Bioresource Project-Wheat (NBRP-Wheat, https://shigen.nig.ac.jp/wheat/komugi/) in Japan and The U.S. National Plant Germplasm System (NPGS, https://www.ars-grin.gov/npgs/) in USA as listed in Table 2. Grain of 38 Korean hexaploid wheat cultivars was harvested in 2016 by RDA National Institute of Crop Sciences, Jeonju, Korea.
3.2. Glutenin Extraction
Glutenin was prepared by the modified method of Singh et al. [41]. Flour (30 mg) was extracted with 1 mL of 50% (v/v) propanol at 65 °C for 30 min to remove gliadins. After centrifugation at 10,000 g for 10 min, the supernatant was removed. These steps were repeated two times with the resulting pellet to remove gliadin completely. The resulting pellet was then extracted with 150 μL of extraction buffer (50% (v/v) propanol, 0.08 M Tris-HCl (pH 8.0) containing 1% (w/v) dithiothreitol (DTT)) at 65 °C for 30 min followed by centrifugation at 10,000 g for 5 min. Glutenins in the supernatant fraction were precipitated with 100 μL of cold acetone. Alternately, cysteine residues in the glutenins were alkylated by the addition of 150 μL of extraction buffer containing 1.4% 4-vinylpyridine (v/v) to the supernatant fraction and incubation at 65 °C for 15 min. After centrifuging at 10,000 g for 2 min, 200 μL of supernatant was precipitated with 135 μL of cold acetone and stored at −20 °C. Before MALDI-TOF-MS analysis, pellets were washed with cold acetone and dried at RT.
3.3. Sample Optimization
To optimize MALDI-TOF-MS resolution, solvent components, dissolving volume, and matrix II components were considered in addition to the effects of 4-vp on sample preparation (Table 1). Solvent components evaluated were (1) 30% acetonitrile (ACN, v/v) containing 0.1% trifluoroacetic acid (TFA) referred to as TA30, (2) 50% ACN (v/v) containing 0.1% TFA referred to as TA50, (3) sinapinic acid (SA) dissolved in 30% ACN (v/v) containing 0.1% TFA at a concentration of 10 μg/μL referred to as SA in TA30, and (4) SA dissolved in 0.1% TFA in 50% ACN (v/v) at a concentration of 10 μg/μL referred to as SA in TA50. After choosing the optimized composition of solvent components, the three dissolving volumes of 10 μL, 50 μL, and 100 μL were compared. Finally, four Matrix II solutions were investigated in which SA was dissolved at a concentration of 20 μg/μL in (1) 30% ACN (v/v); (2) 50% ACN (v/v); (3) 30% ACN (v/v) containing in 0.1% TFA; and (4) 30% ACN (v/v) containing in 0.1% TFA.
3.4. MALDI-TOF-MS
HMW-GS were analyzed by MALDI Microflex LT (Bruker Daltonics, Bremen, Germany) equipped with a 60 Hz nitrogen laser. The parameters of the instrument were used with the following settings; mass range: 60,000 to 110,000 Da, sample rate: 1.00 GS/s, laser shots: 100, laser power: 85%, laser frequency: 80, and detector gain: 13.3X. Spectra were obtained in positive ion mode. Bovine serum albumin (66,463 Da) was used as external standard for mass assignment.
3.5. RP-HPLC
The analysis of HMW-GS by RP-HPLC was described by Jang et al. [33]. RP-HPLC of HMW-GS was performed on a Waters Alliance e2695 equipped with Agilent ZORBAX 300SB-C18 column (5 μm, 4.6 × 250 mm i.d., Agilent Technologies, Santa Clara, CA, USA). The solvents (A) water and (B) ACN, both containing 0.1% (v/v) TFA were used as the mobile phase. Before use, solvents A and B were degassed for 30 min. Glutenin pellets were washed with 0.07% DTT in acetone two times and then dried. Dried glutenin pellets were dissolved completely in 500 μL of 0.1% TFA in 20% ACN and filtered using a 0.45 μm, Whatman PVDF syringe filter. Ten μL of each sample was injected and eluted at 0.8 mL/min using a linear gradient of 23–44% of solvent B for 70 min. The RP-HPLC analysis of HMW-GS was carried out at 60 °C column temperature and monitored at a wavelength of 206 nm.
