Microbial Transformation of Flavonoids by Isaria fumosorosea ACCC 37814
Abstract
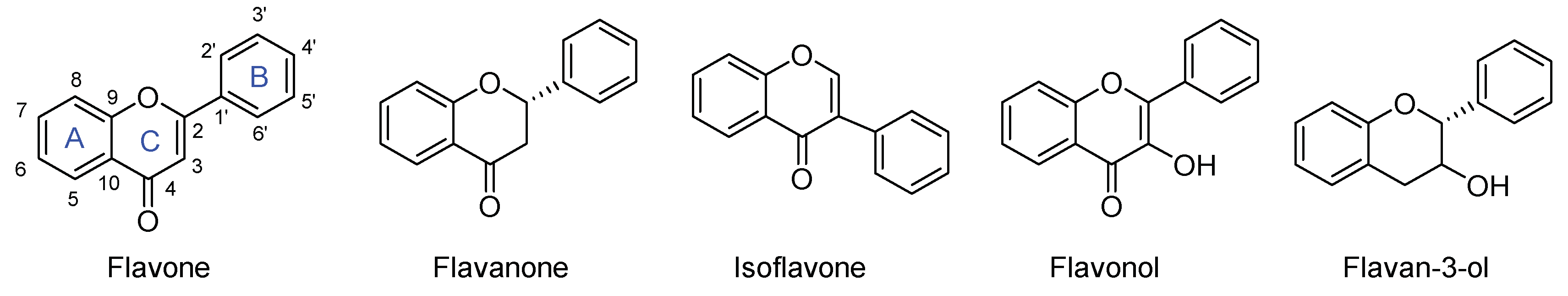
1. Introduction
2. Results and Discussion
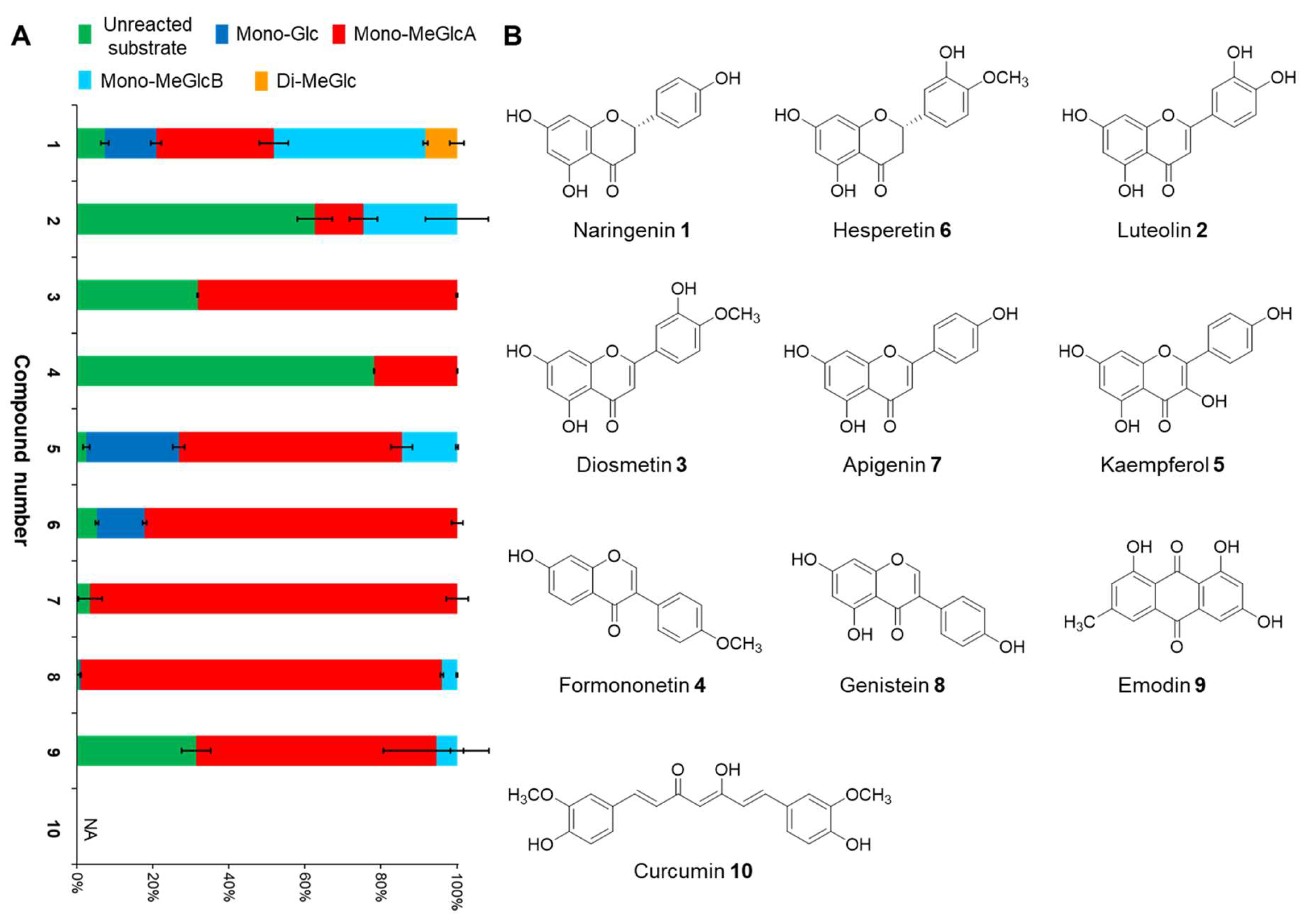
2.1. Biotransformation Assay
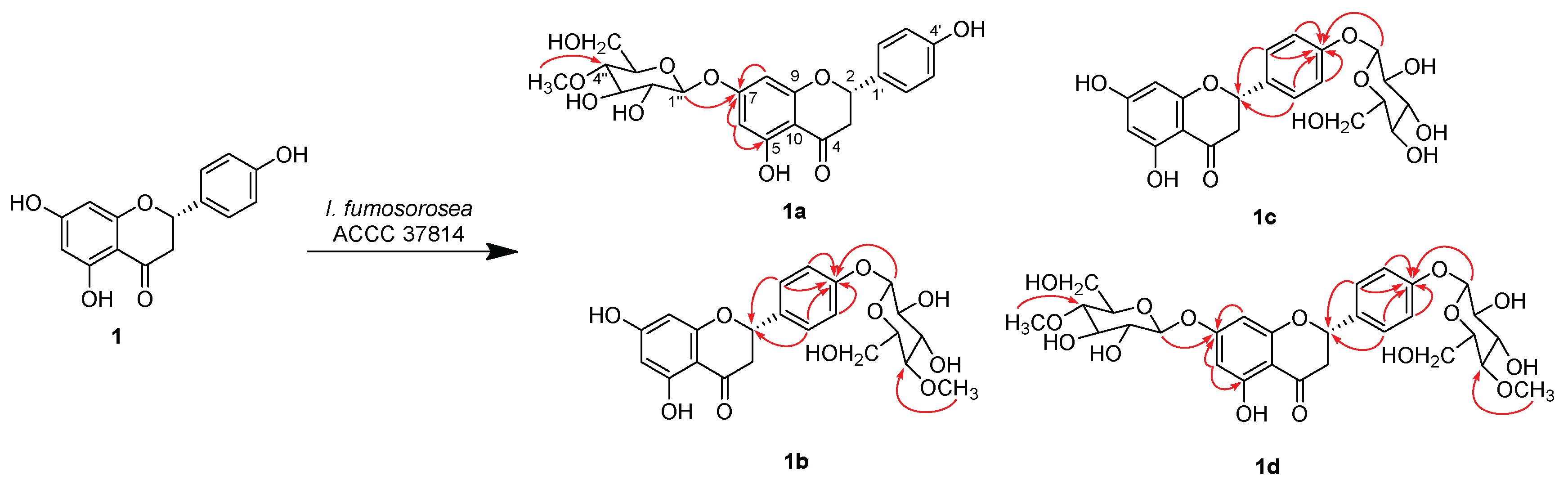
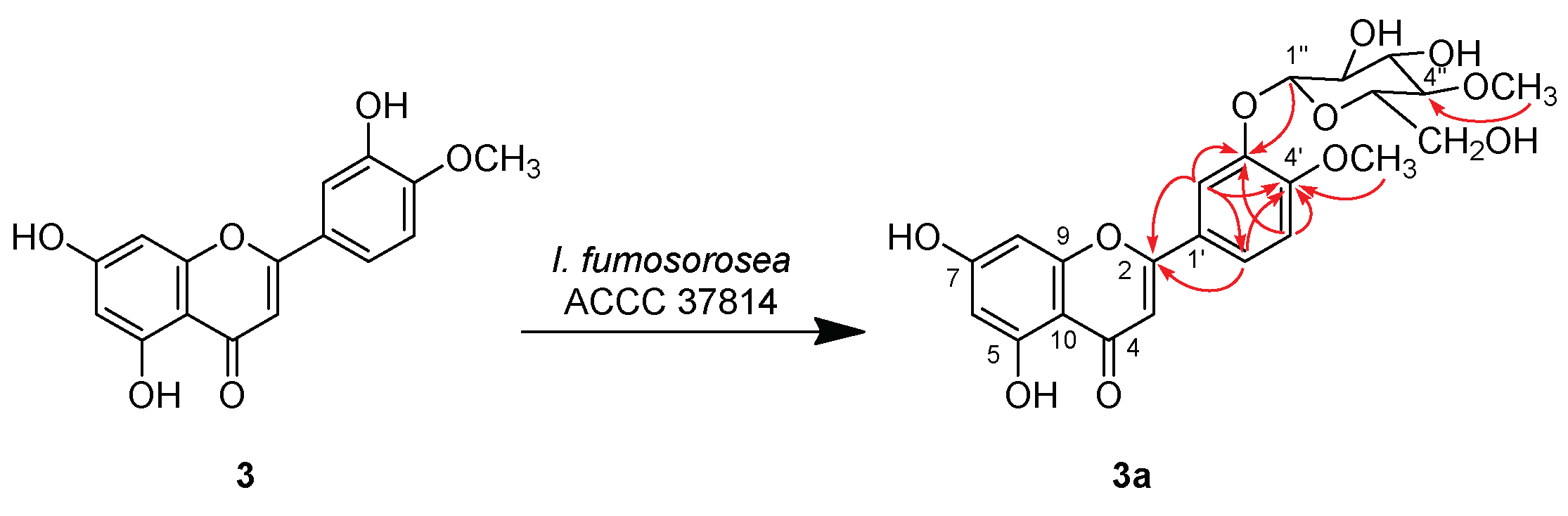
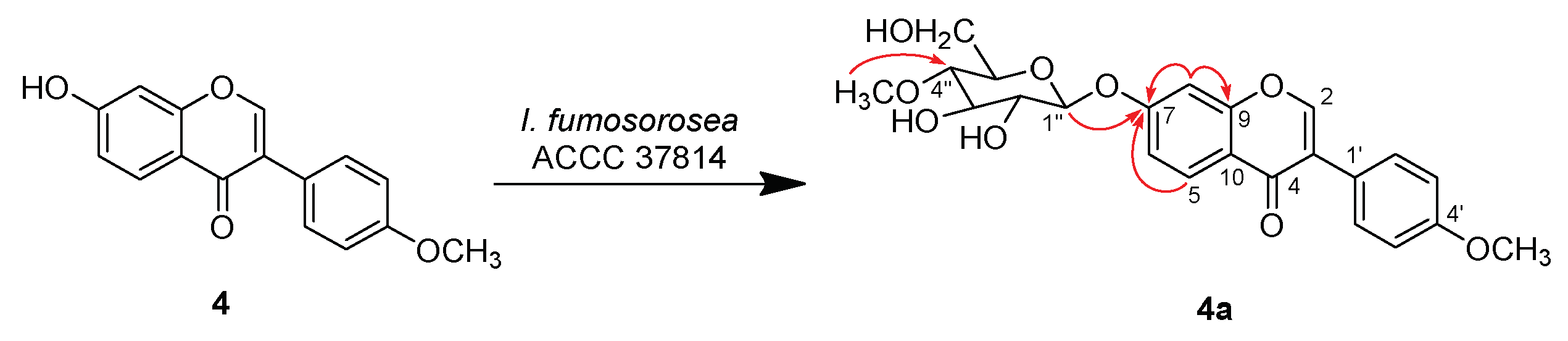
2.2. Structure Characterization
3. Conclusions
4. Materials and Methods
4.1. General Experimental Orocedures
4.2. Culture and Biotransformation Procedures
4.3. Extraction, Isolation and Purification of the Products
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pietta, P.-G. Flavonoids as antioxidants. J. Nat. Prod. 2000, 63, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Veitch, N.C.; Grayer, R.J. Flavonoids and their glycosides, including anthocyanins. Nat. Prod. Rep. 2011, 28, 1626–1695. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J. Dietary flavonoid aglycones and their glycosides: Which show better biological significance? Crit. Rev. Food Sci. Nutr. 2017, 57, 1874–1905. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, H.L.; Yang, J.L.; Gupta, V.K.; Jiang, Y.M. New insights on bioactivities and biosynthesis of flavonoid glycosides. Trends Food Sci. Technol. 2018, 79, 116–124. [Google Scholar] [CrossRef]
- Chen, X. Fermenting next generation glycosylated therapeutics. ACS Chem. Biol. 2011, 6, 14–17. [Google Scholar] [CrossRef]
- Losey, H.C.; Peczuh, M.W.; Chen, Z.; Eggert, U.S.; Dong, S.D.; Pelczer, I.; Kahne, D.; Walsh, C.T. Tandem action of glycosyltransferases in the maturation of vancomycin and teicoplanin aglycones: Novel glycopeptides. Biochemistry 2001, 40, 4745–4755. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Ogawa, K.; Tone, H.; Iguchi, H.; Shomura, T.; Murata, S. Pharmacokinetics of doxorubicin, (2R)-4′-O-tetrahydropyranyl-adriamycin and aclarubicin. Jpn. J. Antibiot. 1986, 39, 1321–1336. [Google Scholar] [PubMed]
