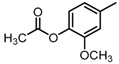
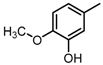
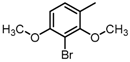
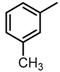
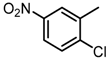
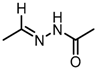
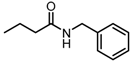
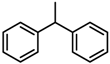
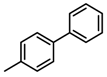
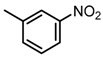
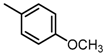
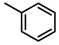
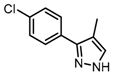
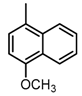
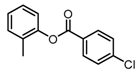
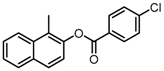
Abstract
SET7, serving as the only histone methyltransferase that monomethylates ‘Lys-4’ of histone H3, has been proved to function as a key regulator in diverse biological processes, such as cell proliferation, transcriptional network regulation in embryonic stem cell, cell cycle control, protein stability, heart morphogenesis and development. What′s more, SET7 is involved inthe pathogenesis of alopecia aerate, breast cancer, tumor and cancer progression, atherosclerosis in human carotid plaques, chronic renal diseases, diabetes, obesity, ovarian cancer, prostate cancer, hepatocellular carcinoma, and pulmonary fibrosis. Therefore, there is urgent need to develop novel SET7 inhibitors. In this paper, based on DC-S239 which has been previously reported in our group, we employed scaffold hopping- and 2D fingerprint-based similarity searches and identified DC-S285 as the new hit compound targeting SET7 (IC50 = 9.3 μM). Both radioactive tracing and NMR experiments validated the interactions between DC-S285 and SET7 followed by the second-round similarity search leading to the identification ofDC-S303 with the IC50 value of 1.1 μM. In cellular level, DC-S285 retarded tumor cell proliferation and showed selectivity against MCF7 (IC50 = 21.4 μM), Jurkat (IC50 = 2.2 μM), THP1 (IC50 = 3.5 μM), U937 (IC50 = 3.9 μM) cell lines. Docking calculations suggested that DC-S303 share similar binding mode with the parent compoundDC-S239. What′s more, it presented good selectivity against other epigenetic targets, including SETD1B, SETD8, G9a, SMYD2 and EZH2. DC-S303 can serve as a drug-like scaffold which may need further optimization for drug development, and can be used as chemical probe to help the community to better understand the SET7 biology.
1. Introduction
In the epigenetic landscape, histone methyltransferases (HMTs) play an essential role in various biological processes including cell cycle progression [1], cell differentiation [2], development [3] as well as other biological processes [4]. Besides, HMTs are involved with the pathogenesis of cancers [5,6,7,8], immune-mediated diseases [9], thus they have been the hot targets for cancer therapy in both academia and industry. HMTs can be categorized into two groups based on structural features: (i) SET domain-containing subfamily, such as SET7 (SET domain-containing lysine methyltransferase 7, also called SETD7, SET9, KMT7), EZH2 (Enhancer of Zest Homologue 2) and SUV39H1 and (ii) non-SET domain containing subfamily, such as DOT1-L (Disruptor of Telomeric silencing 1-Like) [10].
SET7 is the only epigenetic member that specifically monomethylates ‘Lys-4’ of histone H3 and emerging evidences have proved SET7′s unique role in transcriptional regulations [11] (Table 1), DNA repair [12], cell cycle control [12,13,14]. Due to its catalytic activity on diverse non-histone substrates, SET7 also displays a special role in a lot of biological processes (Table 1) and is involved in cell proliferation [15,16], transcriptional network regulation in embryonic stem cell [17,18], cell cycle control [19], protein stability [20,21,22,23], heart morphogenesis and development [24], as well as other biological functions. Notably, its role in regulating p53, whose mutant isoform is an important cancer therapy target [25], remains controversial [26].

Table 1.
SET7 substrates and its biological functions.
In addition, SET7 is reported to be a key regulator in the pathogenies of several diseases in cluding alopecia areata [69], breast cancer [37,70], tumour and cancer progression [71,72,73,74,75,76], atherosclerosis in human carotid plaques [77], chronic renal diseases, diabetes [45,78,79,80,81,82,83,84], obesity [85], ovarian cancer [86], prostate cancer [87], hepatocellular carcinoma [71], and pulmonary fibrosis [57]. Of interest, the role of SET7 in viral infection is being uncovered. Studies revealed that SET7 can facilitate HCV replication in an enzymatic activity-dependent manner through attenuation of the IFN/JAK/STAT pathway [88]. SET7 binds with HIV-1 TAR RNA and monomethylates the HIV viral trans-activator Tat protein at Lys-51 and Lys-71, to enhance HIV transcription [63,89]. Knockdown analysis also proved that SET7 only suppresses Tat transactivation of the viral promoter and has on influence on transcriptional activity of methylation-deficient Tat Shan’s study suggested that SET7 can negatively regulate the anti-viral activity toward vesicular stomatitis virus (VSV) and influenza A by methylation at Lys-88 of virus (IAV) infection of Interferon-induced Transmembrane Protein 3 (IFITM3) [44]. What′s more, SET7 may also have a potential role in HPV (human papillomaviruses) viral survival [90]. Therefore, SET7 may be a promising target for viral infection and potent and selective inhibitor can serve as a useful chemical biology tool to elucidate the viral infection mechanism. The abberant expression patterns and dysfunction of SET7 has also been widely observed in the onset and progression of cancers. In peripheral blood mononuclear cells of patients, the histone modification patterns were greatly altered and the expression of SET7 was elevated [69]. Emerging evidence has also demonstrated its role in solid tumors. Zhang et al. demonstrated that SET7 interacts with transcription factor GATA1 and promotes downstream VEGF transcription and tumor angiogenesis [40]. Inhibition of SET7 activity by the SET7 inhibitor cyproheptadine reduced the estrogen receptor alpha expression in MCF7 cells that is important for cancer progression, phencopying the SET7 knockdown studies [37]. With SET7′s emerging role of therapy targets for cancers, diabetes, hepatocellular carcinoma, alopecia areata, pulmonary fibrosis and viral infections, a potent and selective SET7 inhibitor is in great need to serve as a chemical probe to investigate its delicate biological function and mechanism.
So far, several successful attempts have been made to develop SET7 inhibitors. (R)-PFI-2 [91], as resolved in the crystal complex structure, is bound to the peptide binding pocket with a high affinity (IC50 = 2.0 nM). Using a drug repositioning strategy, Cyproheptadine, a clinically approved antiallergy drug was reported to bind with SET7 peptide binding site as revealed in structural biology studies (IC50 = 1.0 μM) [61].
Computational tools and methods have been widely used for epigenetic inhibitor design and discovery and many successful cases have been reported [92]. Some epigenetic inhibitors have been identified by computational methods targeting PRMT1, DNMT1, DOTL and the protein-protein interfaces like menin-mixed lineage leukemia 1 and EZH2-EED [92]. In our previous study, two selective SET7 inhibitors DC-238 and DC-S239 were identified with an integrated virtual screening method combing pharmacophore and docking as well as chemical modifications [92].
In this paper, a new SET7 inhibitor DC-S285 was identified by scaffold hopping and 2D fingerprint -based similarity search based on the previously identified SET7 inhibitor DC-S239, which was further validated by radioactivity assay and NMR-spectroscopy methods. In cellular level, DC-S285 also inhibited cancer cell proliferation in leukemic cells and breast cancer cells. Therefore, a similarity search was performed in search of more potent inhibitors as well as for its structure-activity relationship (SAR) studies. The predicted binding mode was also investigated in accordance with its SAR. In summary, a novel and selective compound DC-S336 (IC50 = 1.1 μM) was identified by similarity based search.
