Simple Summary
Digital transcriptomics is rapidly emerging as a powerful new technology for modelling the environmental dynamics of the adaptive landscape in diverse lineages. This is particularly valuable in taxa such as turtles and tortoises (order Testudines) which contain a large fraction of endangered species at risk due to anthropogenic impacts on the environment, including pollution, overharvest, habitat degradation, and climate change. Sea turtles (family Cheloniidae) in particular invite a genomics-enabled approach to investigating their remarkable portfolio of adaptive evolution. Our de novo transcriptome assemblies and measurements of tissue- and temperature- specific global gene expression in the loggerhead sea turtle (Caretta caretta) reveal the genomic basis for potential resiliency in this endangered flagship species, and are crucial to future management and conservation strategies with attention to changing climates. We summarize the interactions among differentially expressed genes by producing network visualizations, and highlight the shared biological pathways related to development, migration, immunity, and longevity reported in the avian and reptilian literature. Our original results for loggerhead sea turtles provide a large, new comparative genomic resource for the investigation of genotype–phenotype relationships in amniotes.
Abstract
Background: Digital transcriptomics is rapidly emerging as a powerful new technology for modelling the environmental dynamics of the adaptive landscape in diverse lineages. This is particularly valuable in taxa such as turtles and tortoises (order Testudines) which contain a large fraction of endangered species at risk due to anthropogenic impacts on the environment, including pollution, overharvest, habitat degradation, and climate change. Sea turtles (family Cheloniidae) in particular invite a genomics-enabled approach to investigating their remarkable portfolio of adaptive evolution. The sex of the endangered loggerhead sea turtle (Caretta caretta) is subject to temperature-dependent sex determination (TSD), a mechanism by which exposure to temperatures during embryonic development irreversibly determines sex. Higher temperatures produce mainly female turtles and lower temperatures produce mainly male turtles. Incubation temperature can have long term effects on the immunity, migratory ability, and ultimately longevity of hatchlings. We perform RNA-seq differential expression analysis to investigate tissue- and temperature-specific gene expression within brain (n = 7) and gonadal (n = 4) tissue of male and female loggerhead hatchlings. Results: We assemble tissue- and temperature-specific transcriptomes and identify differentially expressed genes relevant to sexual development and life history traits of broad adaptive interest to turtles and other amniotic species. We summarize interactions among differentially expressed genes by producing network visualizations, and highlight shared biological pathways related to migration, immunity, and longevity reported in the avian and reptile literature. Conclusions: The measurement of tissue- and temperature-specific global gene expression of an endangered, flagship species such as the loggerhead sea turtle (Caretta caretta) reveals the genomic basis for potential resiliency and is crucial to future management and conservation strategies with attention to changing climates. Brain and gonadal tissue collected from experimentally reared loggerhead male and female hatchlings comprise an exceedingly rare dataset that permits the identification of genes enriched in functions related to sexual development, immunity, longevity, and migratory behavior and will serve as a large, new genomic resource for the investigation of genotype–phenotype relationships in amniotes.
1. Introduction
Turtles and tortoises (Testudines) include some of the most endangered groups of vertebrates worldwide, yet have exhibited a remarkable resiliency in their long-term survival and adaptability to environmental change. New technologies for quantifying the differential gene expression in total cellular RNA are allowing us to more precisely model the dynamics of the turtle genotype–phenotype relationship and develop more predictive, genomics-enabled approaches to studies of turtle adaptive evolution and species conservation. Sea turtles (Cheloniidae) in particular have evolved a marine life history that includes an astonishing array of adaptive traits of broad interest to organismal biologists. As hatchlings, loggerhead sea turtles (Caretta caretta) emerge from their nests and enter the ocean, where they will forage as juveniles until maturing into bottom-feeding sub-adults and adults [1]. Adult loggerheads successfully navigate long distances between foraging habitats, wintering sites, and natal beaches to mate, with notable fidelity to migratory routes [2,3,4]. The large geographical range of the loggerhead sea turtle contributes to the species’ vulnerability to fisheries bycatch and habitat degradation. Loggerheads are classified as a vulnerable species with a decreasing population trend by the IUCN Red List (2017).
Loggerheads exhibit temperature-dependent sex determination (TSD), in which incubation temperature influences the primary sex ratio of developing offspring. For loggerheads, temperatures below the pivotal temperature of approximately 29 °C in the middle third of incubation produce mainly male hatchlings, whereas temperatures above 29 °C produce mainly female hatchlings [5,6,7]. With warming climates, there is a risk of skewed sex ratios in loggerhead populations which may negatively impact the persistence of global populations [8]. Elevated incubation temperature has been shown to negatively affect immune function, locomotion, and ultimately survival in turtles [9,10,11,12]. Phenotypic plasticity in nesting phenology may combat short-term effects of environmental warming, but continued adaptation of loggerhead populations to shifting thermal conditions may not be sustainable in the long-term.