- Hofer, B. Recent developments in the enzymatic O-glycosylation of flavonoids. Appl. Microbiol. Biotechnol. 2016, 100, 4269–4281. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, P.; Cui, Y.; Li, K.; Qiao, X.; Zhang, Y.-T.; Li, S.-M.; Cox, R.J.; Wu, B.; Ye, M.; et al. Regio- and stereospecific O-glycosylation of phenolic compounds catalyzed by a fungal glycosyltransferase from Mucor hiemalis. Adv. Synth. Catal. 2017, 359, 995–1006. [Google Scholar] [CrossRef]
- Brglez Mojzer, E.; Knez Hrncic, M.; Skerget, M.; Knez, Z.; Bren, U. Polyphenols: Extraction methods, antioxidative action, bioavailability and anticarcinogenic effects. Molecules 2016, 21, 901. [Google Scholar] [CrossRef]
- Dymarska, M.; Grzeszczuk, J.; Urbaniak, M.; Janeczko, T.; Plaskowska, E.; Stepien, L.; Kostrzewa-Suslow, E. Glycosylation of 6-methylflavone by the strain Isaria fumosorosea KCH J2. PLoS ONE 2017, 12, e0184885. [Google Scholar] [CrossRef]
- Hollman, P.C.; Bijsman, M.N.; van Gameren, Y.; Cnossen, E.P.; de Vries, J.H.; Katan, M.B. The sugar moiety is a major determinant of the absorption of dietary flavonoid glycosides in man. Free Radical Res. 1999, 31, 569–573. [Google Scholar] [CrossRef]
- Wang, W.; Sun, C.; Mao, L.; Ma, P.; Liu, F.; Yang, J.; Gao, Y. The biological activities, chemical stability, metabolism and delivery systems of quercetin: A review. Trends Food Sci. Technol. 2016, 56, 21–38. [Google Scholar] [CrossRef]
- Xie, L.; Zhang, L.; Wang, C.; Wang, X.; Xu, Y.-M.; Yu, H.; Wu, P.; Li, S.; Han, L.; Gunatilaka, A.A.L.; et al. Methylglucosylation of aromatic amino and phenolic moieties of drug-like biosynthons by combinatorial biosynthesis. Proc. Natl. Acad. Sci. USA 2018, 115, E4980–E4989. [Google Scholar] [CrossRef]