2. Results and Discussions
2.1. Scaffold Hopping- and Similarity-Based Virtual Screening
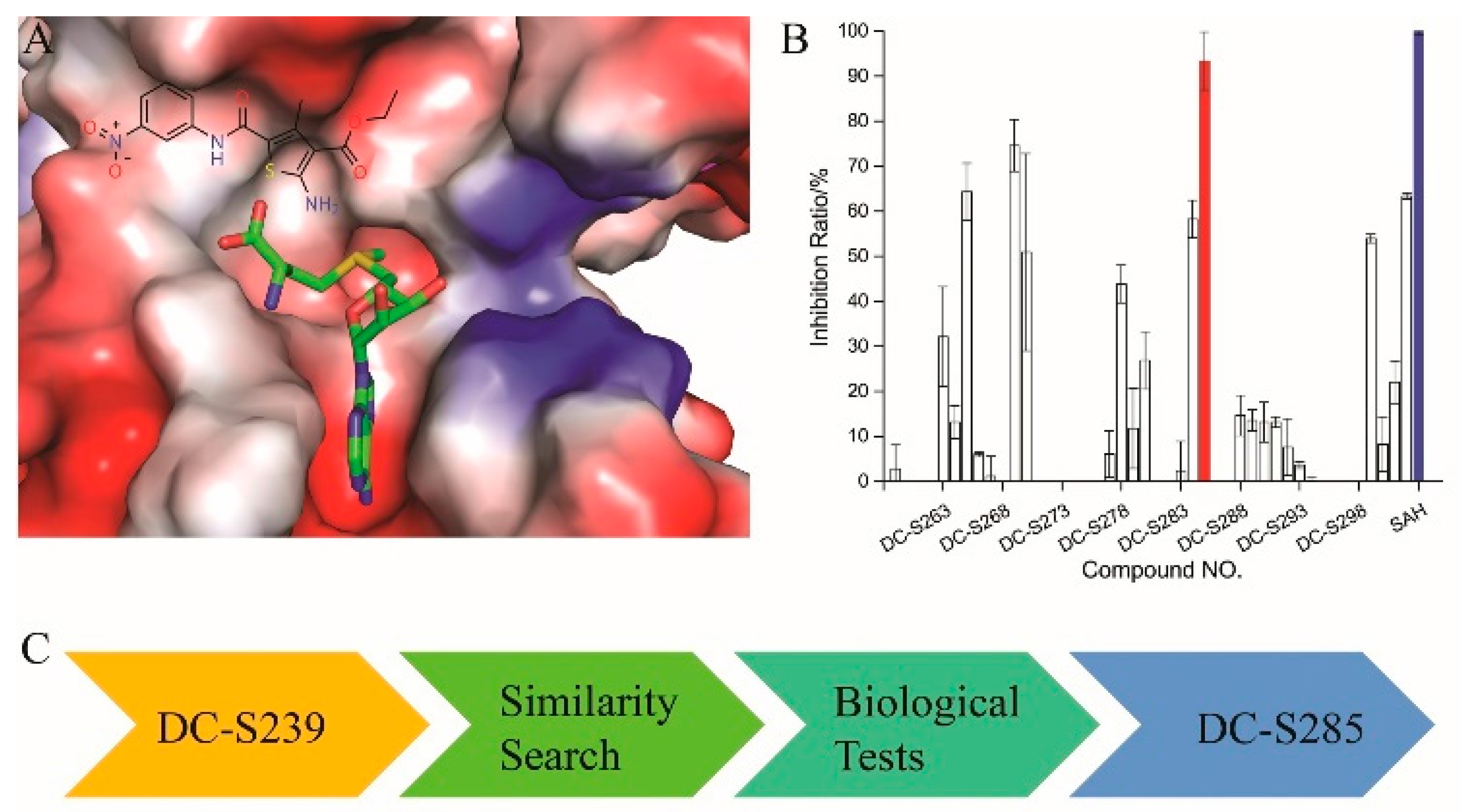
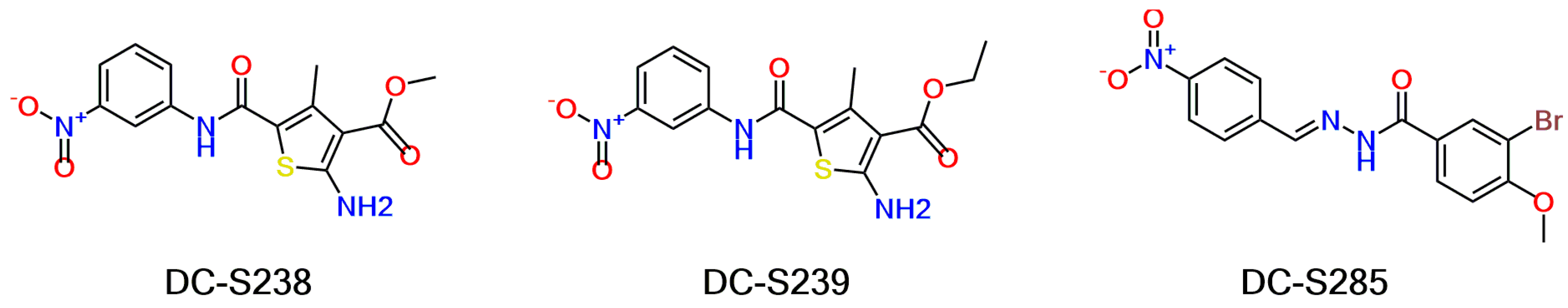
Based on our previous research, two selective SET7 inhibitors, namely DC-S238 (IC50 = 4.9 μM) and DC-S239 (IC50 = 4.6 μM) were obtained (Figure 1A). Herein, with DC-S239 serving as the starting point, combinatorial scaffold hopping [93] and 2D fingerprint based similarity search strategies were employed in order to discover more SET7 inhibitors with novel chemotypes (Figure 2). The overall computational pipeline has been demonstrated (Figure 1C).
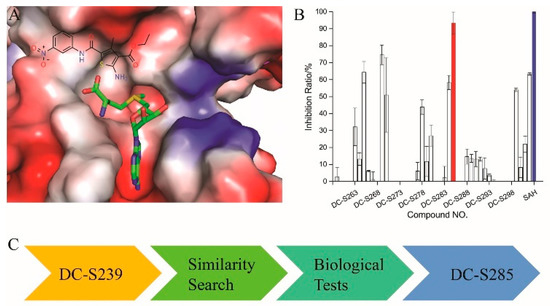
Figure 1.
The flowchart of the combinatorial scaffold hopping and 2D fingerprint based similarity search strategies. (A) Chemical structure of DC-S239 and its predicted binding mode. DC-S239 is shown in sticks and the SET7 protein is shown in electrostatics surface; (B) Inhibition ratio of the compounds in first-round similarity search. DC-S285 is shown in red while the reference compound SAH is depicted in blue; (C) Workflow chart in the study.
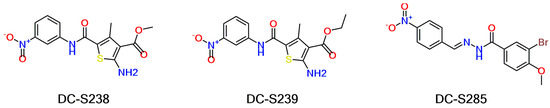
Figure 2.
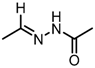
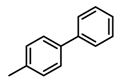
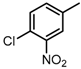
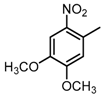
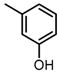
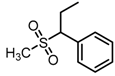
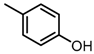
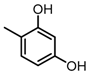
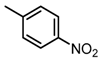
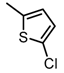
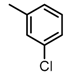
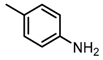
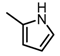
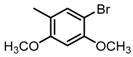
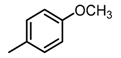
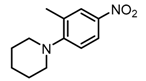
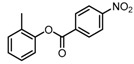
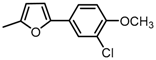
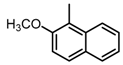
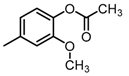
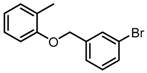
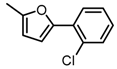
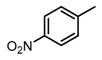
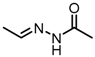
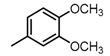
Chemical structures of DC-S238, DC-S239 and DC-S285.
Before performing similarity searches, pre-processing of the Specs molecular database was conducted with Pipeline Pilot, version 7.5 (Pipeline Pilot; Accelrys Software Inc., San Diego, CA, USA). Because a lot of clinical trial failures and unnecessary attritions were due to poor oral bioavailability [94,95], it becomes crucial to estimate druglikeness properties at the early drug design and development stage, and therefore the Specss database was filtered by Rule of Five [96]. Recently, a series of promiscuous, assay-duping molecules, namely ‘pan-assay interference compounds’ (PAINS), were reported due to metal chelation, chemical aggregation, redox activity, compound fluorescence, cysteine oxidation or promiscuous binding against the targets, suggesting that we should get rid of those structures in early state [97,98,99,100]. Therefore, the Specs database was further processed with the PAINS substructure filter developed in our lab using Pipeline Pilot, version 7.5. Then the remaining 182,014 molecules were subjected to moleculardocking to remove low binding affinity compounds or non-binders usingGilde docking software [101] integrated in Maestro 9.0 (Maestro, version 9.0, Schrödinger, LLC, New York, NY, USA, 2009) inXP mode [102] consideringits highest enrichment factor [103] based on our previous tests [92]. Subsequently, top ranking 2000 molecules, with all the previously tested compounds in our previous paper [92] excluded, was subjected to ChemMapper and 2D similarity search. Then 300 compounds of each method were selected, and a total of 520 compounds were obtained after removing the duplicates. In order to cover chemical space with more diverse scaffolds, all the molecules were clustered into 30 groups and finally 44 compounds were selected and purchased from Specs Company (Quezon, Philippines) for biological tests. All the selections were based on the following criteria: (1) In order to get more confident results and more space for future optimization, molecules with simpler structure were chosen; (2) One compound is selected at least in each cluster to get more diverse chemical space; (3) All the molecules with potential reactive functional groups were not our preferences; (4) Molecules with similar structure or dramatic structural differences with DC-S239 were both considered based on our chemical intuitions.
2.2. AlphaLISA-Based Biological Tests
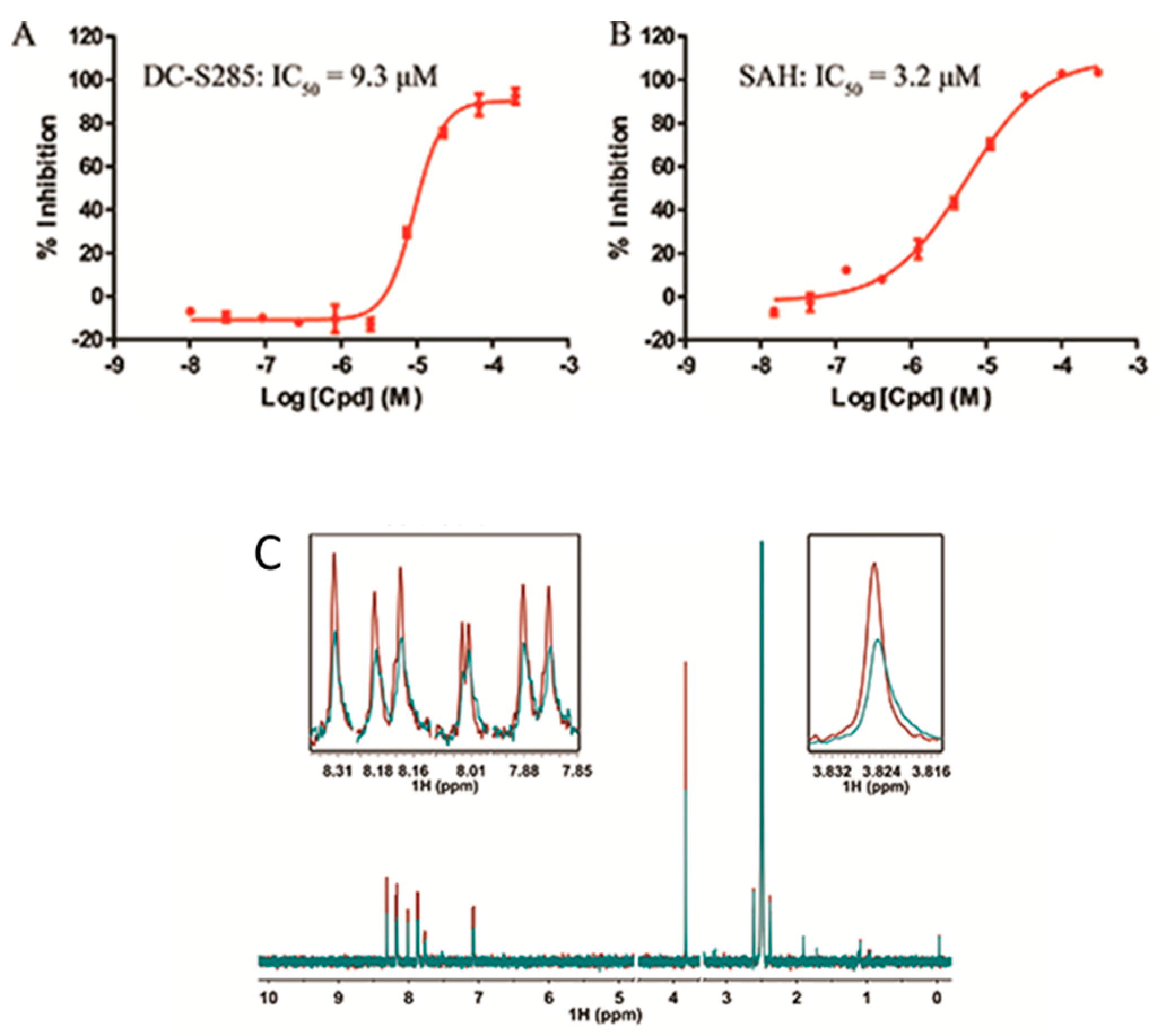
All the 44 candidate compounds cherry-picked from similarity search were evaluated for their biochemical activity against SET7 in vitro based on the AlphaLISA assay. SAH was used as the reference compound (Figure 2B). Among them, eight compounds came out at top against SET7 activity with inhibition rate >50% at 100 μM (Figure 1B), resulting in a hit rate of 18%. Notably, compound DC-S285 presented similar potency as the reference compound SAH at the concentration of 100 μM. Then we measured the inhibitory activity at a range of concentrations for DC-S285. Based from dose-response curve, DC-S285 displayed moderate inhibitory activity against SET7 with an IC50 value of 9.3 μM while the IC50 value of the reference compound SAH was about 3.2 μM (Figure 3A,B).
