Investigating the Effect of Mono- and Dimeric 360A G-Quadruplex Ligands on Telomere Stability by Single Telomere Length Analysis (STELA)
Abstract
1. Introduction
2. Results
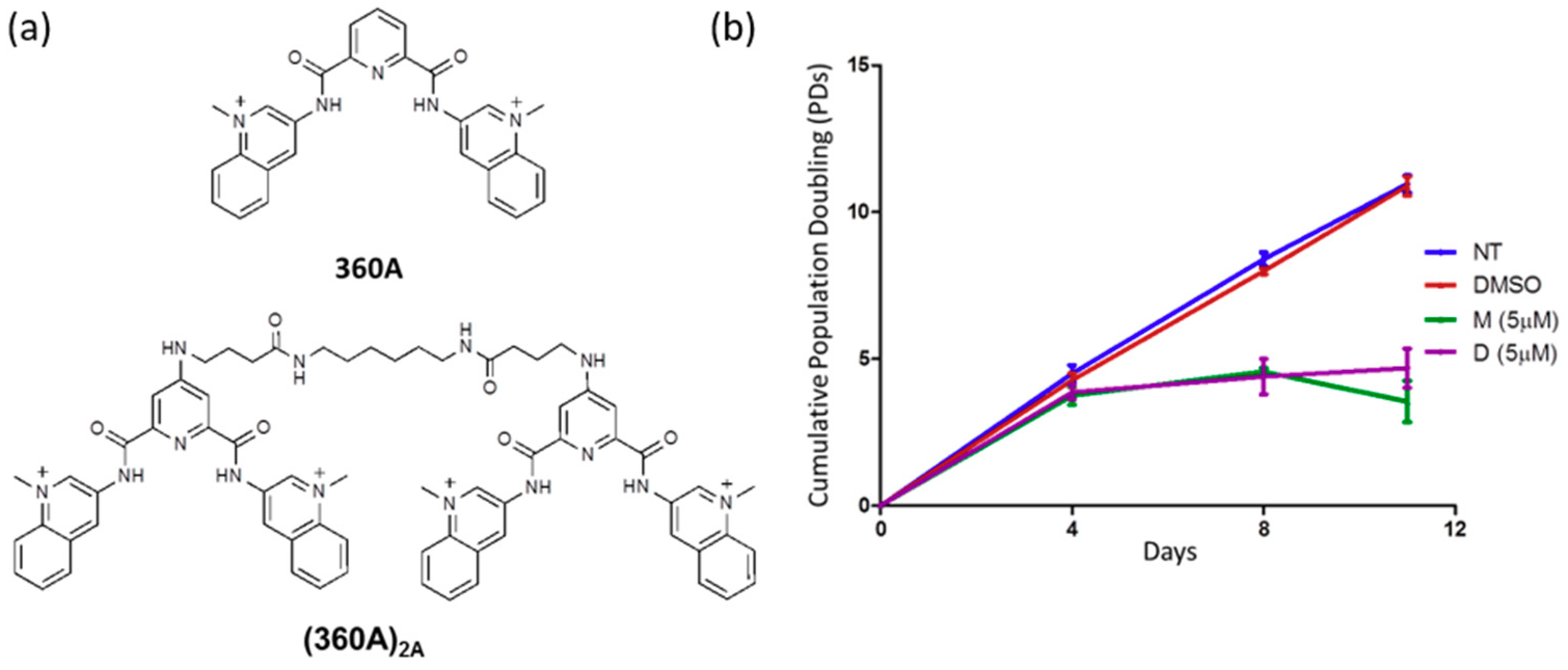
2.1. 360A and (360A)2A Inhibit the Proliferation of A549 Cells with No Effect on Mean Telomere Length
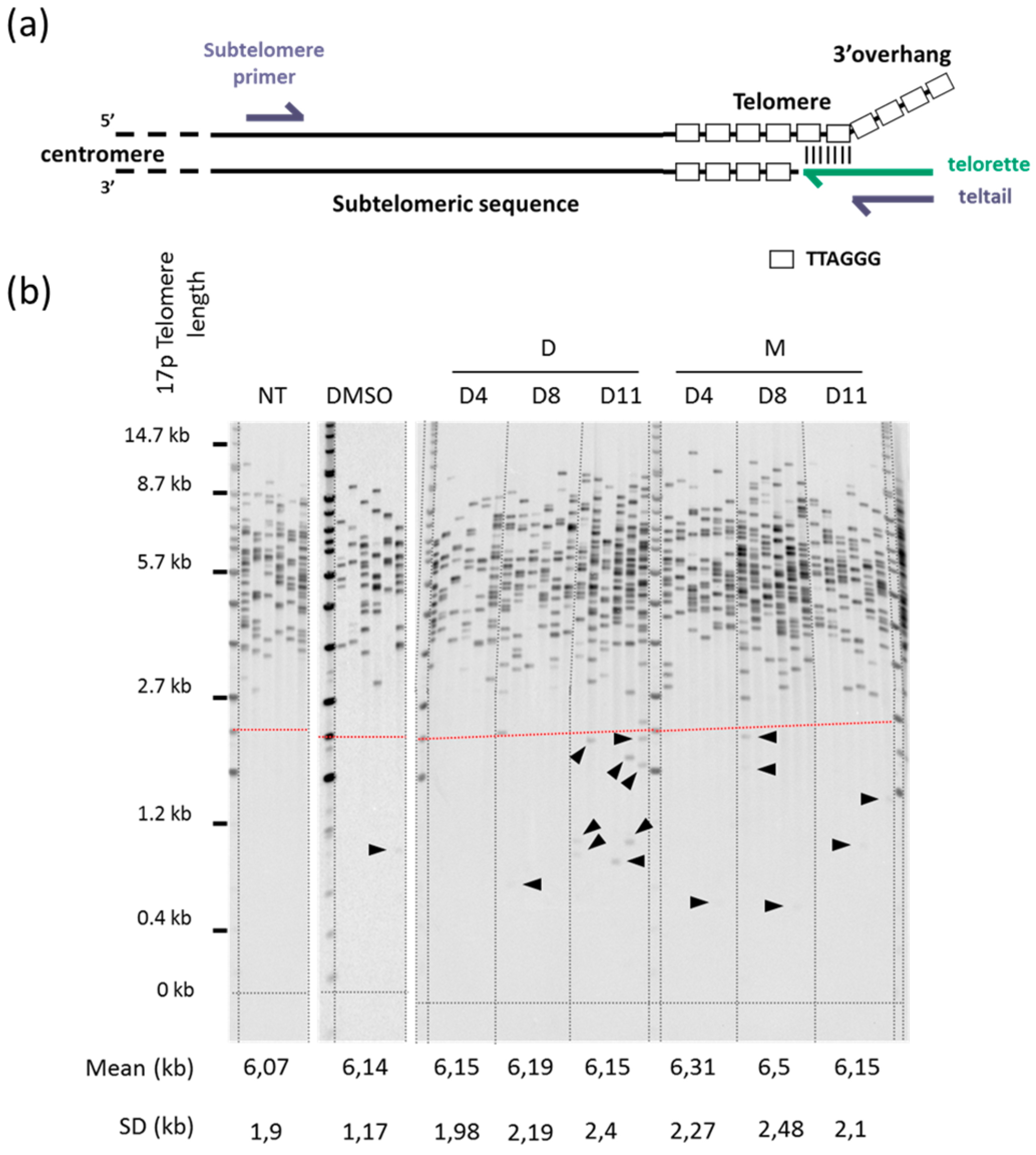
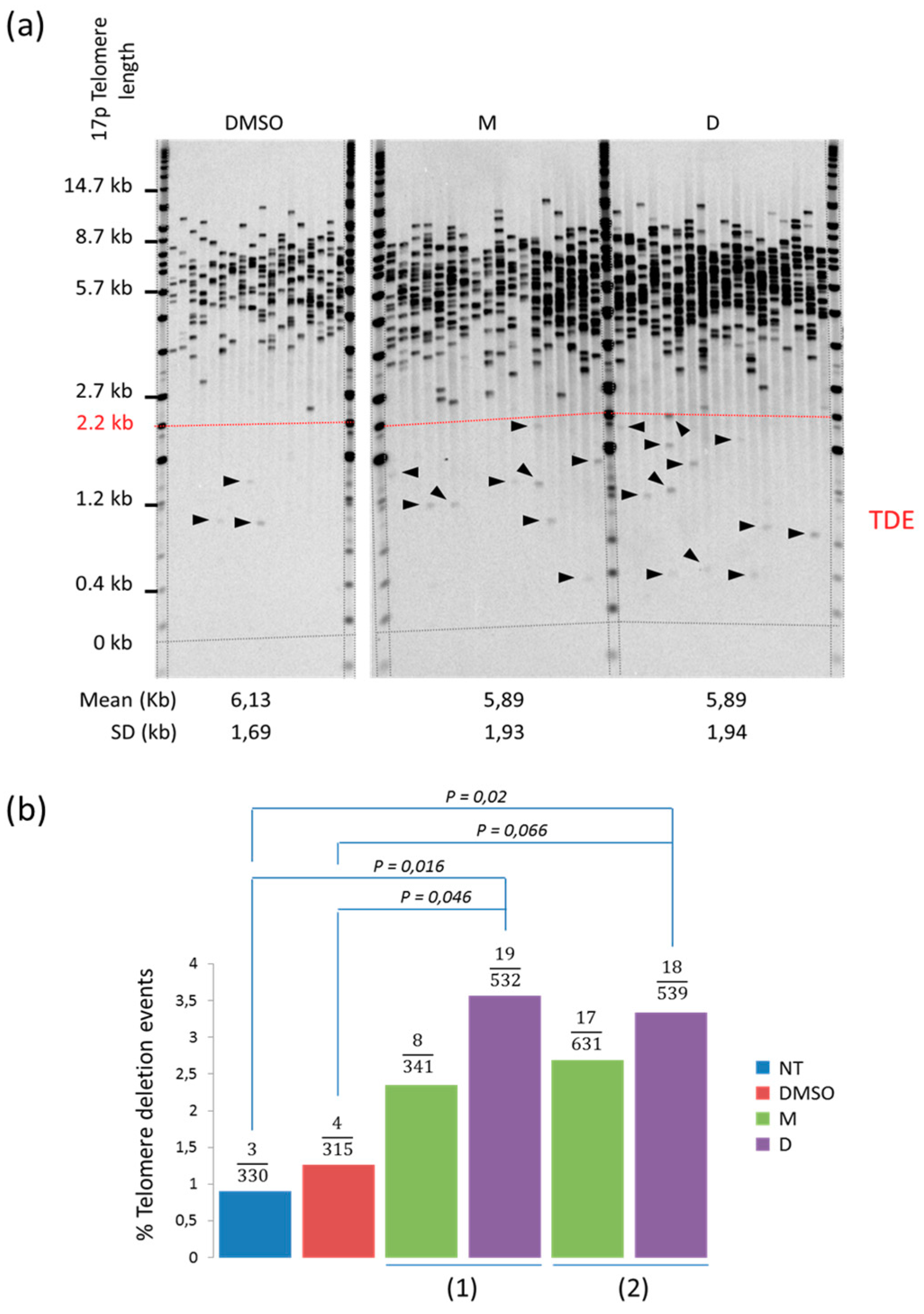
2.2. 360A and (360A)2A Induce Telomere Deletion Events in A549 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Cell Growth Experiments
4.2. DNA Extraction and STELA
4.3. Oligonucleotides
4.4. Chemical Compounds
4.5. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Di Fagagna, F.D.A.; Reaper, P.M.; Clay-Farrace, L.; Fiegler, H.; Carr, P.; Von Zglinicki, T.; Saretzki, G.; Carter, N.P.; Jackson, S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003, 426, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef] [PubMed]
- Olovnikov, A.M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973, 41, 181–190. [Google Scholar] [CrossRef]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Baird, D.M.; Rowson, J.; Wynford-Thomas, D.; Kipling, D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 2003, 33, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Baird, D.M.; Britt-Compton, B.; Rowson, J.; Amso, N.N.; Gregory, L.; Kipling, D. Telomere instability in the male germline. Hum. Mol. Genet. 2006, 15, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Britt-Compton, B.; Rowson, J.; Locke, M.; Mackenzie, I.; Kipling, D.; Baird, D.M. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans. Hum. Mol. Genet. 2006, 15, 725–733. [Google Scholar] [CrossRef]