- Zhan, J.; Gunatilaka, A.A.L. Selective 4′-O-methylglycosylation of the pentahydroxyflavonoid quercetin by Beauveria bassiana ATCC 7159. Biocatal. Biotransform. 2006, 24, 396–399. [Google Scholar] [CrossRef]
- Zeng, J.; Yang, N.; Li, X.M.; Shami, P.J.; Zhan, J. 4′-O-methylglycosylation of curcumin by Beauveria bassiana. Nat. Prod. Commun. 2010, 5, 77–80. [Google Scholar]
- Sordon, S.; Popłonski, J.; Tronina, T.; Huszcza, E. Microbial glycosylation of daidzein, genistein and biochanin A: Two new glucosides of biochanin A. Molecules 2017, 22, 81. [Google Scholar] [CrossRef]
- Kozlowska, E.; Dymarska, M.; Kostrzewa-Suslow, E.; Janeczko, T. Isaria fumosorosea KCH J2 entomopathogenic strain as an effective biocatalyst for steroid compound transformations. Molecules 2017, 22, 1511. [Google Scholar] [CrossRef]
- Fujita, T.; Inoue, K.; Yamamoto, S.; Ikumoto, T.; Sasaki, S.; Toyama, R.; Chiba, K.; Hoshino, Y.; Okumoto, T. Fungal metabolites. Part 11. A potent immunosuppressive activity found in Isaria sinclairii metabolite. J. Antibiot. 1994, 47, 208–215. [Google Scholar] [CrossRef]
- Sakakura, A.; Suzuki, K.; Katsuzaki, H.; Komiya, T.; Imamura, T.; Aizono, Y.; Imai, K. Hanasanagin: A new antioxidative pseudo-di-peptide, 3,4-diguanidinobutanoyl-dopa, from the mushroom, Isaria japonica. Tetrahedron Lett. 2005, 46, 9057–9059. [Google Scholar] [CrossRef]
- Vining, L.C.; Taber, W.A. Isariin, a new depsipeptide from Isaria cretacea. Can. J. Chem. 1962, 40, 1579–1584. [Google Scholar] [CrossRef]
- Sabareesh, V.; Ranganayaki, R.S.; Raghothama, S.; Bopanna, M.P.; Balaram, H.; Srinivasan, M.C.; Balaram, P. Identification and characterization of a library of microheterogeneous cyclohexadepsipeptides from the fungus Isaria. J. Nat. Prod. 2007, 70, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Briggs, L.H.; Fergus, B.J.; Shannon, J.S. Chemistry of fungi-IV: Cyclodepsipeptides from a new species of Isaria. Tetrahedron 1966, 22, 269–278. [Google Scholar] [CrossRef]