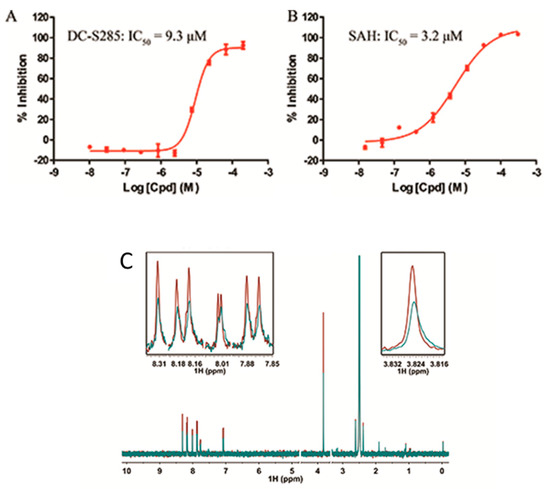
Figure 3.
Activity of DC-S285 and of ligand observed 1D NMR experiments. (A,B). Inhibitory activities of DC-S285 and its reference compound SAH based on the AlphaLISA assay; (C). Carr-Purcell Meiboom-Gill Pulse Sequence reveals that DC-S285 binds to SET7. T1ρ spectra acquired by using DC-S285 (colored in red), 20 μM DC-S285 in the presence of 2.5 μM protein (coloured in blue).
2.3. Validation of DC-S285′s Activity
To rule out the possibility of assay interference of DC-S285, we established the 3H-labeled radioisotope methylation assay. The z′ factor value of the established platform is 0.84, demonstrating its robustness for hit validation. DC-S285 and the reference compounds SAH presented the inhibitory activity with the IC50 values of 19.5 μM and 5.4 μM, respectively, which was in accordance with the AlphaLisa results.
In order to further confirm that compound DC-S285 binds with SET7, Carr-Purcell-Meiboom-Gill sequence (CPMG) experiments were performed for hit validation Strong binding signals were clearly observed in T1ρ (Figure 3C) and the result indicatedthe mutually exclusive binding of DC-S285 and SET7 catalytic domain.
2.4. Cellular Activity of DC-S285
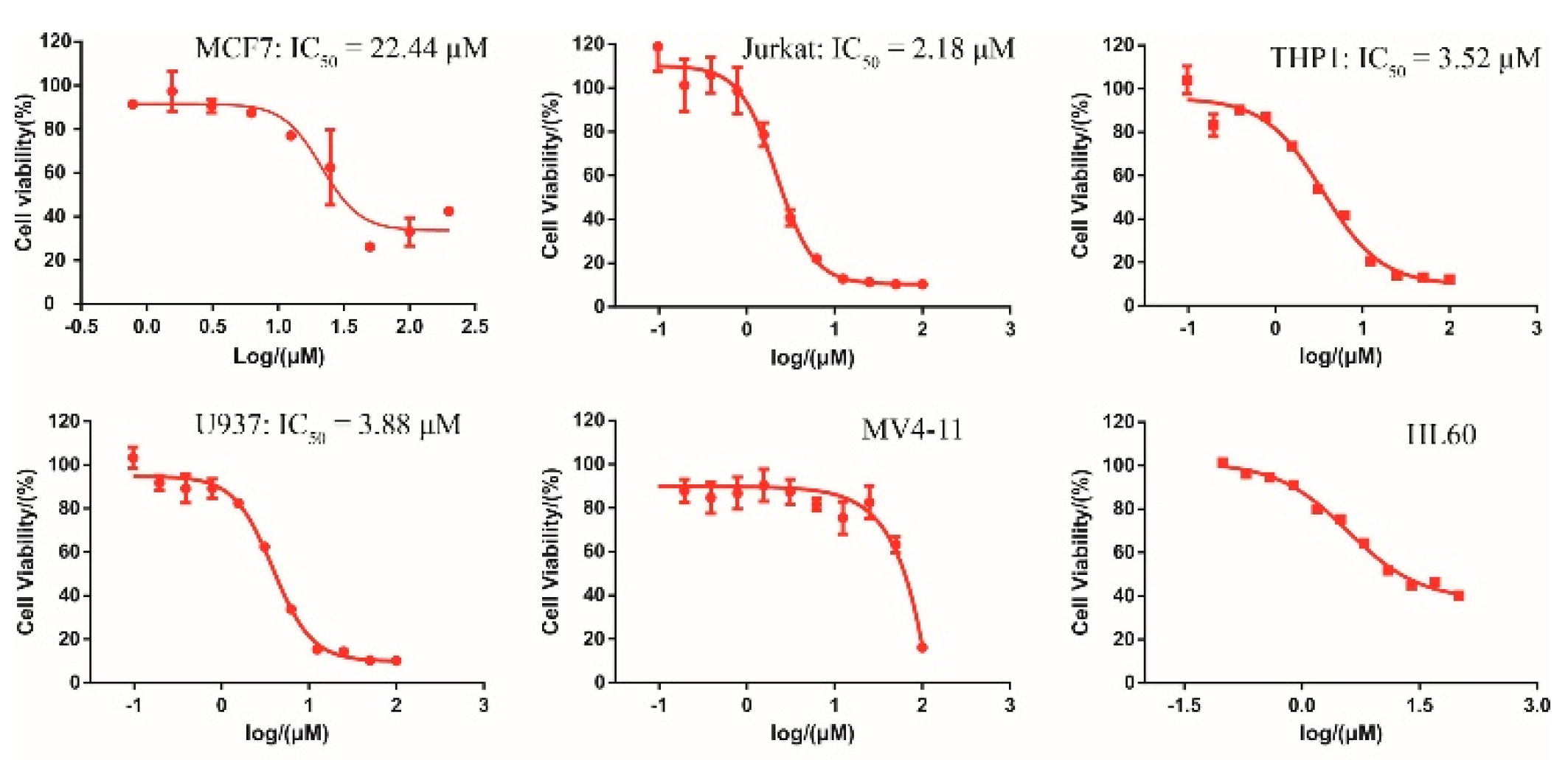
Collectively, both the radioactive and CPMG NMR demonstrated the direct binding between DC-S285 and SET7, wefurther evaluated its antiproliferation activities in several cancer cell lines. As depicted in Figure 4, DC-S285 significantly retard cell proliferation includingMCF7 (breast cancer), and Jurkat, THP1 and U937 leukemia cell lines in a dose-dependent manner at micromolar potencies (Figure 4).
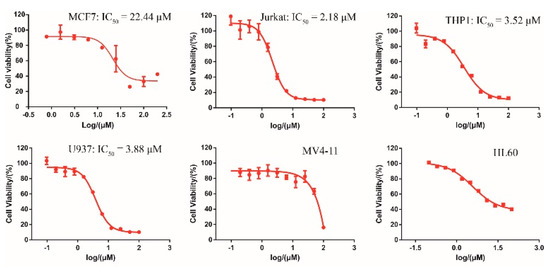
Figure 4.
Cellular activity of DC-S285 against different cell lines.
2.5. Similarity-Based Analog Searching and SAR Analysis
Based on the biological test, the hit, DC-S285, was used as the promising scaffold for the further structure-activity relationship study (SAR). A second round 2D molecular fingerprint based similarity search was conducted using Pipeline Pilot, version 7.5 to explore the SAR and more potent inhibitors.