- Capper, R.; Britt-Compton, B.; Tankimanova, M.; Rowson, J.; Letsolo, B.; Man, S.; Haughton, M.; Baird, D.M. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007, 21, 2495–2508. [Google Scholar] [CrossRef]
- Bugaut, A.; Alberti, P. Understanding the stability of DNA G-quadruplex units in long human telomeric strands. Biochimie 2015, 113, 125–133. [Google Scholar] [CrossRef]
- Granotier, C.; Pennarun, G.; Riou, L.; Hoffschir, F.; Gauthier, L.R.; De Cian, A.; Gomez, D.; Mandine, E.; Riou, J.F.; Mergny, J.L.; et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005, 33, 4182–4190. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; McDonald, K.R.; Zakian, V.A. Telomeres: Structures in need of unwinding. FEBS Lett. 2010, 584, 3760–3772. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Zakian, V. A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef]
- Hansel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Hansel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010, 277, 1118–1125. [Google Scholar] [CrossRef]
- Neidle, S. Quadruplex Nucleic Acids as Novel Therapeutic Targets. J. Med. Chem. 2016, 59, 5987–6011. [Google Scholar] [CrossRef]
- Islam, M.K.; Jackson, P.J.; Rahman, K.M.; Thurston, D.E. Recent advances in targeting the telomeric G-quadruplex DNA sequence with small molecules as a strategy for anticancer therapies. Future Med. Chem. 2016, 8, 1259–1290. [Google Scholar] [CrossRef]