- Baute, R.; Deffieux, G.; Merlet, D.; Baute, M.A.; Neveu, A. New insecticidal cyclodepsipeptides from the fungus Isaria felina. I. Production, isolation and insecticidal properties of isariins b, c and d. J. Antibiot. 1981, 34, 1261–1265. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, S.-S.; Yuan, W.-Y.; Wei, J.-J.; Zhao, Y.-X.; Luo, D.-Q. Carotane-type sesquiterpenes from cultures of the insect pathogenic fungus Isaria fumosorosea. J. Asian Nat. Prod. Res. 2017, 19, 1–7. [Google Scholar] [CrossRef]
- Asai, T.; Chung, Y.M.; Sakurai, H.; Ozeki, T.; Chang, F.R.; Yamashita, K.; Oshima, Y. Tenuipyrone, a novel skeletal polyketide from the entomopathogenic fungus, Isaria tenuipes, cultivated in the presence of epigenetic modifiers. Org. Lett. 2012, 14, 513–515. [Google Scholar] [CrossRef] [PubMed]
- Dymarska, M.; Janeczko, T.; Kostrzewa-Suslow, E. Glycosylation of methoxylated flavonoids in the cultures of Isaria fumosorosea KCH J2. Molecules 2018, 23, 2578. [Google Scholar] [CrossRef]
- Dymarska, M.; Janeczko, T.; Kostrzewa-Suslow, E. Glycosylation of 3-hydroxyflavone, 3-methoxyflavone, quercetin and baicalein in fungal cultures of the genus Isaria. Molecules 2018, 23, 2477. [Google Scholar] [CrossRef] [PubMed]
- Dymarska, M.; Janeczko, T.; Kostrzewa-Suslow, E. Biotransformations of flavones and an isoflavone (daidzein) in cultures of entomopathogenic filamentous fungi. Molecules 2018, 23, 1356. [Google Scholar] [CrossRef]
- Willits, M.G.; Giovanni, M.; Prata, R.T.; Kramer, C.M.; De, L.V.; Steffens, J.C.; Graser, G. Bio-fermentation of modified flavonoids: An example of in vivo diversification of secondary metabolites. Phytochemistry 2004, 5, 31–41. [Google Scholar] [CrossRef]