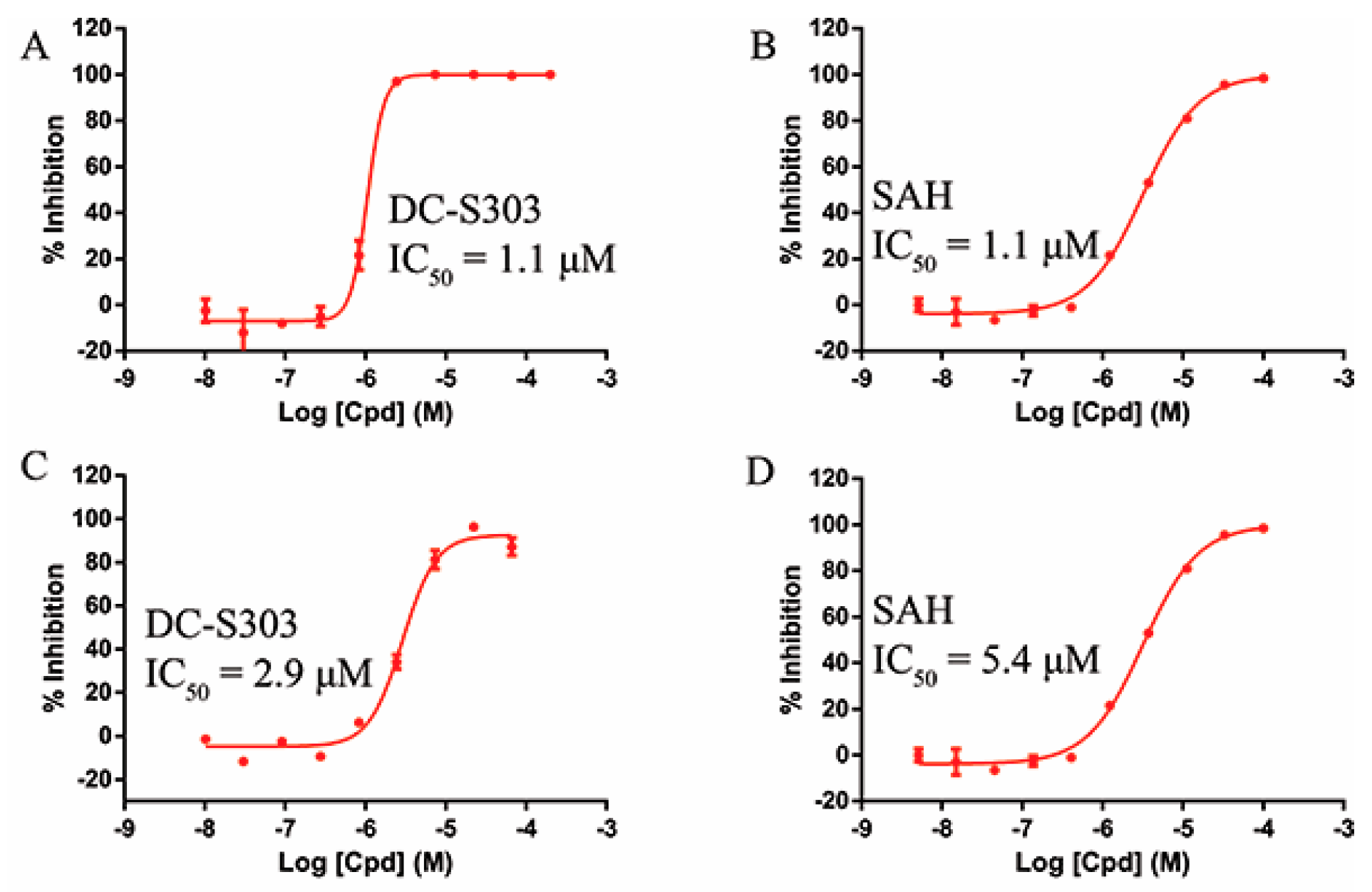
Some functional groups of DC-S285 were removed intentionally for similarity search, making it possible to explore the structure−activity relationship (SAR) of compound DC-S285 and for further chemical modifications. All the molecules were selected based on cluster analysis and selection criteria described before. The biological activity was measured with AlphaLISA assay at 100 μM and the IC50 values of the 10 compounds with inhibition rate over 50% were determined, among which DC-S303 was the most potent SET7 inhibitor with the IC50 value of 1.1 μM (Figure 5).
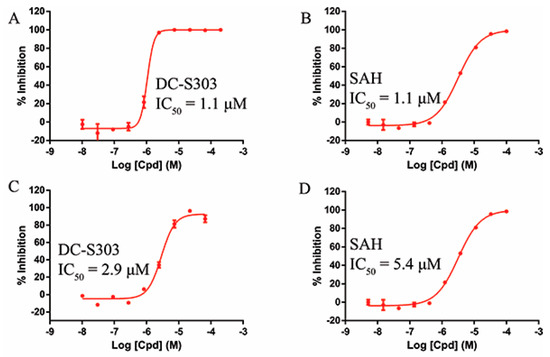
Figure 5.
Enzymatic activity of DC-S303 against SET7. (A) IC50 value of DC-S303 in AlphaLisa assay; (B) IC50 value of the reference compound SAH in AlphaLisa assay; (C) IC50 value of DC-S303 in radioactive assay; (D) IC50 value of the reference compound SAH in radioactive assay.
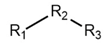
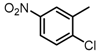
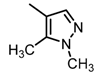
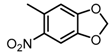
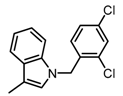
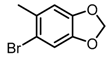
The SAR was summarized in Table 2. It can be concluded that the nitro group in R1 part is essential for activity comparing all the compounds from DC-S303 to DC-S313. If the nitro group is replaced by a chlorine atom, the activity decreases dramatically. What′s more, the meta substituent of the nitro group is more favorable (DC-S303 and DC-S304) while DC-S305 displays no activity against SET7. DC-S301 presented moderate inhibitory activity with IC50 value of13 μM, indicating the possibility that the benzene ring can be substituted by other aromatic ring with similar size. With the nitro group substituted at the meta position and chlorine atom at the ortho position, the comparison of compounds DC-S315, DC-S317, DC-S318, DC-S324, DC-S327 indicates that if the aromatic ring is not directly linked with R2 part or there is no aromatic ring linked with R2 part, the activities against SET7 decrease. DC-S314 is an exception possibly because of the flexible alkane chain meaning that it can adapt a suitable conformation to bind with SET7. Moreover, a single aromatic ring with a proper substituent will contribute to better activity. For example, if R3 is the benzene ring or a bromine substituted one, the activity is much higher than other ones (DC-S328 and DC-S333). The furan ring can contribute as an aromatic ring, but less favorable than benzene ring (DC-S329 with IC50 value = 92 μM). And it can be concluded that the diphenyl ring is the best candidate for R3 based on compound DC-S303. When R2 and R3 are fixed (from DC-S365 to DC-S364), nitro group at para position with a different R2 group from previous discussions contribute to better activity like DC-S334, but not for other substitution groups in benzene ring or aryl linkers. The rest of this table supports that the linker A is the best suitable choice.

Table 2.
Structure-Activity Relationship (SAR) of DC-S303 and its derivatives.
2.6. Selectivity of DC-S303
A qualified lead compound or chemical probe should feature not only potent binding affinity, but also goodselectivity. Considering that besides SET7, there are some other methyltransferases that share the same cofactor and similar substrate pocket, we further tested the inhibition ratios of DC-S303 against other epigenetic targets, including SETD1B, SETD8, G9a, SMYD2 and EZH2 in vitro (Table 3). The results suggested that this compound displayed moderate selectivity against epigenetic targets that underscored its value for further optimization.

Table 3.
Selectivity of DC-S303 over other epigenetic targets.
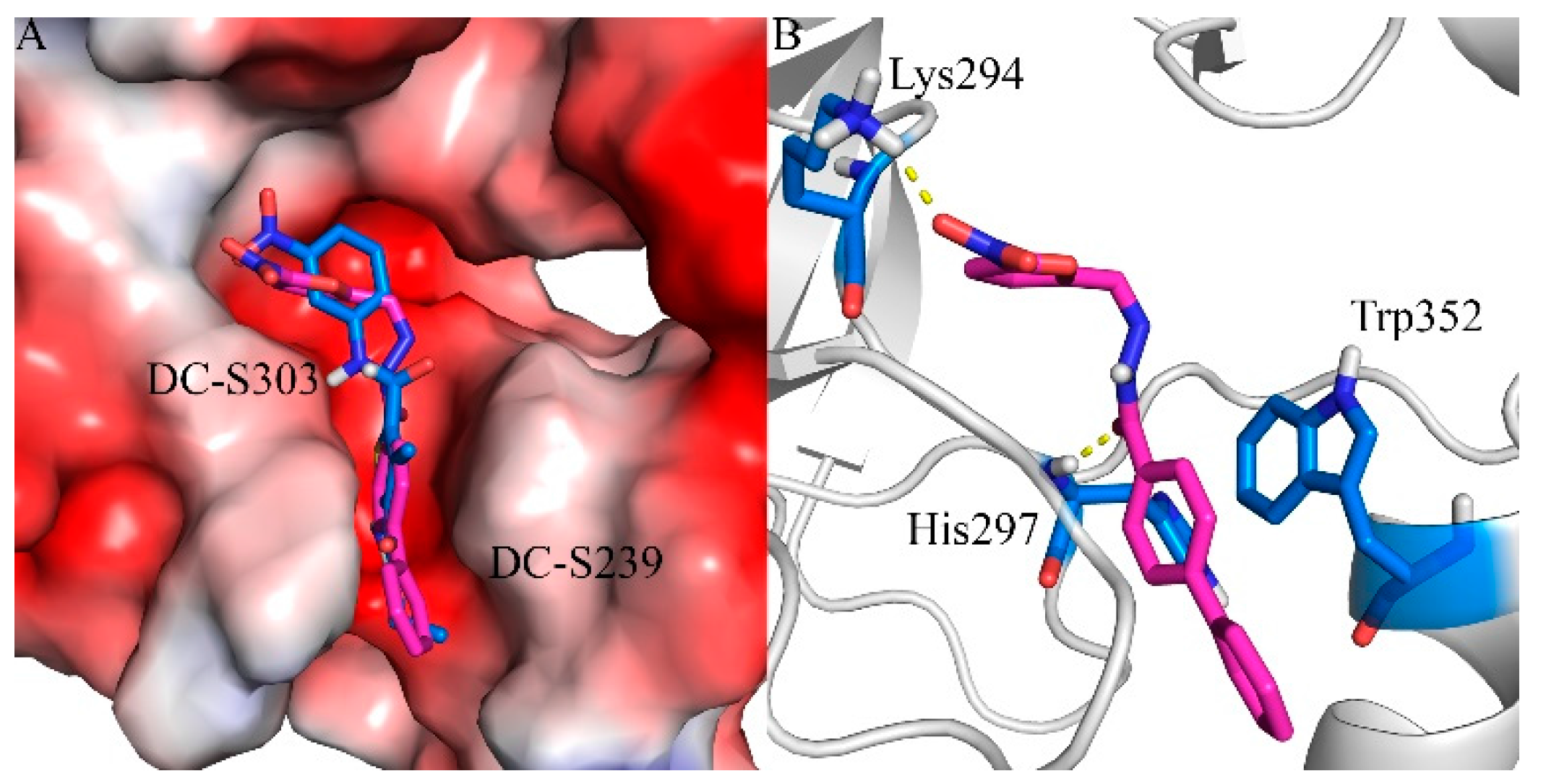
2.7. Binding Mode Prediction of DC-S303
In order to predict the putative binding mode, a docking calculation was conducted as mentioned before. The proposed binding mode (Figure 6) suggests that it shares similar binding with the previous reported compound DC-S239 at the SAM binding region. It forms a key hydrogen bond with residue Lys294, which is reported to be a potential factor for selective SET7 inhibitor design. The linking benzene of the diphenyl group forms π-π stacking interactions with Trp352, stabilizing its binding into the SAM pocket. The hydrogen bond between DC-S303 and SET7 contributes to the orientation by pulling the middle of this compound.