Previous studies in turtles and other reptiles have identified differentially expressed genes at female and male promoting temperatures [13,14,15,16], and enrichment of sex-specific and heat shock proteins were detected recently in loggerheads [17,18]. We present a summary of the application of an RNA-seq analysis pipeline to a rare dataset consisting of brain and gonadal tissue from experimentally reared female and male loggerhead hatchlings. Through enrichment analyses, we identify biological processes and significantly differentially expressed genes potentially relevant to sexual development, migration, longevity, and immunity within our study against the reptile and avian literature. An improved understanding of differential gene expression at a critical developmental period in loggerhead sea turtles may reveal genes that may contain genetic variation important to adaptive potential in changing environments thereby extending the predictive value of genomic resources for managing loggerheads [19].
2. Methods
2.1. Hatchling Sample Collection and RNA Extraction
A visual summary of our sampling design and analytical workflow to identify significantly differentially expressed genes related to TSD, migration, and longevity via genome-wide transcriptomic analysis of loggerhead brain and gonadal tissue from male and female turtles is displayed in Supplemental file S6.
A total of 11 C. caretta hatchlings were used in our transcriptomics study, which were originally used as part of a pivotal temperature experiment from the Northern Recovery Unit of the Southeastern United States [20]. In the summer of 2011, 7 eggs from which brain tissue would be harvested were salvaged from the same nest, and similarly, 4 eggs from which gonadal tissue would be harvested were collected from the same nest in the summer of 2013. We were unable to collect brain and gonadal tissue from the same animals because other samples had been assigned to different collaborative studies [21,22,23] to maximize the information derived from these rare samples. The eggs were collected from “doomed” nests that would not have been successful if left in place due to erosion and or predation. We followed the accepted protocols of the American Veterinary Medical Association (AVMA) Guidelines on Euthanasia (June 2007 edition, current at the same of sample collection and consistent with the 2020 edition), and used decapitation followed immediately by pithing to euthanize hatchlings. Full permit information is listed in the Acknowledgements.
Salvaged eggs were transported <12 h after laying for laboratory incubation at the NCSU Center for Marine Sciences and Technology (CMAST) and reared in temperature-controlled incubators above and below 29 °C for expected female and male development, respectively [20]. From the total sample of 11 individuals, brain tissue was collected from 4 male and 3 female hatchlings, and gonad tissue was collected from 2 male and 2 female hatchlings. Sex was confirmed histologically at CMAST by previously published methods [20,21,22,23,24].
Brain and gonad tissue were immediately placed into RNAlater reagent (Qiagen, Inc., Germantown, MD, USA) for high quality RNA stabilization after dissection. Tissues were homogenized and purified, and cDNA was extracted using Qiagen TissueRuptor, RNeasy, and Omniscript Reverse Transcription (RT) kits. RNA samples were quantified via an Agilent Bioanalyzer prior to Illumina TruSeq library preparation. Brain samples were single-end sequenced on an Illumina HiSeq 2000 (1 × 100 bases) and gonad samples were paired-end sequenced on an Illumina HiSeq 2500 (2 × 100 bases) at the Medical University of South Carolina (MUSC) Center for Genomic Medicine.
2.2. De Novo Transcriptome Assembly
The quality of RNA sequenced ends was assessed using FastQC (http://www.bioin-formatics.babraham.ac.uk/projects/fastqc/, accessed on 19 October 2021), and low-quality reads were discarded. Reads were then trimmed via Trimmomatic (0.32) (-phred33) with LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, and MINLEN:36 for brain and MINLEN:70 for gonads [25]. De novo transcriptome assembly for brain and gonad reads was completed using the Trinity software package (r20140717) using separate references for brain and gonads with default parameters and a minimum contig length of 400 [26], resulting in one brain and one gonad assembly.
2.3. Differential Gene Expression and Enrichment Analyses
Differential gene expression in brain and gonad tissues among male and female log-gerheads were evaluated using the RSEM-EBSeq pipeline [27,28]. Transcript reads were provided to RSEM (1.2.18) and aligned to the transcriptome assembly via Bowtie2 (--bowtie2) using default parameters [27,29].
Clustered heatmaps were constructed via the analyze_diff_expr.pl script (-P 0.001 -C 2 –max_DE_genes_per_comparison 50) from Trinity. Transcripts were annotated via NCBI BLASTX (2.2.31) using non-redundant proteins from vertebrates with -max_target_seqs 3 and -evalue 0.0001, and significantly differentially expressed transcripts (q-value < 0.05, in which q-value refers to FDR corrected p-values) were retained.