3.6. Wheat DNA Extraction and PCR Analysis
Genomic DNA was extracted from 50 mg of wheat flour for the standard cultivars using a DNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Genomic DNA was quantified using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MS, USA) and diluted to 50 ng/μL. PCR was performed in a reaction volume of 25 μL using 200 ng of genomic DNA, 1.25 U of GoTaq DNA polymerase (Promega, Madison, WI, USA), 1x Green GoTaq reaction buffer (containing 1.5 mM MgCl2), 200 μM of dNTP mix (Bioneer, Daejeon, South Korea) and 10 pmole of forward and reverse primers. Bx7 coding region primers were forward 5′-CGCAACAGCCAGGACAATT-3′, and reverse 5′-AGAGTTCTATCACTGCCTGGT-3′ [53]. Left junction primers were: forward 5′-ACGTGTCCAAGCTTTGGTTC-3′ and reverse 5′-GATTGGTGGGTGGATACAGG-3′; and right junction primers were forward 5′-CCACTTCCAAGGTGGGACTA-3′ and reverse 5′-TGCCAACACAAAAGAAGCTG-3′ [54]. Amplification conditions for the PCR reaction were an initial cycle at 95 °C for 5 min, followed by 34 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 2 min, followed by a final extension at 72 °C for 5.25 min. PCR products were resolved on 1.5–2.0% agarose gels in 0.5x TBE buffer, stained with ethidium bromide and visualized under UV.
4. Conclusions
MALDI-TOF-MS is an accurate and high-throughput method for determining the HMW-GS composition of wheat varieties. Analyses can be completed in about one minute per sample and require only minimal amounts of protein and solvents. Eighteen HMW-GS in 24 standard wheat varieties grown in countries throughout the world were distinguished using the method optimized in this study. Additionally, varieties previously found to have the 1Bx7 subunit were shown to contain one of two different subunits that differed by 600 Da in molecular weight. The optimized method was further used to identify 15 different HMW-GS in 38 Korean wheat cultivars and will be valuable for the future analysis of large numbers of wheat breeding lines.
Supplementary Materials
The Supplementary Material for this article can be found online, Figure S1: Amplification of DNA from Korean cultivars containing 1Bx7 group 1 or 1 Bx7 group 2 HMW-GS and standard cultivars containing 1Bx7OE. (A) PCR primers span the region of the 18 bp insertion in 1Bx7OE; (B) PCR primers from the left junction of the retroelement and duplicated region of Bx7OE; (C) PCR primers from right junction of the retroelement and duplicated region of Bx7OE. Glenlea and IT166460 were used as positive controls for Bx7OE.The 100bp Plus DNA Ladder is shown in lanes (M), Table S1: Comparison of molecular weights of alkylated and non-alkylated 1Dy12, 1By8, 1Bx7 and 1Dx2 HMW-GS from Chinese Spring determined by MALDI-TOF-MS, Table S2: Comparison of molecular weights of HMW-GS predicted from mature proteins deduced from gene sequences with actual molecular weights determined by MALDI-TOF-MS in Korean cultivars.