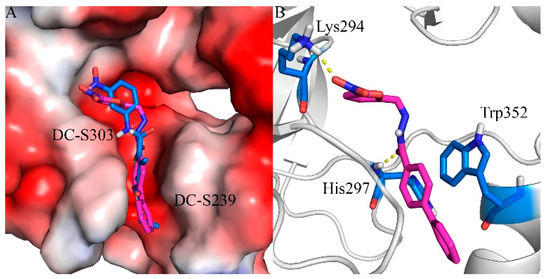
Figure 6.
Predicted binding mode of DC-S303 against SET7. (A) Binding conformation alignment of DC-S303 and DC-S239. All the compounds are shown in sticks and the protein is shown in surface; (B) Putative binging mode of DC-S239 and important polar interactions. The compound is shown in magentas sticks and key residues are displayed in blue sticks.
3. Materials and Methods
3.1. Virtual Screening: Ligand Database Preparation
The Specs commercial database, containing approximately 200,000 molecules, was filtered using Pipeline Pilot, version 7.5 (Pipeline Pilot; Accelrys Software Inc., San Diego, CA, USA) based on Lipinski′s Rule of Five [96] in order to get promising molecules with good drug-likeproperties. What′s more, the ‘pan-assay interference compounds’ (PAINS) were also removed using the substructure filter protocol developed in our lab with Pipeline Pilot, version 7.5 [97,98,99,100]. As for those molecules for docking calculations, they were subjected to LigPrep to generate all stereoisomers and different protonation states with Epik [104].
3.2. Virtual Screening: ProteinPreparation
The protein structure was processed as previously described [92]. Twenty-nine X-ray crystal structures of SET7 are available in the PDB database, and the SET domain of these structures were aligned for comparison and root mean square deviation (RMSD) calculation, which is conserved in the SET domain-containing family. We obtained a maximum root mean-square derivation (RMSD) value of 0.37, suggesting that the three-dimensional structure of the SET domain in SET7 is conserved. Considering the structure resolution and integrity, the crystal structure of SET7 complex with SAM (PDB ID: 1N6A) was chosen for docking. The protein was prepared with Protein Preparation Wizard Workflow, as provided in Maestro, with a pH value of 7.0 ± 2.0. Other parameters were set as the default.
3.3. Virtual Screening: 2D Molecular FingerprintBased Similarity Search
2D molecular fingerprint based similarity search was conducted with similarity search protocol implemented in Pipeline Pilot, version 7.5. Top-ranked 300 molecules were selected for further investigation. As for the SAR part of DC-S285, the similarity search is also performed as described in our previous published paper.
3.4. Virtual Screening: Scaffold Hopping Based Similarity Search
To get compounds with similar 3D conformations, scaffold hopping was performed with ChemMapper web server (http://lilab.ecust.edu.cn/chemmapper/) [93] against Specs molecule library. Other parameters were set as default.
3.5. SET7 Inhibition Assays
AlphaLISA assays were applied to determine the inhibitory activity targeting SET7. The purified SET7 protein was incubated in modified Tris buffer in 384-well plates (Perkin Elmer, Cat. No. 6007299, Waltham, MA, USA) at room temperature for 15 min. The compounds were transferred to the assay plate using Echo in 100% DMSO, and substrate solution was added to each well to start the reaction. Acceptor and donor beads were added and incubated for 60 min at room temperature, shielded from light. The endpoint was evaluated with EndSpire in Alpha mode. The experimental data was fitted in GraphPad Prism 5 to obtain inhibition values using the equation as follows:
As for 3Hradioactive methylation assay, SAH was used as the reference compound and the compounds were tested in 10 concentrations in duplicate in modified Tris buffer (1× assay buffer). The following materials were purchased: SET7 (Enzo, Cat. No. ALX-201-178, New York, NY, USA), [3H]-SAM (PerkinElmer Inc., Lot. No. 1790854), SAH (Sigma, Cat. No. A9384-25MG, St. Louis, MO, USA) and 384-well Flashplate (Perkin Elmer, Cat. No. SMP410A001PK,). The compounds were transferred to the assay plate by Echo 550 with 3-fold dilution in 100% DMSO where the final DMSO concentration is 1%. Then the enzyme solution was transferred to the assay plate and 1× assay buffer was transferred for low control. After 15 min incubation at room temperature, peptide and [3H]-SAM mix solution was transferred to each well to start the reaction. The cold SAM in 1× buffer was added to each well to stop the reaction after 60 min incubation at room temperature. 25 μL of the whole reaction system per well was transferred to Flashplate which was washed with dH2O and 0.1% Tween-20 for three times after 60 min incubation. The data was read on Microbeta. All the inhibition assays are performed in duplicate.
3.6. Enzymatic Selectivity Assays
For SETD1B, G9a and SMYD2, the activities against those targets were tested in modified Tris buffer (1× assay buffer). All the solid compounds were dissolved to 20 mM in 100% DMSO. The materials used were SETD1B (Active Motif, Cat. No. M1083, Carlsbad, CA, USA), G9a (BPS, Cat. No. 51001, San Diego, CA, USA), SMYD2 (Active Motif, Cat. No. 31323, c), G9a (BPS, Cat. No. 51001,) and 384-well plate (Perkin Elmer, Cat. No. 6007299). Both the enzyme solution and the substrate solution were prepared in 1× assay buffer enzyme solution or 1× assay buffer for low control was transferred to the assay plate. Then substrate mix solution was added to each well to start the solution for 15 min incubation at room temperature. For SETD1B, G9a, the incubation time was adjusted to 60 min and for SMYD2, the incubation time was set to 240 min. For detection, acceptor and donor beads were added and incubated for 60 min at room temperature, shielded from light. The endpoint was evaluated with EnSpire in Alpha mode.
The radioactive methylation inhibition assay of SETD8 was performed in modified Tris buffer. H3K27me peptide and [3H]-SAM (PerkinElmer, Cat No. NET1551MC) were added in 1x buffer as the substrate solution. The enzyme solution was incubated at room temperature for 15 min before substrate solution was added to each well to start the reaction. Cold SAM (Sigma, Cat. No. 7007-100MG) was added in 1x buffer to prepare the stop mix, and was added to stop the reaction; finally 10 μL of the reaction system was transferred to flashplate (PerkinElmer Inc., Cat. No. SMP410A001PK) and incubated at room temperature for a minimum of 2 h. The plate was washed three times with dH2O and 0.1% Tween-20, and the radioactivity signal was detected by liquid scintillation counting (MicroBeta, PerkinElmer). SAH was used as the reference compound.
The radioactive methylation inhibition assay of EZH2 was performed in modified Tris buffer. H3K27me peptide and [3H]-SAM (Perkin Elmer Inc. Waltham, MA, USA, Lot. No. 1731619) were added in 1x buffer as the substrate solution. The enz the yme solution was incubated at room temperature for 15 min before substrate solution was added to each well to start the reaction. Cold SAM (Sigma, Cat. NO. 7007) was added in 1× buffer to prepare stop mix (final concentration 0.5 mM), and was added to stop the reaction; finally 25 μL of the reaction system was transferred to a FlashPlate (Perkin Elmer, Cat. No. SMP410A001PK) and incubated at room temperature for a minimum of 1 h. The plate was washed three times with dH2O and 0.1% Tween-20, and the radioactivity was determined by liquid scintillation counting (MicroBeta). GSK-126 was used as the reference compound.
3.7. NMR Experiment
Ligand observed T1ρ NMR experiment were applied to investigate ligand-protein interactions. All NMR spectra were acquired at 25 °C on a 600 MHz Bruker Avance III spectrometer equipped with a cryogenically cooled probe (Bruker Biospin, Ettlingen, Germany). Samples containing 20 μM DC-S285, and 20 μM DC-S285 in the presence of 2.5 μM SET7 were dissolved in Tris-HCl buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.4, 5% DMSO, 95% D2O) and then used in NMR data acquisition. T1ρ spectra were recorded by using the pulse sequence of solvent-suppressed 1D 1H CPMG (cpmgPr1d). The 90° pulse length was adjusted to about 11.80 μs. A total of four dummy scans and 64 free induction decays (FIDs) were collected into 64 K acquisition points, covering a spectral width of 12 kHz (20 ppm) and giving an acquisition time (ACQ) of 2.73 s. STD data was acquired using 4 dummy scans and a relaxation delay of 3 s, followed by a 40 dB pulsed irradiation at frequency of −1.0 ppm or 33 ppm alternatively. The total acquisition time for STD spectrum was 23 min with 128 FIDs.
3.8. Cell Culture and Cell Viability Assay
MCF7, HL60, MV4-11, K562, Kasumi-1, U937, THP1, and Jurkat cell lines were purchased from American Type Culture Collection (Manassas, VA, USA). Fetal bovine serum was purchased from Life Technologies. MCF7 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum at 37 °C in an incubator with 5% CO2 atmosphere. HL60 and MV4-11 cells were cultured in 1640 medium supplemented with 10% fetal bovine serum. K562, Kasumi-1 and HL60 were cultured in 96 plate at 10,000/well for 2 h. DC-S285 was dissolved in DMSO (Sigma) and then stored at 4 °C. The cells were incubated with DC-S285 at different concentrations ranging from 0 to 100 µM for approximately 72 h. The activities of DC-S239 against MCF7, HL60, MV4-11, K562, Kasumi-1, U937, THP1, and Jurkat were measured by the alamarBlue assays and MTT assays.