- Kim, J.H.; Shin, K.H.; Ko, J.H.; AHn, J.-H. Glucosylation of flavonols by Escherichia coli expressing glucosyltransferase from rice (Oryza sativa). J. Biosci. Bioeng. 2006, 102, 135–137. [Google Scholar] [CrossRef]
- He, X.-Z.; Li, W.-S.; Blount, J.W.; Dixon, R.A. Regioselective synthesis of plant (iso)flavone glycosides in Escherichia coli. Appl. Microbiol. Biotechnol. 2008, 90, 253–260. [Google Scholar] [CrossRef]
- Ko, J.H.; Kim, B.G.; Ahn, J.-H. Glycosylation of flavonoids with a glycosyltransferase from Bacillus cereus. FEMS Microbiol. Lett. 2006, 258, 263–268. [Google Scholar]
- Kim, J.H.; Kim, B.G.; Kim, J.A.; Park, Y.; Lee, Y.J.; Lim, Y.; Ahn, J.-H. Glucosylation of flavonoids with E. coli expressing glycosyltransferase from Xanthomonas campestris. J. Microbiol. Biotechnol. 2007, 17, 539–542. [Google Scholar]
- Ahn, B.C.; Kim, B.G.; Jeon, Y.M.; Lee, E.J.; Lim, Y.; Ahn, J.-H. Formation of flavone di-O-glucosides using a glycosyltransferase from Bacillus cereus. J. Microbiol. Biotechnol. 2009, 19, 387–390. [Google Scholar] [CrossRef]
- Yoon, J.-A.; Kim, B.-G.; Lee, W.J.; Lim, Y.; Chong, Y.; Ahn, J.-H. Production of a novel quercetin glycoside through metabolic engineering of Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 78, 4256–4262. [Google Scholar] [CrossRef]
- Kim, B.G.; Yang, S.M.; Kim, S.Y.; Cha, M.N.; Ahn, J.-H. Biosynthesis and production of glycosylated flavonoids in Escherichia coli: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 2979–2988. [Google Scholar] [CrossRef]
- Xie, K.; Dou, X.; Chen, R.; Chen, D.; Fang, C.; Xiao, Z.; Dai, J. Two novel fungal phenolic UDP glycosyltransferases from Absidia coerulea and Rhizopus japonicus. Appl. Environ. Microbiol. 2017, 83, e03103-16. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1–10, 1a–1d, 2a, 2b, 3a and 4a are available from the authors. |


| No. | 1a | 1b | 1c | 1d | ||||
|---|---|---|---|---|---|---|---|---|
| δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) | |
| 2 | 78.7, CH | 5.50, brd (12.7) | 78.0, CH | 5.52, brd (12.2) | 78.1, CH | 5.52, brd (12.2) | ||
| 3 | 42.0, CH2 | 3.35, m 2.73, dt (17.4, 3.6) | 42.0, CH2 | 3.27, m 2.73, dt (17.1, 3.2) | 3.27, m 2.73, dt (17.1, 3.2) | 42.1, CH2 | 3.36, m 2.80, dd (17.1, 2.8) | |
| 4 | 197.2, C | 196.1, C | 195.9, C | 197.0, C | ||||
| 5 | 162.9, C | 163.5, C | 163.5, C | 162.9, C | ||||
| 6 | 95.4, CH | 6.15, brs | 95.9, CH | 5.89, brs | 96.0, CH | 5.87, brs | 95.4, CH | 6.17, brs |
| 7 | 165.1, C | 166.8, C | 167.3, C | 165.1, C | ||||
| 8 | 96.5, CH | 6.13, brs | 95.0, CH | 5.88, brs | 95.2, CH | 5.86, brs | 96.5, CH | 6.14, brs |
| 9 | 162.8, C | 162.8, C | 162.7, C | 162.6, C | ||||
| 10 | 103.2, C | 101.7, C | 101.6, C | 103.3, C | ||||
| 1′ | 128.6, C | 131.9, C | 132.0, C | 131.7, C | ||||
| 2′ | 128.4, CH | 7.33, d (8.2) | 128.0, CH | 7.43, d (8.2) | 128.0, CH | 7.43, d (8.2) | 128.1, CH | 7.45, d (8.2) |
| 3′ | 115.2, CH | 6.80, d (8.2) | 116.1, CH | 7.06, d (8.2) | 116.2, CH | 7.06, d (8.2) | 116.2, CH | 7.06, d (8.2) |
| 4′ | 157.8, C | 157.4, C | 157.5, C | 157.5, C | ||||
| 5′ | 115.2, CH | 6.80, d (8.2) | 116.1, CH | 7.06, d (8.2) | 116.2, CH | 7.06, d (8.2) | 116.2, CH | 7.06, d (8.2) |
| 6′ | 128.4, CH | 7.33, d (8.2) | 128.0, CH | 7.43, d (8.2) | 128.0, CH | 7.43, d (8.2) | 128.1, CH | 7.45, d (8.2) |
| 1′′ | 99.1, CH | 5.01, d (7.8) | 99.9, CH | 4.91, d (7.7) | 100.3, CH | 4.88, d (7.4) | 99.1, CH | 5.01, d (7.8) |
| 2′′ | 73.2, CH | 3.24, t (8.1) | 73.4, CH | 3.24, m | 73.2, CH | 3.24, m | 73.2, CH | 3.24, t (8.1) |
| 3′′ | 76.0, CH | 3.40, m | 76.3, CH | 3.41, m | 77.1, CH | 3.33, m | 76.0, CH | 3.40, m |
| 4′′ | 78.8, CH | 3.01, t (9.3) | 79.0, CH | 3.03, t (9.4) | 69.7, CH | 3.16, t (9.0) | 78.8, CH | 3.01, t (9.3) |