- Pennarun, G.; Granotier, C.; Gauthier, L.R.; Gomez, D.; Hoffschir, F.; Mandine, E.; Riou, J.F.; Mergny, J.L.; Mailliet, P.; Boussin, F.D. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene 2005, 24, 2917–2928. [Google Scholar] [CrossRef]
- Pennarun, G.; Granotier, C.; Hoffschir, F.; Mandine, E.; Biard, D.; Gauthier, L.R.; Boussin, F.D. Role of ATM in the telomere response to the G-quadruplex ligand 360A. Nucleic Acids Res. 2008, 36, 1741–1754. [Google Scholar] [CrossRef]
- Gauthier, L.R.; Granotier, C.; Hoffschir, F.; Etienne, O.; Ayouaz, A.; Desmaze, C.; Mailliet, P.; Biard, D.S.; Boussin, F.D. Rad51 and DNA-PKcs are involved in the generation of specific telomere aberrations induced by the quadruplex ligand 360A that impair mitotic cell progression and lead to cell death. Cell. Mol. Life Sci. 2012, 69, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.; O’Donohue, M.F.; Wenner, T.; Douarre, C.; Macadre, J.; Koebel, P.; Giraud-Panis, M.J.; Kaplan, H.; Kolkes, A.; Shin-ya, K.; et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006, 66, 6908–6912. [Google Scholar] [CrossRef] [PubMed]
- Tahara, H.; Shin-Ya, K.; Seimiya, H.; Yamada, H.; Tsuruo, T.; Ide, T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3’ telomeric overhang in cancer cells. Oncogene 2006, 25, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
- Hampel, S.M.; Pepe, A.; Greulich-Bode, K.M.; Malhotra, S.V.; Reszka, A.P.; Veith, S.; Boukamp, P.; Neidle, S. Mechanism of the antiproliferative activity of some naphthalene diimide G-quadruplex ligands. Mol. Pharmacol. 2013, 83, 470–480. [Google Scholar] [CrossRef]