4. Conclusions
As a key member of the histone methyltransferase family, SET7 has been reported to play diverse biological roles, including cell proliferation, transcriptional network regulation in embryonic stem cell, cell cycle control, protein stability, heart morphogenesis and development. The dysfunction of SET7 is involved in the pathogenesis of several diseases including alopecia areata, breast cancer, tumor and cancer progression, atherosclerosis in human carotid plaques, chronic renal diseases, diabetes, obesity, ovarian cancer, prostate cancer, hepatocellular carcinoma, and pulmonary fibrosis. Despite its controversial role in multiple diseases, the aberrant expression patterns has been observed in the onset and progression of cancers. In peripheral blood mononuclear cells of patients, the histone modification patterns were altered and the expression of SET7 was elevated [69]. Emerging evidence has also demonstrated its role in solid tumors. Zhang et al. demonstrated that SET7 interacts with transcription factor GATA1 and promotes downstream VEGF transcription and tumor angiogenesis [40]. Inhibition of SET7 activity by the SET7 inhibitor cyproheptadine reduced the estrogen receptor alpha expression in MCF7 cells that is important for cancer progression, phencopying the SET7 knockdown studies [37].
Therefore, there is urgent need to develop novel SET7 inhibitors for further detailed chemical biology investigations as well as drug design for cancer therapy.
Several attempts have been made to develop SET7 inhibitors. Both (R)-PFI-2 and cyproheptadine were reported to bind with the peptide binding site while other inhibitors lack potencyor selectivity. Recently, we have reported the first cofactor-competitive SET7 inhibitor, DC-S239, with the help of structure-based drug design methods and chemical modifications. However, the chemotypes of current SET7 inhibitors are limited and no SET7 inhibitors have progressed into clinical trials. Thus, potent, selective SET7 inhibitors with novel scaffolds still need to be developed.
In this paper, the previous identified DC-S239 was used as the starting point for scaffold hopping and 2D fingerprint based similarity search leading to the identification of DC-S285, Both radioactive methylation assays and CPMG assays validate the binding between DC-S285 and SET7. In addition, in cellular studies, DC-S285 could significantly inhibit cancer cell proliferation in a dose-dependent manner with micromolar potencies.
Then in the second round similarity search based on DC-S285, the more potent compound DC-S303 was identified with an IC50 value of 1.1 μM. In vitro selectivity profiling demonstrated its moderate selectivity against other methyltransferases. Combined with molecular docking result, we carried out the SAR study that shed light on future medicinal chemistry optimizations. This promising compound will facilitate the SET7 related biology studies and provide a good scaffold for future drug design and development.
Acknowledgments
We are extremely grateful to National Centre for Protein Science Shanghai (Shanghai Science Research Center, Protein Expression and Purification system) for their instrument support and technical assistance. We gratefully acknowledge Xiaojie Lu, Jingqiu Liu, Shien Liu, Kongkai Zhu for their support and help. We gratefully acknowledge financial support from the Ministry of Science and Technology of China (National Key R&D Program of China (2017YFB0202600 to H.D.)); the National Natural Science Foundation of China (21472208 and 81430084 to C.L.).
Author Contribution
Kai Xian Chen, Cheng Luo, Hong Ding and Fan Wang Meng conceived and designed the research; Fan Wang Meng performed the computational calculations; Hong Ding, Yu-Chih Liu, Chen Hua Zhang conducted the biological assays and the corresponding data analysis; Fu Lin Lian and Nai Xia Zhang were in charge of the NMR experiments; Cheng Luo, Hong Ding, Wen Chao Lu, Jun Chi Hu and Fan Wang Meng wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tajima, K.; Yae, T.; Javaid, S.; Tam, O.; Comaills, V.; Morris, R.; Wittner, B.S.; Liu, M.Z.; Engstrom, A.; Takahashi, F.; et al. SETD1A modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat. Commun. 2015, 6, 8257. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Hung, M.C.; Li, L.Y. EZH2: A pivotal regulator in controlling cell differentiation. Am. J. Transl. Res. 2012, 4, 364–375. [Google Scholar] [PubMed]
- Martens, M.B.; Frega, M.; Classen, J.; Epping, L.; Bijvank, E.; Benevento, M.; van Bokhoven, H.; Tiesinga, P.; Schubert, D.; Nadif Kasri, N. Euchromatin histone methyltransferase 1 regulates cortical neuronal network development. Sci. Rep. 2016, 6, 35756. [Google Scholar] [CrossRef] [PubMed]
- Herz, H.M.; Garruss, A.; Shilatifard, A. SET for life: Biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 2013, 38, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Jung, M. New lysine methyltransferase drug targets in cancer. Nat. Biotechnol. 2012, 30, 622–623. [Google Scholar] [CrossRef] [PubMed]
- Arrowsmith, C.H.; Bountra, C.; Fish, P.V.; Lee, K.; Schapira, M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 384–400. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Issa, J.P.J.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Tough, D.F.; Tak, P.P.; Tarakhovsky, A.; Prinjha, R.K. Epigenetic drug discovery: Breaking through the immune barrier. Nat. Rev. Drug Discov. 2016, 15, 835–853. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.T.; Zhang, Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011, 25, 1345–1358. [Google Scholar] [CrossRef] [PubMed]
- Keating, S.T.; El-Osta, A. Transcriptional regulation by the Set7 lysine methyltransferase. Epigenetics 2013, 8, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhou, J.; Liu, X.; Lu, D.; Shen, C.; Du, Y.; Wei, F.Z.; Song, B.; Lu, X.; Yu, Y.; et al. Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc. Natl. Acad. Sci. USA 2013, 110, 5516–5521. [Google Scholar] [CrossRef] [PubMed]
- Carr, S.M.; Munro, S.; Kessler, B.; Oppermann, U.; La Thangue, N.B. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. Embo J. 2011, 30, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.; Carr, S.M.; La Thangue, N.B. Diversity within the pRb pathway: Is there a code of conduct? Oncogene 2012, 31, 4343–4352. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, Q.; Lu, Q.; Jia, Z.; Chen, P.; Ma, K.; Wang, W.; Zhou, C. ISL-1 promotes pancreatic islet cell proliferation by forming an ISL-1/Set7/9/PDX-1 complex. Cell Cycle 2015, 14, 3820–3829. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Son, M.J.; Kim, W.K.; Oh, K.J.; Park, A.; Lee da, S.; Han, B.S.; Lee, S.C.; Bae, K.H. Methyltransferase and demethylase profiling studies during brown adipocyte differentiation. BMB Rep. 2016, 49, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Tuano, N.K.; Okabe, J.; Ziemann, M.; Cooper, M.E.; El-Osta, A. Set7 mediated interactions regulate transcriptional networks in embryonic stem cells. Nucleic Acids Res. 2016, 44, 9206–9217. [Google Scholar] [CrossRef] [PubMed]
- Castano, J.; Morera, C.; Sese, B.; Boue, S.; Bonet-Costa, C.; Marti, M.; Roque, A.; Jordan, A.; Barrero, M.J. SETD7 Regulates the Differentiation of Human Embryonic Stem Cells. PLoS ONE 2016, 11, e0149502. [Google Scholar] [CrossRef] [PubMed]
- Carr, S.M.; La Thangue, N.B. Cell cycle control by a methylation-phosphorylation switch. Cell Cycle 2011, 10, 733–734. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gaughan, L.; Stockley, J.; Wang, N.; McCracken, S.R.; Treumann, A.; Armstrong, K.; Shaheen, F.; Watt, K.; McEwan, I.J.; Wang, C.; et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res. 2011, 39, 1266–1279. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Geng, H.; Xue, C.H.; Beer, T.M.; Qian, D.Z. Functional regulation of hypoxia inducible factor-1 alpha by SET9 lysine methyltransferase. Bba-Mol. Cell Res. 2015, 1853, 881–891. [Google Scholar]
- Yang, J.; Huang, J.; Dasgupta, M.; Sears, N.; Miyagi, M.; Wang, B.; Chance, M.R.; Chen, X.; Du, Y.; Wang, Y.; et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 21499–21504. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.S.; Suzuki, T.; Dohmae, N.; Hayami, S.; Unoki, M.; Yoshimatsu, M.; Toyokawa, G.; Takawa, M.; Chen, T.; Kurash, J.K.; et al. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011, 71, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.D.; Kim, E.; Koun, S.; Ham, H.J.; Rhee, M.; Kim, M.J.; Huh, T.L. Proper Activity of Histone H3 Lysine 4 (H3K4) Methyltransferase Is Required for Morphogenesis during Zebrafish Cardiogenesis. Mol. Cells 2015, 38, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.; Vousden, K.H. p53 mutations in cancer. Nat. Cell Biol. 2013, 15, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Toledo, F.; Wahl, G.M. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 2006, 6, 909–923. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cao, R.; Xia, L.; Erdjument-Bromage, H.; Borchers, C.; Tempst, P.; Zhang, Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 2001, 8, 1207–1217. [Google Scholar] [CrossRef]