| 5′′ | 75.6, CH | 3.43, m | 75.6, CH | 3.38, m | 76.6, CH | 3.26, m | 75.6, CH | 3.43, m |
| 6′′ | 60.1, CH2 | 3.60, brd (11.8) 3.47, m | 60.2, CH2 | 3.63, dd (11.8, 4.4) 3.49, m | 60.7, CH2 | 3.69, btd (11.8) 3.45, m | 60.1, CH2 | 3.60, brd (11.8) 3.47, m |
| 4′′-OCH3 | 59.6, CH3 | 3.44, s | 59.7, CH3 | 3.45, s | 59.6, CH3 | 3.44, s | ||
| 1′′′ | 99.9, CH | 4.91, d (7.7) | ||||||
| 2′′′ | 73.4, CH | 3.24, m | ||||||
| 3′′′ | 76.3, CH | 3.41, m | ||||||
| 4′′′ | 79.0, CH | 3.03, t (9.4) | ||||||
| 5′′′ | 75.6, CH | 3.38, m | ||||||
| 6′′′ | 60.2, CH2 | 3.63, dd (11.8, 4.4) 3.49, m | ||||||
| 4′′′-OCH3 | 59.7, CH3 | 3.45, s | ||||||
| 5-OH | 12.01, s | 12.13, s | 12.15, s | 12.02, s | ||||
| 4′-OH | 9.62, s | |||||||
| No. | 2a | 2b | 3a | 4a | ||||
|---|---|---|---|---|---|---|---|---|
| δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) | |
| 2 | 163.4, C | 163.1, C | 163.0, C | 153.6, CH | 8.44, s | |||
| 3 | 103.0, CH | 6.79, s | 104.0, CH | 6.82, s | 103.7, CH | 6.89, s | 123.4, C | |
| 4 | 181.7, C | 181.7, C | 181.7, C | 174.6, C | ||||
| 5 | 161.4, C | 161.4, C | 161.4, C | 127.0, CH | 8.06, d (8.8) | |||
| 6 | 98.9, CH | 6.18, brs | 98.9, CH | 6.20, brs | 99.1, CH | 6.17, brs | 115.6, CH | 7.15, dd (8.8, 1.8) |
| 7 | 164.3, C | 164.3, C | no show | 161.3, C | ||||
| 8 | 94.1, CH | 6.49, brs | 94.0, CH | 6.50, brs | 94.2, CH | 6.49, brs | 103.3, CH | 7.24, d (1.8) |
| 9 | 157.3, C | 157.3, C | 157.4, C | 157.0, C | ||||
| 10 | 103.6, C | 103.8, C | 103.4, C | 118.5, C | ||||
| 1′ | 121.1, C | 124.7, C | 122.9, C | 124.0, C | ||||
| 2′ | 114.4, C | 7.74, d (2.2) | 113.6, C | 7.50, brs | 112.8, C | 7.70, d (2.0) | 130.1, CH | 7.53, d (8.4) |
| 3′ | 145.7, C | 146.9, C | 146.5, C | 115.6, CH | 7.00, d (8.4) | |||
| 4′ | 151.4, C | 148.4, C | 152.1, C | 159.0, C | ||||
| 5′ | 116.6, CH | 6.95, d (8.5) | 115.8, CH | 7.22, d (8.5) | 112.4, C | 7.16, d (8.6) | 115.6, CH | 7.00, d (8.4) |