Author Contributions
J.-Y.L. designed and conducted experiments and wrote the manuscript, Y.-R.J., J.-R.S., and S.W.K. conducted experiments, S.B.A. and K.C. contributed to the interpretation of data and writing of the manuscript, S.-H.L. conducted experiments and contributed to the experimental design and writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Institute of Agricultural Science (RDA PJ014838) and the Next-Generation BioGreen 21 Program (RDA PJ013159 and RDA PJ013149), Korea.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Payne, P.I. Genetics of wheat storage proteins and the effect of allelic variation on breadmaking quality. Annu. Rev. Plant. Physiol. 1987, 38, 141–153. [Google Scholar] [CrossRef]
- Payne, P.I.; Holt, L.M.; Law, C.N. Structural and genetical studies on the high-molecular-weight subunits of wheat glutenin. Theor. Appl. Genet. 1981, 60, 229–236. [Google Scholar] [CrossRef]
- Shewry, P.R.; Halford, N.G.; Tatham, A.S. High molecular weight subunits of wheat glutenin. J. Cereal. Sci. 1992, 15, 105–120. [Google Scholar] [CrossRef]
- Payne, P.I.; Lawrence, G.J. Catalogue of alleles for the complex gene loci Glu-A1, Glu-B1 and Glu-D1 which code for high-molecular weight subunits of glutenin in hexaploid wheat. Cereal. Res. Commun. 1983, 11, 29–35. [Google Scholar]
- Barro, F.; Rooke, L.; Bekes, F.; Gras, P.; Tatham, A.S.; Fido, R.; Lazzeri, P.A.; Shewry, P.R.; Barcelo, P. Transformation of wheat with HMW subunit genes results in improved functional properties. Nat. Biotechnol. 1997, 15, 1295–1299. [Google Scholar] [CrossRef]
- Gianibelli, M.C.; Larroque, O.R.; MacRitchie, F.; Wrigley, C.W. Biochemical, genetic, and molecular characterization of wheat glutenin and its component subunits. Cereal. Chem. 2001, 78, 635–646. [Google Scholar] [CrossRef]
- Vawser, M.J.; Cornish, G.B. Over-expression of HMW glutenin subunit Glu-B1 7x in hexaploid wheat varieties (Triticum aestivum). Crop Pasture Sci. 2004, 55, 577–588. [Google Scholar] [CrossRef]
- Branlard, G.; Dardevet, M. Diversity of grain protein and bread wheat quality. II. Correlation between high molecular weight subunits of glutenin and flour quality characteristics. J. Cereal. Sci. 1985, 3, 345–354. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Y.; Gao, X.; Wang, K.; Wang, S.; Zhang, Y.; He, Z.; Ma, W.; Yan, Y. Comparative proteome analysis of glutenin synthesis and accumulation in developing grains between superior and poor quality bread wheat cultivars. J. Sci. Food Agric. 2012, 92, 106–115. [Google Scholar] [CrossRef]
- Butow, B.J.; Ma, W.; Gale, K.R.; Cornish, G.B.; Rampling, L.; Larroque, O.; Morell, M.K.; Bekes, F. Molecular discrimination of Bx7 alleles demonstrates that a highly expressed high molecular weight glutenin allele has a major impact on wheat flour dough strength. Theor. Appl. Genet. 2003, 107, 1524–1532. [Google Scholar] [CrossRef]
- D’Ovidio, R.; Masci, S.; Porceddu, E.; Kasarda, D. Duplication of the high molecular weight glutenin subunit gene in bread wheat (Triticum aestivum L.) cultivar ‘Red River 68′. Plant Breed 1997, 116, 525–531. [Google Scholar] [CrossRef]
- Lukow, O.M.; Forsyth, S.A.; Payne, P.I. Over-production of HMW glutenin subunits coded on chromosome 1B in common wheat, Triticum aestivum. J. Genet. Breed. 1992, 46, 187–192. [Google Scholar]
- Radovanovic, N.; Cloutier, S.; Brown, D.; Humphreys, D.G.; Lukow, O.M. Genetic variance for gluten strength contributed by high molecular weight glutenin proteins. Cereal Chem. 2002, 79, 843–849. [Google Scholar] [CrossRef]