- Nishioka, K.; Chuikov, S.; Sarma, K.; Erdjument-Bromage, H.; Allis, C.D.; Tempst, P.; Reinberg, D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Gene Dev. 2002, 16, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Dhayalan, A.; Kudithipudi, S.; Rathert, P.; Jeltsch, A. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem. Biol. 2011, 18, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kassner, I.; Barandun, M.; Fey, M.; Rosenthal, F.; Hottiger, M.O. Crosstalk between SET7/9-dependent methylation and ARTD1-mediated ADP-ribosylation of histone H1.4. Epigenet. Chromatin 2013, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Kassner, I.; Andersson, A.; Fey, M.; Tomas, M.; Ferrando-May, E.; Hottiger, M.O. SET7/9-dependent methylation of ARTD1 at K508 stimulates poly-ADP-ribose formation after oxidative stress. Open Biol. 2013, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Oppenheimer, H.; Kumar, A.; Meir, H.; Schwartz, I.; Zini, A.; Haze, A.; Kandel, L.; Mattan, Y.; Liebergall, M.; Dvir-Ginzberg, M. Set7/9 Impacts COL2A1 Expression Through Binding and Repression of SirT1 Histone Deacetylation. J. Bone Miner. Res. 2014, 29, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Esteve, P.O.; Chin, H.G.; Benner, J.; Feehery, G.R.; Samaranayake, M.; Horwitz, G.A.; Jacobsen, S.E.; Pradhan, S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. USA 2009, 106, 5076–5081. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hevi, S.; Kurash, J.K.; Lei, H.; Gay, F.; Bajko, J.; Su, H.; Sun, W.T.; Chang, H.; Xu, G.L.; et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009, 41, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Lezina, L.; Aksenova, V.; Ivanova, T.; Purmessur, N.; Antonov, A.V.; Tentler, D.; Fedorova, O.; Garabadgiu, A.V.; Talianidis, I.; Melino, G.; et al. KMTase Set7/9 is a critical regulator of E2F1 activity upon genotoxic stress. Cell Death Differ. 2014, 21, 1889–1899. [Google Scholar] [CrossRef] [PubMed]
- Kontaki, H.; Talianidis, I. Lysine methylation regulates E2F1-induced cell death. Mol. Cell 2010, 39, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, K.; Jia, D.; Kapoor-Vazirani, P.; Powell, D.R.; Collins, R.E.; Sharma, D.; Peng, J.M.; Cheng, X.D.; Vertino, P.M. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell 2008, 30, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Calnan, D.R.; Webb, A.E.; White, J.L.; Stowe, T.R.; Goswami, T.; Shi, X.; Espejo, A.; Bedford, M.T.; Gozani, O.; Gygi, S.P.; et al. Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging (Albany NY) 2012, 4, 462–479. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniyan, N.; Ananthanarayanan, M.; Suchy, F.J. Direct methylation of FXR by Set7/9, a lysine methyltransferase, regulates the expression of FXR target genes. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G937–G947. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, J.; Lin, J.; Zhou, L.; Song, Y.; Wei, B.; Luo, X.; Chen, Z.; Chen, Y.; Xiong, J.; et al. The transcription factor GATA1 and the histone methyltransferase SET7 interact to promote VEGF-mediated angiogenesis and tumor growth and predict clinical outcome of breast cancer. Oncotarget 2016, 7, 9859–9875. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Wu, H.; Cheng, S.Y.; Gao, D.; Zhang, L.; Zhao, Y. Set7 mediated Gli3 methylation plays a positive role in the activation of Sonic Hedgehog pathway in mammals. eLife 2016, 5, e15690. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, Z.; Xu, C.; Leng, X.; Cao, H.; Ouyang, G.; Xiao, W. Repression of hypoxia-inducible factor alpha signaling by Set7-mediated methylation. Nucleic Acids Res. 2015, 43, 5081–5098. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Kim, K.I. Regulation of HIF-1 alpha stability by lysine methylation. BMB Rep. 2016, 49, 245–246. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Han, Q.; Nie, J.; Cao, X.; Chen, Z.; Yin, S.; Gao, Y.; Lin, F.; Zhou, X.; Xu, K.; et al. Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation. J. Biol. Chem. 2013, 288, 35093–35103. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Gao, D.; Zhang, W.; Liu, S.; Yang, S.; Li, X. Puerarin suppresses high glucose-induced MCP-1 expression via modulating histone methylation in cultured endothelial cells. Life Sci. 2015, 130, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, C.; Li, X.; Cui, P.; Li, Q.; Guo, Q.; Han, H.; Liu, S.; Sun, G. Involvement of Histone Lysine Methylation in p21 Gene Expression in Rat Kidney In Vivo and Rat Mesangial Cells In Vitro under Diabetic Conditions. J. Diabetes Res. 2016, 2016, 3853242. [Google Scholar] [CrossRef] [PubMed]
- Brasacchio, D.; Okabe, J.; Tikellis, C.; Balcerczyk, A.; George, P.; Baker, E.K.; Calkin, A.C.; Brownlee, M.; Cooper, M.E.; El-Osta, A. Hyperglycemia Induces a Dynamic Cooperativity of Histone Methylase and Demethylase Enzymes Associated with Gene-Activating Epigenetic Marks That Coexist on the Lysine Tail. Diabetes 2009, 58, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Ea, C.K.; Baltimore, D. Regulation of NF-kappa B activity through lysine monomethylation of p65. Proc. Natl. Acad. Sci. USA 2009, 106, 18972–18977. [Google Scholar] [CrossRef] [PubMed]
- Masatsugu, T.; Yamamoto, K. Multiple lysine methylation of PCAF by Set9 methyltransferase. Biochem Biophys. Res. Commun. 2009, 381, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Spaeth, J.M.; Walker, E.M.; Stein, R. Impact of Pdx1-associated chromatin modifiers on islet beta-cells. Diabetes Obes. Metab. 2016, 18 (Suppl. 1), 123–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shi, Q.; Guo, T.; Yang, Z.; Jia, Z.; Chen, P.; Zhou, C. PDX1 and ISL1 differentially coordinate with epigenetic modifications to regulate insulin gene expression in varied glucose concentrations. Mol. Cell. Endocrinol. 2016, 428, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Maganti, A.V.; Maier, B.; Tersey, S.A.; Sampley, M.L.; Mosley, A.L.; Ozcan, S.; Pachaiyappan, B.; Woster, P.M.; Hunter, C.S.; Stein, R.; et al. Transcriptional Activity of the Islet beta Cell Factor Pdx1 Is Augmented by Lysine Methylation Catalyzed by the Methyltransferase Set7/9. J. Biol. Chem. 2015, 290, 9812–9822. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Li, S.D.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.H.; et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1 alpha. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Huq, M.D.; Tsai, N.P.; Khan, S.A.; Wei, L.N. Lysine trimethylation of retinoic acid receptor-alpha: A novel means to regulate receptor function. Mol. Cell. Proteom. 2007, 6, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, Y.; Gao, Y.; Geng, P.; Lu, Y.; Liu, X.; Yao, R.; Hou, P.; Liu, D.; Lu, J.; et al. JMJD3 promotes SAHF formation in senescent WI38 cells by triggering an interplay between demethylation and phosphorylation of RB protein. Cell Death Differ. 2015, 22, 1630–1640. [Google Scholar] [CrossRef] [PubMed]
- He, S.Y.; Owen, D.R.; Jelinsky, S.A.; Lin, L.L. Lysine Methyltransferase SETD7 (SET7/9) Regulates ROS Signaling through mitochondria and NFE2L2/ARE pathway. Sci. Rep.-UK 2015, 5, 14368. [Google Scholar] [CrossRef] [PubMed]
- Elkouris, M.; Kontaki, H.; Stavropoulos, A.; Antonoglou, A.; Nikolaou, K.C.; Samiotaki, M.; Szantai, E.; Saviolaki, D.; Brown, P.J.; Sideras, P.; et al. SET9-Mediated Regulation of TGF-beta Signaling Links Protein Methylation to Pulmonary Fibrosis. Cell Rep. 2016, 15, 2733–2744. [Google Scholar] [CrossRef] [PubMed]
- Vasileva, E.; Fedorova, O.; Shuvalov, O.; Daks, A.; Petukhov, A.; Barlev, N. The role of methyltransferase Set7/9 interaction with RNA-binding protein Sam68. Int. J. Mol. Med. 2015, 36, S46. [Google Scholar]
- Liu, X.Y.; Wang, D.L.; Zhao, Y.; Tu, B.; Zheng, Z.X.; Wang, L.N.; Wang, H.Y.; Gu, W.; Roeder, R.G.; Zhu, W.G. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1). Proc. Natl. Acad. Sci. USA 2011, 108, 1925–1930. [Google Scholar] [CrossRef] [PubMed]
- Albacker, C.E.; Storer, N.Y.; Langdon, E.M.; DiBiase, A.; Zhou, Y.; Langenau, D.M.; Zon, L.I. The Histone Methyltransferase SUV39H1 Suppresses Embryonal Rhabdomyosarcoma Formation in Zebrafish. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Couture, J.F.; Collazo, E.; Hauk, G.; Trievel, R.C. Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 2006, 13, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Kouskouti, A.; Scheer, E.; Staub, A.; Tora, L.; Talianidis, I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell 2004, 14, 175–182. [Google Scholar] [CrossRef]
- Pagans, S.; Kauder, S.E.; Kaehlcke, K.; Sakane, N.; Schroeder, S.; Dormeyer, W.; Trievel, R.C.; Verdin, E.; Schnolzer, M.; Ott, M. The Cellular Lysine Methyltransferase Set7/9-KMT7 Binds HIV-1 TAR RNA, Monomethylates the Viral Transactivator Tat, and Enhances HIV Transcription. Cell Host Microbe 2010, 7, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.Y.; Li, X.X.; Han, H.B.; Li, C.Y.; Liu, S.J.; Gao, W.H.; Sun, G.D. Histone Lysine Methylation in TGF-beta 1 Mediated p21 Gene Expression in Rat Mesangial Cells. Biomed. Res. Int. 2016, 2016, 6927234. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Doi, S.; Nakashima, A.; Irifuku, T.; Yamada, K.; Kokoroishi, K.; Ueno, T.; Doi, T.; Hida, E.; Arihiro, K.; et al. Inhibition of SET Domain-Containing Lysine Methyltransferase 7/9 Ameliorates Renal Fibrosis. J. Am. Soc. Nephrol. 2016, 27, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Madapura, M.P.; Bhat, U.A.; Rao, M.R.S. Mapping of Post-translational Modifications of Transition Proteins, TP1 and TP2, and Identification of Protein Arginine Methyltransferase 4 and Lysine Methyltransferase 7 as Methyltransferase for TP2. J. Biol. Chem. 2015, 290, 12101–12122. [Google Scholar] [CrossRef] [PubMed]