| 6′ | 122.0, CH | 7.64, dd (8.5, 2.2) | 118.5, CH | 7.51, brd (9.0) | 121.0, CH | 7.64, dd (8.6, 2.0) | 130.1, CH | 7.53, d (8.4) |
| 1′′ | 101.5, CH | 4.93, d (7.8) | 100.7, CH | 4.92, d (7.8) | 99.4, CH | 5.17, d (7.8) | 99.6, CH | 5.14, d (7.8) |
| 2′′ | 73.5, CH | 3.34, m | 73.4, CH | 3.34, m | 73.4, CH | 3.30, m | 73.3, CH | 3.30, m |
| 3′′ | 75.8, CH | 3.52, m | 75.8, CH | 3.52, m | 76.6, CH | 3.46, m | 76.2, CH | 3.46, m |
| 4′′ | 79.3, CH | 3.04, t (9.2) | 79.0, CH | 3.06, t (9.3) | 79.1, CH | 3.05, t (9.3) | 78.9, CH | 3.06, t (9.1) |
| 5′′ | 75.7, CH | 3.46, m | 75.6, CH | 3.46, m | 75.6, CH | 3.52, m | 75.7, CH | 3.52, m |
| 6′′ | 60.4, CH2 | 3.70, brs (11.4) 3.56, m | 60.2, CH2 | 3.66, brs (11.4) 3.52, m | 60.2, CH2 | 3.64, brs (11.0) 3.52, m | 60.2, CH2 | 3.66, brs (11.0) 3.52, m |
| 4′-OCH3 | 55.8, CH3 | 3.86, s | 55.2, CH3 | 3.79, s | ||||
| 4′′-OCH3 | 59.7, CH3 | 3.47, s | 59.7, CH3 | 3.47, s | 59.7, CH3 | 3.47, s | 59.7, CH3 | 3.47, s |
| 5-OH | 12.96, s | 12.90, s | 12.90, s | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dou, F.; Wang, Z.; Li, G.; Dun, B. Microbial Transformation of Flavonoids by Isaria fumosorosea ACCC 37814. Molecules 2019, 24, 1028. https://doi.org/10.3390/molecules24061028
Dou F, Wang Z, Li G, Dun B. Microbial Transformation of Flavonoids by Isaria fumosorosea ACCC 37814. Molecules. 2019; 24(6):1028. https://doi.org/10.3390/molecules24061028
Chicago/Turabian StyleDou, Fangmin, Zhi Wang, Guiying Li, and Baoqing Dun. 2019. "Microbial Transformation of Flavonoids by Isaria fumosorosea ACCC 37814" Molecules 24, no. 6: 1028. https://doi.org/10.3390/molecules24061028
APA StyleDou, F., Wang, Z., Li, G., & Dun, B. (2019). Microbial Transformation of Flavonoids by Isaria fumosorosea ACCC 37814. Molecules, 24(6), 1028. https://doi.org/10.3390/molecules24061028