- Wieser, H. Chemistry of gluten proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef]
- An, X.; Zhang, Q.; Yan, Y.; Li, Q.; Zhang, Y.; Wang, A.; Pei, Y.; Tian, J.; Wang, H.; Hsam, S.L.K.; et al. Cloning and molecular characterization of three novel LMW-i glutenin subunit genes from cultivated einkorn (Triticum monococcum L.). Theor. Appl. Genet. 2006, 113, 383–395. [Google Scholar] [CrossRef]
- Bietz, J.A. Separation of cereal proteins by reversed-phase high-performance liquid chromatography. J. Chromatogr. A 1983, 255, 219–238. [Google Scholar] [CrossRef]
- Courcoux, P.; Serot, T.; Larre, C.; Popineau, Y. Characterization and identification of wheat cultivars by multi-dimensional analysis of reversed-phase high-performance liquid chromatograms. J. Chromatogr. A 1992, 596, 225–232. [Google Scholar] [CrossRef]
- Cozzolino, R.; Giorgi, S.D.; Fisichella, S.; Garozzo, D.; Lafiandra, D.; Palermo, A. Matrix-assisted laser desorption/ionization mass spectrometric peptide mapping of high molecular weight glutenin subunits 1Bx7 and 1Dy10 in Cheyenne cultivar. Rapid Commun. Mass Spectrom. 2001, 15, 778–787. [Google Scholar] [CrossRef]
- Dong, K.; Hao, C.; Wang, A.; Cai, M.; Yan, Y. Characterization of HMW glutenin subunits in bread and tetraploid wheats by reversed-phase high-performance liquid chromatography. Cereal Res. Commun. 2009, 37, 65–73. [Google Scholar] [CrossRef]
- Gao, L.; Ma, W.; Chen, J.; Wang, K.; Li, J.; Wang, S.; Bekes, F.; Appels, R.; Yan, Y. Characterization and comparative analysis of wheat high molecular weight glutenin subunits by SDS-PAGE, RP-HPLC, HPCE, and MALDI-TOF-MS. J. Agric. Food Chem. 2010, 58, 2777–2786. [Google Scholar] [CrossRef]
- Naeem, H.A.; Sapirstein, H.D. Ultra-fast separation of wheat glutenin subunits by reversed-phase HPLC using a superficially porous silica-based column. J. Cereal Sci. 2007, 46, 157–168. [Google Scholar] [CrossRef]
- Yan, X.; Liu, W.; Yu, Z.; Han, C.; Zeller, F.J.; Hsam, S.L.K.; Yan, Y. Rapid separation and identification of wheat HMW glutenin subunits by UPLC and comparative analysis with HPLC. Aust. J. Crop Sci. 2014, 8, 140–147. [Google Scholar]
- Zhang, Q.; Dong, Y.; An, X.; Wang, A.; Zhang, Y.; Li, X.; Gao, L.; Xia, X.; He, Z.; Yan, Y. Characterization of HMW glutenin subunits in common wheat and related species by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). J. Cereal. Sci. 2008, 47, 252–261. [Google Scholar] [CrossRef]
- Yan, Y.; Hsam, S.L.K.; Yu, J.; Zeller, F.J. Allelic variation of HMW glutenin subunits in Ae. tauschii accessions detected by SDS-PAGE, A-PAGE and capillary electrophoresis. Euphytica 2003, 130, 377–385. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, X.L.; He, Z.H.; Ma, W.; Appels, R.; Peña, R.J.; Xia, X.C. Characterization of low-molecular-weight glutenin subunit Glu-B3 genes and development of STS markers in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2009, 118, 525–539. [Google Scholar] [CrossRef]
- Wang, L.H.; Li, G.Y.; Peña, R.J.; Xia, X.C.; He, Z.H. Development of STS markers and establishment of multiplex PCR for Glu-A3 alleles in common wheat (Triticum aestivum L.). J. Cereal Sci. 2010, 51, 305–312. [Google Scholar] [CrossRef]
- Zhang, W.; Gianibelli, M.C.; Rampling, L.R.; Gale, K.R. Characterisation and marker development for low molecular weight glutenin genes from Glu-A3 alleles of bread wheat (Triticum aestivum L). Theor. Appl. Genet. 2004, 108, 1409–1419. [Google Scholar] [CrossRef]