- Rizzo, A.; Salvati, E.; Porru, M.; D’Angelo, C.; Stevens, M.F.; D’Incalci, M.; Leonetti, C.; Gilson, E.; Zupi, G.; Biroccio, A. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res. 2009, 37, 5353–5364. [Google Scholar] [CrossRef] [PubMed]
- Saintome, C.; Alberti, P.; Guinot, N.; Lejault, P.; Chatain, J.; Mailliet, P.; Riou, J.F.; Bugaut, A. Binding properties of mono- and dimeric pyridine dicarboxamide ligands to human telomeric higher-order G-quadruplex structures. Chem. Commun. 2018, 54, 1897–1900. [Google Scholar] [CrossRef]
- Sfeir, A.J.; Chai, W.; Shay, J.W.; Wright, W.E. Telomere-end processing the terminal nucleotides of human chromosomes. Mol. Cell 2005, 18, 131–138. [Google Scholar] [CrossRef]
- Sidibe, A.; Hamon, F.; Largy, E.; Gomez, D.; Teulade-Fichou, M.P.; Trentesaux, C.; Riou, J.F. Effects of a halogenated G-quadruplex ligand from the pyridine dicarboxamide series on the terminal sequence of XpYp telomere in HT1080 cells. Biochimie 2012, 94, 2559–2568. [Google Scholar] [CrossRef]
- Baird, D.M. New developments in telomere length analysis. Exp. Gerontol. 2005, 40, 363–368. [Google Scholar] [CrossRef]
- Mendoza, O.; Bourdoncle, A.; Boule, J.B.; Brosh, R.M., Jr.; Mergny, J.L. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef]
- Crabbe, L.; Verdun, R.E.; Haggblom, C.I.; Karlseder, J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 2004, 306, 1951–1953. [Google Scholar] [CrossRef] [PubMed]
- Wold, M.S. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997, 66, 61–92. [Google Scholar] [CrossRef] [PubMed]