Gene ontology (GO) enrichment was performed via OmicsBox (1.2.4) [30]. To perform enrichment analysis with OmicsBox, Blastx-fast was executed through CloudBlast with default parameters using non-redundant protein sequences (nr_v5). To further functionally characterize the protein families, domains, and sites associated with the assembled sequences, InterProScan was run within OmicsBox, enabling by default the use of ProfileScan, HMMSmart, HMMTigr, Gene3D, SLFD, SuperFamily, and MobiDBLite databases. Following annotation, Fisher’s Exact Test (FDR < 0.05) was implemented to identify significantly enriched terms with default OmicsBox settings.
Significant GO terms (FDR < 0.05 and e-value < 1.0 × 10−6) were retrieved and visualized via REViGO with default settings (allowed similarity = medium; 0.70) [31]. To functionally cluster transcript annotations, DAVID (6.8) was used on significantly differentially expressed transcripts with default settings [32]. Protein–protein interaction networks of significantly differentially expressed genes were constructed via Cytoscape (3.8.0) and the stringApp plugin (1.5.1) with a confidence score 0.7 to select for high confidence interactions and Chelonia mydas as the species of interest [33]. The Network Analyzer tool within Cytoscape was used to calculate node degree per gene and identify hub genes and other network statistics. Genes with a degree greater than 3 were visualized within constructed interaction networks. Functional enrichment was assessed via the STRING enrichment function.
3. Results
To characterize differential gene expression relevant to the development of broad life history traits, including immunity, migratory behavior, and longevity in loggerhead sea turtles (Caretta caretta) subjected to temperatures below and above the pivotal temperature (29 °C) for sex determination, we