- Zhang, X.; Jin, H.; Zhang, Y.; Liu, D.; Li, G.; Xia, X.; He, Z.; Zhang, A. Composition and functional analysis of low-molecular-weight glutenin alleles with Aroona near-isogenic lines of bread wheat. BMC Plant Biol. 2012, 12, 243. [Google Scholar] [CrossRef]
- Zhao, X.L.; Xia, X.C.; He, Z.H.; Gale, K.R.; Lei, Z.S.; Appels, R.; Ma, W. Characterization of three low-molecular-weight Glu-D3 subunit genes in common wheat. Theor. Appl. Genet. 2006, 113, 1247–1259. [Google Scholar] [CrossRef]
- Bean, S.R.; Lookhart, G.L. Ultrafast capillary electrophoresis analysis of cereal storage proteins and its application to protein characterization and cultivar differentiation. J. Agric. Food Chem. 2000, 48, 344–353. [Google Scholar] [CrossRef]
- Bietz, J.A.; Schmalzried, E. Capillary electrophoresis of wheat gliadin: Initial studies and application to varietal identification. LWT-Food Sci. Technol. 1995, 28, 174–184. [Google Scholar] [CrossRef]
- Yan, Y.; Surlan-Momirovic, G.; Prodanovic, S.; Zoric, D.; Liu, G. Capillary zone electrophoresis analysis of gliadin proteins from Chinese and Yugoslav winter wheat cultivars. Euphytica 1999, 105, 197–204. [Google Scholar] [CrossRef]
- Jang, Y.R.; Beom, H.R.; Altenbach, S.B.; Lee, M.K.; Lim, S.H.; Lee, J.Y. Improved method for reliable HMW-GS identification by RP-HPLC and SDS-PAGE in common wheat cultivars. Molecules 2017, 22, 1055. [Google Scholar] [CrossRef] [PubMed]
- Burnouf, T.; Bietz, J.A. Reversed-phase high-performance liquid chromatography of durum wheat gliadins: Relationships to durum wheat quality. J. Cereal Sci. 1984, 2, 3–14. [Google Scholar] [CrossRef]
- Alberghina, G.; Cozzolino, R.; Fisichella, S.; Garozzo, D.; Savarino, A. Proteomics of gluten: Mapping of the 1Bx7 glutenin subunit in Chinese Spring cultivar by matrix-assisted laser desorption/ionization. Rapid Commun. Mass Sp. 2005, 19, 2069–2074. [Google Scholar] [CrossRef]
- Cunsolo, V.; Foti, S.; Saletti, R.; Gilbert, S.; Tatham, A.S.; Shewry, P.R. Structural studies of the allelic wheat glutenin subunits 1Bx7 and 1Bx20 by matrix-assisted laser desorption/ionization mass spectrometry and high-performance liquid chromatography/electrospray ionization mass spectrometry. J. Mass Spectrom. 2004, 39, 66–78. [Google Scholar] [CrossRef]
- Dworschak, R.G.; Ens, W.; Standing, K.G.; Preston, K.R.; Marchylo, B.A.; Nightingale, M.J.; Stevenson, S.G.; Hatcher, D.W. Analysis of wheat gluten proteins by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 1998, 33, 429–435. [Google Scholar] [CrossRef]
- Foti, S.; Maccarrone, G.; Saletti, R.; Roepstorff, P.; Gilbert, S.; Tatham, A.S.; Shewry, P.R. Verification of the cDNA deduced sequence of glutenin subunit 1Dx5 and an Mr 58000 repetitive peptide by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). J. Cereal Sci. 2000, 31, 173–183. [Google Scholar] [CrossRef]
- Garozzo, D.; Cozzolino, R.; Giorgi, S.D.; Fisichella, S.; Lafiandra, D. Use of hydroxyacetophenones as matrices for the analysis of high molecular weight glutenin mixtures by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Sp. 1999, 13, 2084–2089. [Google Scholar] [CrossRef]
- Muccilli, V.; Cunsolo, V.; Saletti, R.; Foti, S.; Masci, S.; Lafiandra, D. Characterization of B- and C-type low molecular weight glutenin subunits by electrospray ionization mass spectrometry and matrix-assisted laser desorption/ionization mass spectrometry. Proteomics 2005, 5, 719–728. [Google Scholar] [CrossRef]