- Verdun, R.E.; Karlseder, J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 2006, 127, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Dejardin, J.; Kingston, R.E. Purification of proteins associated with specific genomic Loci. Cell 2009, 136, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Sato, K.; Kibe, T.; Seimiya, H.; Nakamura, A.; Yukawa, M.; Tsuchiya, E.; Ueno, M. Expression of mutant RPA in human cancer cells causes telomere shortening. Biosci. Biotechnol. Biochem. 2010, 74, 382–385. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, I.P.; Mailliet, P.; Hossard, V.; Riou, J.-F.; Bugaut, A.; Roger, L. Investigating the Effect of Mono- and Dimeric 360A G-Quadruplex Ligands on Telomere Stability by Single Telomere Length Analysis (STELA). Molecules 2019, 24, 577. https://doi.org/10.3390/molecules24030577
Hwang IP, Mailliet P, Hossard V, Riou J-F, Bugaut A, Roger L. Investigating the Effect of Mono- and Dimeric 360A G-Quadruplex Ligands on Telomere Stability by Single Telomere Length Analysis (STELA). Molecules. 2019; 24(3):577. https://doi.org/10.3390/molecules24030577
Chicago/Turabian StyleHwang, In Pyo, Patrick Mailliet, Virginie Hossard, Jean-Francois Riou, Anthony Bugaut, and Lauréline Roger. 2019. "Investigating the Effect of Mono- and Dimeric 360A G-Quadruplex Ligands on Telomere Stability by Single Telomere Length Analysis (STELA)" Molecules 24, no. 3: 577. https://doi.org/10.3390/molecules24030577
APA StyleHwang, I. P., Mailliet, P., Hossard, V., Riou, J.-F., Bugaut, A., & Roger, L. (2019). Investigating the Effect of Mono- and Dimeric 360A G-Quadruplex Ligands on Telomere Stability by Single Telomere Length Analysis (STELA). Molecules, 24(3), 577. https://doi.org/10.3390/molecules24030577