performed RNA sequencing and de novo assembly of the transcriptomes of female and male loggerhead hatchlings using brain (4 male, 3 female) and gonadal (2 male, 2 female) tissue. Significantly enriched gene ontology terms were identified, and potential association of differentially expressed transcripts with temperature-dependent sex determination (TSD), migration, longevity, and immune function were identified by comparison with genes previously reported in the reptile and avian literature, including those for reference genome assemblies published for turtles [34,35].
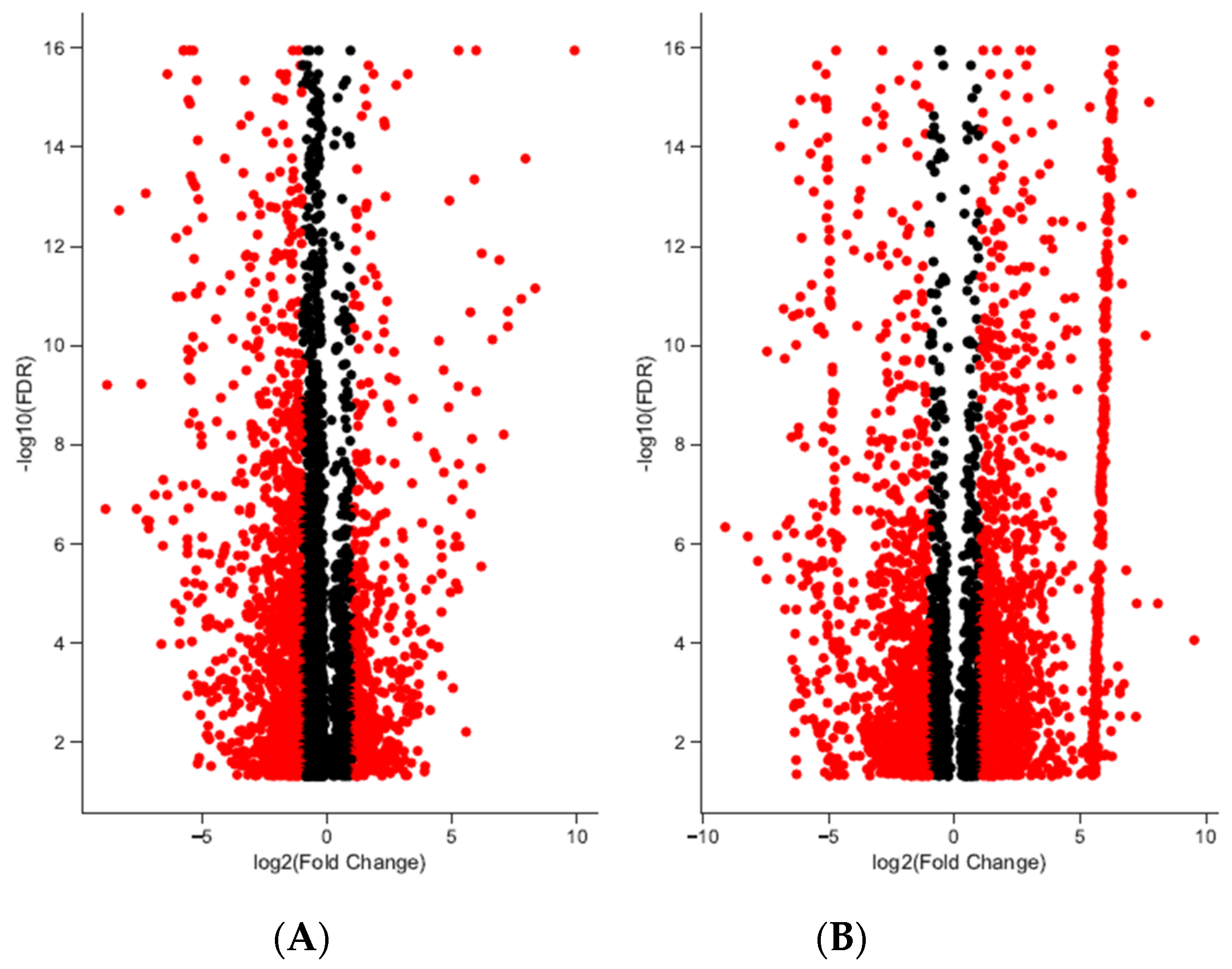
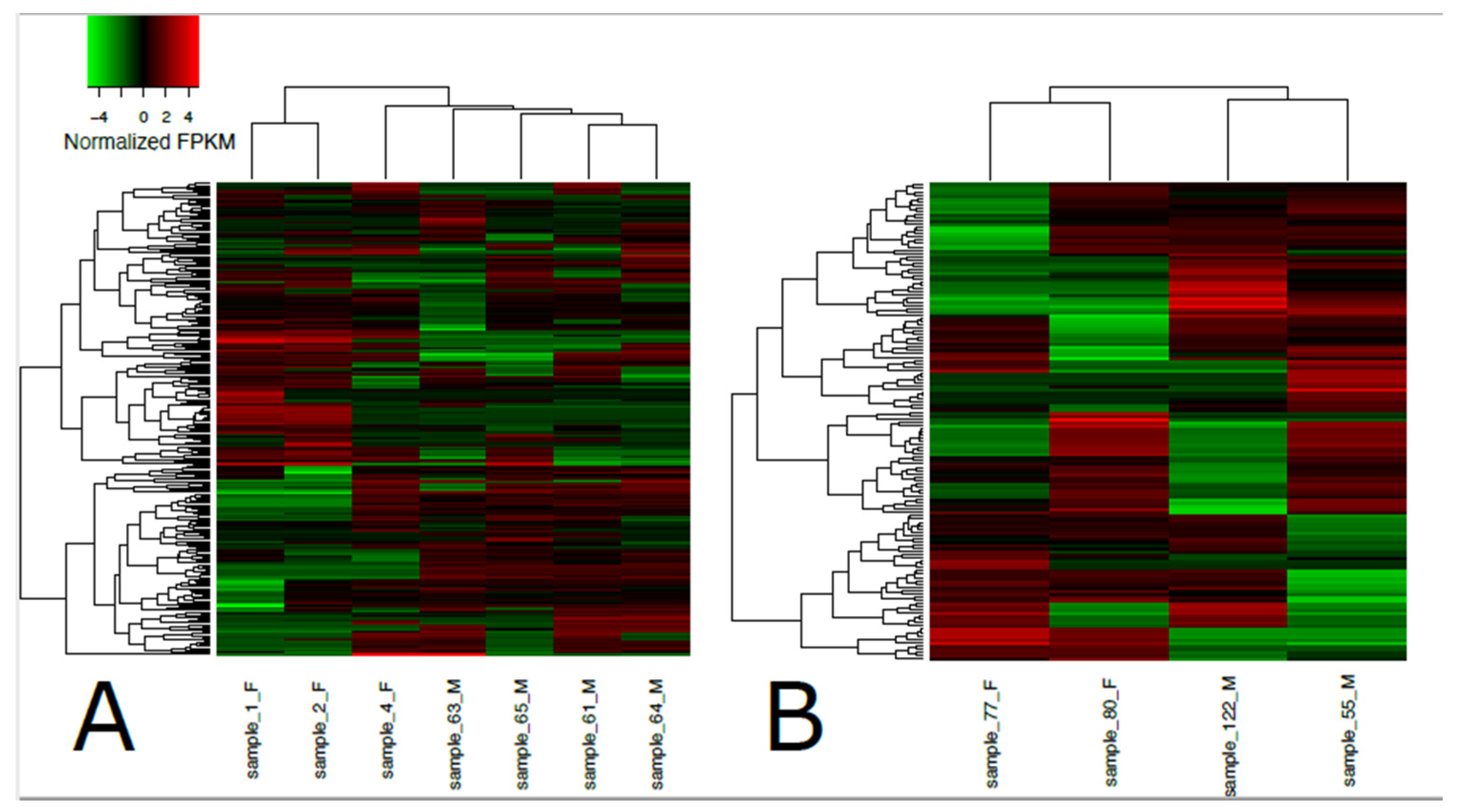
A total of 192,076 transcripts were identified for loggerhead brain tissue, and 150,489 transcripts were identified for gonadal tissue (Table 1). Approximately 96.31% and 97.14% of transcript reads passed trimming quality filters with an average length of 1399 and 1733 bp, respectively, for brain and gonad. In brain tissue from male and female loggerheads, 1513 and 774 transcripts were found to be significantly downregulated (PostFC < 0.5) and upregulated (PostFC > 2), respectively, at false discovery rate (FDR) < 0.001; in gonad, 872 transcripts were found to be downregulated, while 1270 were found to be upregulated (FDR < 0.001, PostFC < 0.5 and > 2) (Figure 1). Selected candidate genes potentially relevant to TSD, immunity, migratory behavior, and other broad life history traits that are significantly differentially expressed in female and male loggerhead brain and gonadal tissue are displayed in Table 2. Hierarchically clustered heatmaps of tran-scripts with at least 4-fold differential expression (FDR < 0.001) while extracting the top 50 differential expression features per pairwise comparison (--max_DE_genes_per_compar-ison 50) are displayed in Figure 2 for loggerhead brain and gonad.