- Singh, N.K.; Sheperd, K.W.; Cornish, G.B. A simplified SDS-PAGE procedure for separating LMW subunits of glutenin. J. Cereal Sci. 1991, 14, 203–208. [Google Scholar] [CrossRef]
- Wang, K.; Ma, J.; Islam, S.; Yan, Y.; Appels, R.; Yan, G.; Ma, W. Detection of cysteine residue numbers in wheat gluten proteins by MALDI-TOF. Int. J. Mass Spectrom. 2015, 392, 91–95. [Google Scholar] [CrossRef]
- Williams, T.L.; Andrzejewski, D.; Lay, J.O.; Jr, L.; Musser, S.M. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J. Am. Soc. Mass Spectrom. 2003, 14, 342–351. [Google Scholar] [CrossRef]
- Branlard, G.; Dardevet, M.; Amiour, N.; Igrejas, G. Allelic diversity of HMW and LMW glutenin subunits and omega-gliadins in French bread wheat (Triticum aestivum L.). Genet. Resour. Crop Evol. 2003, 50, 669–679. [Google Scholar] [CrossRef]
- Cho, S.W.; Roy, S.K.; Chun, J.B.; Cho, K.; Park, C.S. Overexpression of the Bx7 high molecular weight glutenin subunit on the Glu-B1 locus in a Korean wheat landrace. Plant. Biotechnol. Rep. 2017, 11, 97–105. [Google Scholar] [CrossRef]
- Dupont, F.M.; Chan, R.; Lopez, R. Molar fractions of high-molecular-weight glutenin subunits are stable when wheat is grown under various mineral nutrition and temperature regimens. J. Cereal Sci. 2007, 45, 134–139. [Google Scholar] [CrossRef]
- Kazman, M.E.; Lein, V. Cytological and SDS-PAGE characterization of 1994-95-grown European wheat cultivars. Annu. Wheat New. 1996, 42, 86–92. [Google Scholar]
- Liu, L.; Ikeda, T.M.; Branlard, G.; Pena, R.J.; Rogers, W.F.; Lerner, S.E.; Kolman, M.A.; Xia, X.; Wang, L.; Ma, W. Comparison of low molecular weight glutenin subunits identified by SDS-PAGE, 2-DE, MALDI-TOF-MS and PCR in common wheat. BMC Plant Biol. 2010, 10, 124–148. [Google Scholar] [CrossRef]
- Park, C.S.; Kang, C.S.; Jeung, J.U.; Woo, S.H. Influence of allelic variations in glutenin on the quality of pan bread and white salted noodles made from Korean wheat cultivars. Euphytica 2011, 180, 235–250. [Google Scholar] [CrossRef]
- Liu, L.; Wang, A.; Appels, R.; Ma, J.; Xia, X.; Lan, P.; He, Z.; Bekes, F.; Yan, Y.; Ma, W. A MALDI-TOF based analysis of high molecular weight glutenin subunits for wheat breeding. J. Cereal Sci. 2009, 50, 295–301. [Google Scholar] [CrossRef]
- Hua, C.; Takata, K.; Yang-Fen, Z.; Ikeda, T.M.; Yanaka, M.; Nagamine, T.; Fujimaki, H. Novel high molecular weight glutenin subunits at the Glu-D1 locus in wheat landraces from the Xinjiang district of China and relationship with winter habit. Breed. Sci. 2005, 55, 459–463. [Google Scholar] [CrossRef]
- Shewry, P.R.; Field, J.M.; Faulks, A.J.; Parmar, S.; Miflin, B.J.; Dietler, M.D.; Lew, E.J.-L.; Kasarda, D.D. The purification and N-terminal amino acid sequence analysis of the high molecular weight gluten polypeptides of wheat. Biochim. Biophys. Acta 1984, 788, 23–34. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, W.; Gale, K.R. Multiplex-PCR typing of high molecular weight glutenin alleles in wheat. Euphytica 2003, 134, 51–60. [Google Scholar] [CrossRef]
- Ragupathy, R.; Naeem, H.A.; Reimer, E.; Lukow, O.M.; Sapirstein, H.D.; Cloutier, S. Evolutionary origin of the segmental duplication encompassing the wheat Glu-B1 locus encoding the overexpressed Bx7 (Bx7OE) high molecular weight glutenin subunit. Theor. Appl. Genet. 2008, 116, 283–296. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).