- Oudhoff, M.J.; Braam, M.J.S.; Freeman, S.A.; Wong, D.; Rattray, D.G.; Wang, J.; Antignano, F.; Snyder, K.; Refaeli, I.; Hughes, M.R.; et al. SETD7 Controls Intestinal Regeneration and Tumorigenesis by Regulating Wnt/beta-Catenin and Hippo/YAP Signaling. Dev. Cell 2016, 37, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Wu, X.N.; Shi, T.T.; Xu, H.T.; Yi, J.; Shen, H.F.; Huang, M.F.; Shu, X.Y.; Wang, F.F.; Peng, B.L.; et al. Regulation of Transcription Factor Yin Yang 1 by SET7/9-mediated Lysine Methylation. Sci. Rep.-UK 2016, 6, 21718. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liang, G.; Wu, X.; Wang, S.; Zhang, P.; Su, Y.; Yin, H.; Tan, Y.; Zhang, J.; Lu, Q. Abnormal epigenetic modifications in peripheral blood mononuclear cells from patients with alopecia areata. Br. J. Dermatol. 2012, 166, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.C.; Zhang, J.L.; Tian, T.; Fu, X.; Wang, W.J.; Li, S.N.; Shi, T.T.; Suo, A.L.; Ruan, Z.P.; Guo, H.; et al. SET7/9 inhibits oncogenic activities through regulation of Gli-1 expression in breast cancer. Tumor Biol. 2016, 37, 9311–9322. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Yang, S.S.; Hu, J.W.; Yu, C.Q.; He, M.X.; Cai, Z.L. Increased Expression of SETD7 Promotes Cell Proliferation by Regulating Cell Cycle and Indicates Poor Prognosis in Hepatocellular Carcinoma. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Takeuchi, T.; Nagayama, T.; Furihata, M. T-Cadherin Modulates Tumor-Associated Molecules in Gallbladder Cancer Cells. Cancer Investig. 2010, 28, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, Y.; Koda, Y.; Byeon, S.J.; Shimada, S.; Nishikawaji, T.; Sakamoto, A.; Chen, Y.X.; Kojima, K.; Kawano, T.; Eishi, Y.; et al. Reduced expression of SET7/9, a histone mono-methyltransferase, is associated with gastric cancer progression. Oncotarget 2016, 7, 3966–3983. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Nam, H.J.; Lee, J.; Park, D.Y.; Kim, C.; Yu, Y.S.; Kim, D.; Park, S.W.; Bhin, J.; Hwang, D.; et al. Methylation-dependent regulation of HIF-1 alpha stability restricts retinal and tumour angiogenesis. Nat. Commun. 2016, 7, 10347. [Google Scholar] [CrossRef] [PubMed]
- Lezina, L.; Aksenova, V.; Fedorova, O.; Malikova, D.; Shuvalov, O.; Antonov, A.V.; Tentler, D.; Garabadgiu, A.V.; Melino, G.; Barlev, N.A. KMT Set7/9 affects genotoxic stress response via the Mdm2 axis. Oncotarget 2015, 6, 25843–25855. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.C.; Wang, D.L.; Liu, X.Y.; Gu, B.; Du, Y.P.; Wei, F.Z.; Cao, L.L.; Song, B.Y.; Lu, X.P.; Yang, Q.Y.; et al. SET7/9 regulates cancer cell proliferation by influencing beta-catenin stability. Faseb J. 2015, 29, 4313–4323. [Google Scholar] [CrossRef] [PubMed]
- Greissel, A.; Culmes, M.; Burgkart, R.; Zimmermann, A.; Eckstein, H.H.; Zernecke, A.; Pelisek, J. Histone acetylation and methylation significantly change with severity of atherosclerosis in human carotid plaques. Cardiovasc. Pathol. 2016, 25, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Reddy, M.A.; Deshpande, S.; Jia, Y.; Park, J.T.; Lanting, L.L.; Jin, W.; Kato, M.; Xu, Z.G.; Das, S.; et al. Epigenetic Histone Modifications Involved in Profibrotic Gene Regulation by 12/15-Lipoxygenase and Its Oxidized Lipid Products in Diabetic Nephropathy. Antioxid. Redox. Signal. 2016, 24, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Paneni, F.; Costantino, S.; Battista, R.; Castello, L.; Capretti, G.; Chiandotto, S.; Scavone, G.; Villano, A.; Pitocco, D.; Lanza, G.; Volpe, M.; et al. Adverse epigenetic signatures by histone methyltransferase Set7 contribute to vascular dysfunction in patients with type 2 diabetes mellitus. Circ. Cardiovasc. Genet. 2015, 8, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Okabe, J.; Orlowski, C.; Balcerczyk, A.; Tikellis, C.; Thomas, M.C.; Cooper, M.E.; El-Osta, A. Distinguishing Hyperglycemic Changes by Set7 in Vascular Endothelial Cells. Circ. Res. 2012, 110, 1067–1121. [Google Scholar] [CrossRef] [PubMed]
- Goru, S.K.; Kadakol, A.; Pandey, A.; Malek, V.; Sharma, N.; Gaikwad, A.B. Histone H2AK119 and H2BK120 mono-ubiquitination modulate SET7/9 and SUV39H1 in type 1 diabetes-induced renal fibrosis. Biochem. J. 2016, 473, 3937–3949. [Google Scholar] [CrossRef] [PubMed]
- Costantino, S.; Paneni, F.; Virdis, A.; Volpe, M.; Taddei, S.; Cosentino, F. Chromatin changes by methyltransferase Set7 induce inflammatory adhesion molecules and endothelial dysfunction in small visceral arteries from insulin resistant subjects. Eur. Heart J. 2015, 36, 112–113. [Google Scholar]
- Ciccarelli, M.; Vastolo, V.; Albano, L.; Lecce, M.; Cabaro, S.; Liotti, A.; Longo, M.; Oriente, F.; Russo, G.L.; Macchia, P.E.; et al. Glucose-induced expression of the homeotic transcription factor Prep1 is associated with histone post-translational modifications in skeletal muscle. Diabetologia 2016, 59, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Al-Haddad, R.; Karnib, N.; Assaad, R.A.; Bilen, Y.; Emmanuel, N.; Ghanem, A.; Younes, J.; Zibara, V.; Stephan, J.S.; Sleiman, S.F. Epigenetic changes in diabetes. Neurosci. Lett. 2016, 625, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Son, M.J.; Kim, W.K.; Park, A.; Oh, K.J.; Kim, J.H.; Han, B.S.; Kim, I.C.; Chi, S.W.; Park, S.G.; Lee, S.C.; et al. Set7/9, a methyltransferase, regulates the thermogenic program during brown adipocyte differentiation through the modulation of p53 acetylation. Mol. Cell. Endocrinol. 2016, 431, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xie, M.; Shi, Y.; Luo, B.H.; Gong, G.H.; Li, J.N.; Wang, J.P.; Zhao, W.J.; Zi, Y.; Wu, X.Y.; et al. MicroRNA-153 functions as a tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer cells. Oncol. Rep. 2015, 34, 111–120. [Google Scholar] [CrossRef] [PubMed]
- D’Adamo, C.R.; D’Urso, A.; Ryan, K.A.; Yerges-Armstrong, L.M.; Semba, R.D.; Steinle, N.I.; Mitchell, B.D.; Shuldiner, A.R.; McArdle, P.F. A Common Variant in the SETD7 Gene Predicts Serum Lycopene Concentrations. Nutrients 2016, 8, 82. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Wan, Y.S.; Wang, J.; Zhao, P.; Yuan, Y.; Wang, L.; She, Y.L.; Broering, R.; Lu, M.J.; Ye, L.B.; et al. Set7 Facilitates Hepatitis C Virus Replication via Enzymatic Activity-Dependent Attenuation of the IFN-Related Pathway. J. Immunol. 2015, 194, 2757–2768. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Ramage, H.; Boehm, D.; Dirk, L.M.A.; Sakane, N.; Hanada, K.; Pagans, S.; Kaehlcke, K.; Aull, K.; Weinberger, L.; et al. The HIV-1 Tat Protein Is Monomethylated at Lysine 71 by the Lysine Methyltransferase KMT7. J. Biol. Chem. 2016, 291, 16240–16248. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.H.; Peng, K.L.; Jhang, H.C.; Lin, C.H.; Wu, S.Y.; Chiang, C.M.; Lee, S.C.; Yu, W.C.Y.; Juan, L.J. The HPV E6 oncoprotein targets histone methyltransferases for modulating specific gene transcription. Oncogene 2012, 31, 2335–2349. [Google Scholar] [CrossRef] [PubMed]
- Barsyte-Lovejoy, D.; Li, F.L.; Oudhoff, M.J.; Tatlock, J.H.; Dong, A.P.; Zeng, H.; Wu, H.; Freeman, S.A.; Schapira, M.; Senisterra, G.A.; et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc. Natl. Acad. Sci. USA 2014, 111, 12853–12858. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.W.; Cheng, S.F.; Ding, H.; Liu, S.; Liu, Y.; Zhu, K.K.; Chen, S.J.; Lu, J.Y.; Xie, Y.Q.; Li, L.J.; et al. Selective Histone Methyltransferase SET7 Inhibitors by Pharmacophore- and Docking-Based Virtual Screening. J. Med. Chem. 2015, 58, 8166–8181. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.Y.; Cai, C.Q.; Liu, X.F.; Ku, X.; Jiang, H.L.; Gao, D.Q.; Li, H.L. ChemMapper: A versatile web server for exploring pharmacology and chemical structure association based on molecular 3D similarity method. Bioinformatics 2013, 29, 1827–1829. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.P.; Hersey, A.; Montanari, D.; Overington, J. Probing the links between in vitro potency, ADMET and physicochemical parameters. Nat. Rev. Drug Discov. 2011, 10, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Van de Waterbeemd, H.; Gifford, E. ADMET in silico modelling: Towards prediction paradise? Nat. Rev. Drug Discov. 2003, 2, 192–204. [Google Scholar] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [PubMed]
- Whitty, A. Growing PAINS in academic drug discovery. Future Med. Chem. 2011, 3, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.; Walters, M.A. Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B. Observations on screening-based research and some concerning trends in the literature. Future Med. Chem. 2010, 2, 1529–1546. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pK (a) prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).