Table 1.
Raw reads retrieved per sample and number of reads aligned to assembled transcriptomes for loggerhead brain and gonadal tissues.
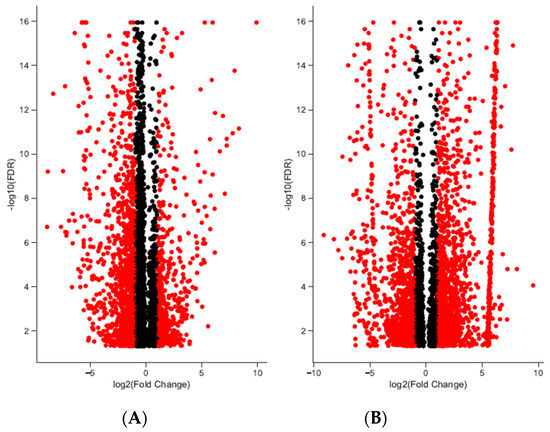
Figure 1.
Differentially expressed transcripts in brain and gonadal tissue in loggerhead sea turtles reared below and above 29 °C. Significant differentiation from fold change (FC) of two is indicated in red (FDR < 0.001). In brain (A), 774 and 1513 transcripts are significantly up- and downregulated with fold change greater than 2; 1270 and 872 transcripts with fold change greater than 2 are significantly up- and downregulated in gonads (B).

Table 2.
Genes with potential relevance to TSD, immunity, migratory behavior, and longevity in brain and gonadal tissue of loggerheads. Select genes with significant differential expression (FDR corrected p-value (q-value) < 0.05) and fold change greater than 2 among loggerheads reared below and above 29 °C are displayed.
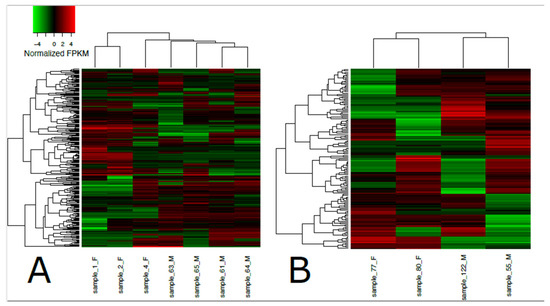
Figure 2.
Heatmap of differentially expressed transcripts in loggerhead brain (A) and gonadal (B) tissue clustered by sex. Sex is indicated in sample names for females (F) and males (M). Significantly expressed transcripts (FDR < 0.001) with at least 4-fold differential expression are displayed in normalized FPKM values using the top 50 differential expression features within pairwise comparisons.
Gene Ontology Enrichment and Functional Annotation Clustering
To identify pathways and biological processes related to TSD, migration, longevity, and immunity, Gene Ontology (GO) enrichment was performed via OmicsBox 1.2.4. A total of 3629 transcripts were assigned GO terms for brain, and 3289 GO terms were assigned for gonad, of which 2149 and 994 biological process GO terms displayed significant (FDR < 0.05) enrichment in brain and gonadal tissue, respectively. Selected biological processes relevant to sexual development are displayed in Table 3. Biological processes related to cellular metabolism and DNA metabolism (GO:0006259) were significantly en-riched in brain, as were the terms DNA repair (GO:0006281) and the cell cycle (GO:0007049) in gonadal tissue (Supplemental file S1). Overall, the enrichment of terms relevant to transport (GO:0006810) and biosynthetic processes in brain and the enrichment of chromosomal organization (GO:0051276), and signal transduction (GO:0007165) in gonadal tissue suggests the enrichment of processes important for embryonic development. REViGO was used to summarize enriched GO terms by reducing functional redundancies, producing treemaps indicating that protein phosphorylation and metabolism-related biological processes are enriched in the loggerhead brain and gonads (Supplemental files S2 and S3).

Table 3.
Significantly enriched biological processes in brain and gonadal tissue of loggerheads. Selected biological process Gene Ontology (GO) terms with significant enrichment between loggerheads reared below and above 29 °C are displayed. Q-value refers to p-values adjusted for the false discovery rate (FDR).
Functional annotation was performed via DAVID to identify shared enrichment of biological functions among significantly differentially expressed genes (q-value < 0.01). Functional analysis identified 1 enriched KEGG pathway (cmy04512:ECM-receptor interaction) in gonadal tissue and 2 KEGG pathways (cmy03460:Fanconi anemia pathway 13, cmy03440:Homologous recombination) in brain tissue with Benjamini-Hochberg < 0.05 (Supplemental file S4). In brain, shared functions of DNA repair and replication exists within the Fanconi anemia pathway 18 and Homologous recombination clusters, including shared enrichment of genes relevant to DNA polymerase and topoisomerase. The ECM receptor interaction cluster identified in gonads consists primarily of genes associated with collagen, laminin, and integrin, indicating an enrichment in signal transduction pathways and cellular communication in these tissues. The DAVID functional annotation clustering tool was used to further describe similarity among enriched functional annotations terms, identifying 11 clusters from brain and 8 clusters from gonadal tissue, and returning enrichment scores describing the degree of gene co-association; the clusters with highest enrichment scores for brain were associated with DNA repair and transport, and clusters relevant to metabolism were identified in gonads (Supplemental file S4).
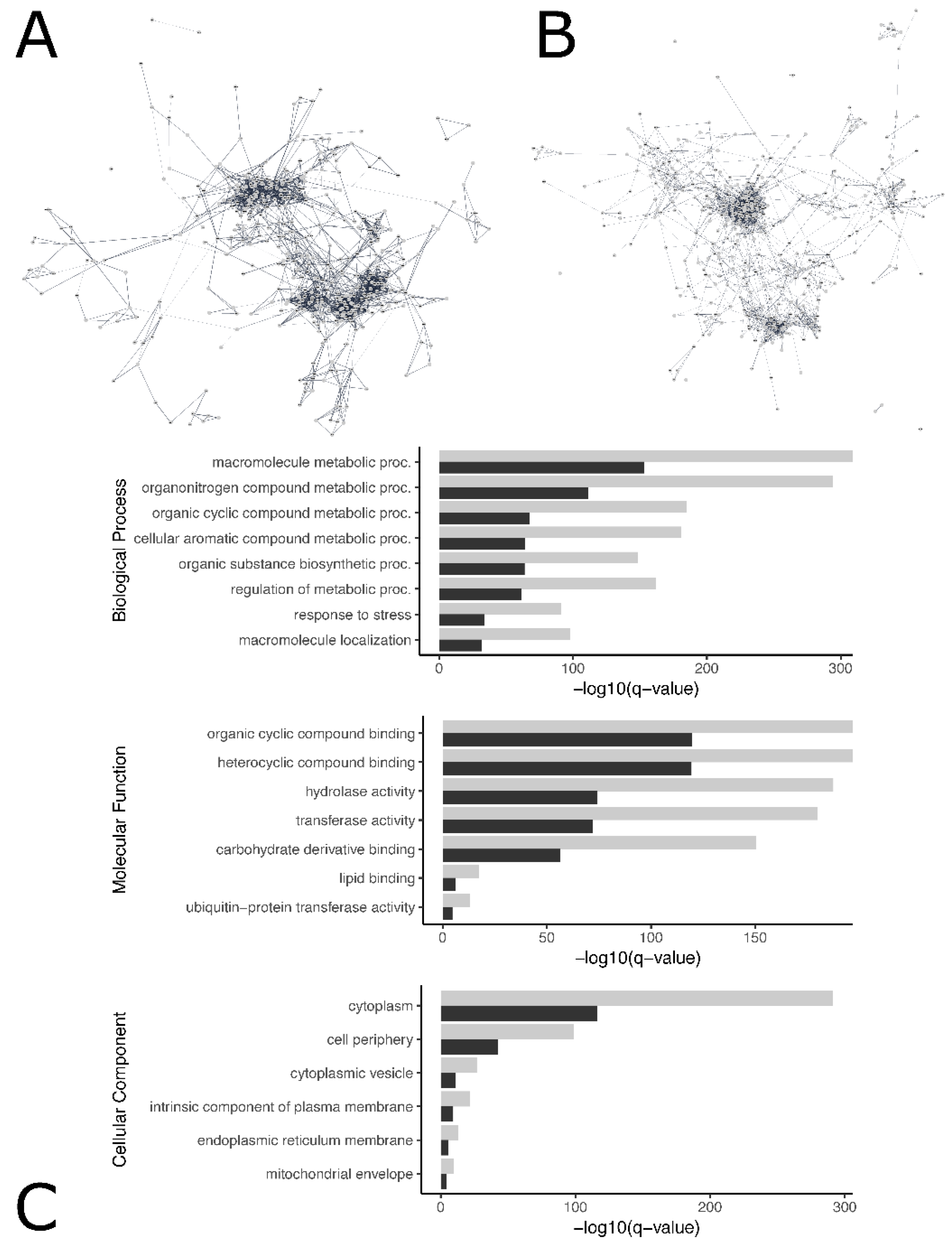
Protein–protein interaction networks of significantly differentially expressed genes among female and male loggerhead brain and gonadal tissue were constructed via Cytoscape to identify networks of interconnected genes potentially involved in the same biological pathways, identify genes of high degree likely to be important in multiple pathways relevant to temperature-specific sexual development, and visualize protein interactions (Figure 3). Constructed networks are publicly available on the Network Data Exchange (NDEx) Public Database, displaying gene-specific information including measures of centrality, degree, and associated functional enrichment terms (brain: https://www.ndexbio.org/viewer/networks/68a25356-18d2-11ec-9fe4-0ac135e8bacf, accessed on 19 October 2021; gonad: https://www.ndexbio.org/viewer/networks/e3fedc13-18cf-11ec-9fe4-0ac135e8bacf, accessed on 19 October 2021). A network with a total of 2502 nodes with 3309 edges was constructed to represent protein–protein interaction in brain tissue. The genes PDCD11, CDK1, and BRIX1 had the highest degrees observed. For gonads, a network of 2296 nodes and 3653 edges was created; CDK1 and CDC20 displayed the highest degrees. Node statistics for brain and gonad protein–protein interaction networks are displayed in Supplemental file S5. The stringApp was used to extract functional enrichment terms associated with groups of genes within the constructed protein–protein interaction networks, yielding significant enrichment in the coiled coil (KW-0175) in both gonads (q-value < 5.86 × 10−7) and brain (q-value < 2.9 × 10−6), and transferase (KW-0808, q-value < 1.5 × 10−3) and ECM-receptor interaction KEGG pathway (cmy04512, q-value < 3.47 × 10−2) in gonads (Supplemental file S5).
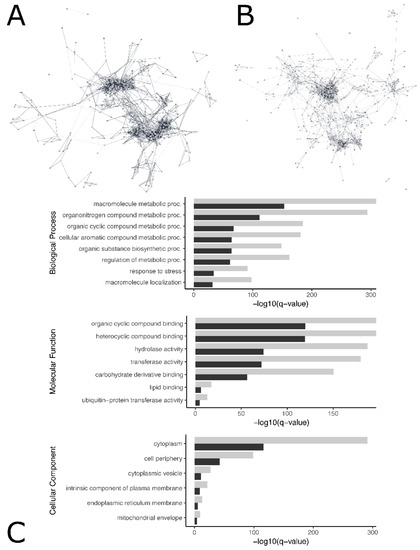
Figure 3.
Visual representation of protein–protein interaction in brain (A) and gonads (B) for genes with degree greater than 3. Interactive visualizations of the constructed networks are publicly available for brain (https://www.ndexbio.org/viewer/networks/e3fedc13-18cf-11ec-9fe4-0ac135e8bacf, accessed on 19 October 2021) and gonads (https://www.ndexbio.org/viewer/networks/e3fedc13-18cf-11ec-9fe4-0ac135e8bacf, accessed on 19 October 2021). Gene-specific information, including association with enrichment terms can be viewed by selecting individual genes in the network. Larger degree indicates greater number of physical interactions between proteins associated with a gene and its neighbors in the network, and genes with large degree may be relatively important to multiple biological pathways. Topological parameters measuring node degree distribution, centrality, and other metrics were calculated and displayed per gene in Supplemental file S5 for gonads and brain. Significant biological processes, molecular function, and cellular process Gene Ontology (GO) terms associated with differentially expressed genes in brain (grey) and gonadal (black) tissue (C).
4. Discussion
Biological Significance of Differentially Expressed Genes among Female and Male Loggerheads
We have performed de novo transcriptome assembly and differential gene expression analyses using brain and gonadal tissue from loggerhead hatchlings experimentally reared below and above the pivotal temperature for sex determination (29 °C). Significantly differentially expressed genes in gonadal tissue were generally enriched for signal transduction, while brain tissue was enriched in terms related to protein binding, transport, metabolism, and biosynthesis.
With special attention to genes potentially involved in the traits of TSD, migratory behavior, longevity, and immunity, we have identified significantly differentially ex-pressed genes of interest in loggerheads reared below and above the pivotal temperature for sex determination. Here, we highlight these genes from our analyses on loggerhead brain and gonadal tissue that have also been reported in the reptile and avian literature as pertinent.
Temperature-dependent sex determination (TSD).
Loggerheads display TSD, in which individuals reared above and below the pivotal temperature of 29 °C mainly develop into females and males, respectively. In both brain and gonads, the gene HSD17B7 was found to be significantly differentially expressed. HSD17B7 has previously been potentially associated with TSD and has a role in the synthesis of sex steroids [34,36,37].
Genes differentially expressed in gonads relevant to TSD.
Candidate sex determining genes such as FOXL2, RSPO1, and ESR1 were significantly enriched in gonadal tissue and tend to be highly expressed during female promoting temperatures in various turtle species [38,39,40,41]. However, the exact roles of certain genes that are important to gonadogenesis, such as CBX2, EMX2, GATA4, and LHX9, are relatively uncharacterized in TSD [14]. Our analysis further identified SERPINH1, AMHR2, and BCR as differentially expressed in loggerhead gonads, with potential functions in gonadogensis as suggested previously [42]. AMHR2 is a TSD regulator, and BCR has been associated with stress and immune response. CIRBP was previously found to be involved in TSD in the snapping turtle (Chelydra serpentina) [42].
Genes differentially expressed in brain relevant to TSD.
The genes KDM3A, important in ovary and testicle formation in turtles that exhibit TSD, an estrogen receptor interactor GREB1 [43], a gene related to fetal sex in humans GRB10, a germ cell-specific factor necessary for meiotic progression TOPAZ1 [44], and M1AP, a gene implicated in male germ cell development [45], were found to be significantly enriched in brain tissue. The role of KDM6B in TSD has been previously described in loggerheads, and JARID2 has been found to be relevant to TSD via RNA-seq analysis of brain tissue in loggerheads and gonadal tissue in Alligator mississippiensis [16,17,46,47,48]. STIP1 has been associated with heat shock proteins under thermal stress in loggerhead brain tissue, and the genes A2ML1 and ASB16 have also been found to be differentially expressed in the loggerhead brain during heat stress [17].
Migration.
From birth, loggerhead hatchlings migrate long distances from coastal areas to the open sea [49]. Loggerheads display natal homing behavior; many mature loggerheads forage in neritic zones close to natal nesting sites, and female loggerheads return to their natal nesting region to reproduce [1,50]. Previous evidence suggests that sea turtles are able to detect and distinguish among magnetic fields, enabling navigation [51]. Philopatry and the ability to successfully navigate long distances has been observed in many species [52]. The genes MICAL1, involved in FAD binding, and IFT20, both of which were significantly enriched in loggerhead brain, may play a role in photoreceptor activity and magnetic receptions in birds [53]. Additionally in the loggerhead brain or gonadal tissue, CACNA1B, SRPK2, COL1A2, NRXN1, ARPP21, and ITPR1 were previously found to be significantly differentially expressed among juvenile and adult migrating willow warblers [54]. NPAS2 and DRD4 are associated with avian migratory behavior; specifically, NPAS2 is associated with circadian rhythm, and DRD4 is a candidate for exploratory behavior [55]. VPS13A, previously linked to avian migration, was enriched in loggerhead brain tissue [56].
Longevity and immunity.
The lifespan of a loggerhead can surpass 70 years (https://www.fisheries.noaa.gov/species/loggerhead-turtle, accessed on 19 October 2021), and the resiliency of the loggerhead immune system contributes to loggerhead longevity. Previously, RHOF has been implicated in normal B-cell function and is upregulated in malignant lymphoid tissue [57]. SHISA6 regulates stem cell differentiation in spermatogonia by inhibition of Wnt [58]. UBE3D facilitates protein degradation and may be relevant to prevention of cellular senescence. Genes involved in tumor neovascularization and tumor suppression such as SART3 and NEDD4, were found to be significantly differentially expressed in the loggerhead brain or gonads. There was significant differential expression of genes regulating apoptosis (CCAR2, GRB10), the cell cycle (SIPA1, CCAR2), and DNA repair (RAD54B), in addition to macrophage stimulation (MST1R, CSF1R) and other genes involved in immune cell signaling (TLR3, IRAK2, BCAP29).
5. Conclusions
We have characterized the differential expression of genes with potential relevance to TSD, migration, longevity, and immunity in brain and gonadal tissue of female and male loggerhead sea turtles. Significantly differentially expressed genes associated with gonadogenesis, synthesis of sex hormones, and gamete formation were identified as potentially relevant to TSD. A search of differentially expressed genes in male and female loggerheads identified genes in the avian and reptile literature with roles in magnetoreception, migratory behavior, cellular senescence, DNA repair, and immunity, among other functions.
We observed significant enrichment of biological processes specifically related to TSD and sexual development, indicating that distinct groups of genes in brain and gonads are differentially regulated in loggerheads experimentally reared below and above the pivotal temperature for sex determination in C. caretta. Through functional clustering and protein–protein interaction enrichment analyses, enrichment of the ECM-receptor pathway was identified in both brain and gonadal tissue, suggesting that extracellular communication may play an important role in sex determination.
Through summarization of differentially expressed genes in male and female loggerheads, this work has produced a set of candidate genes with particular relevance to mechanism of TSD. Further study of these genes may yield a clearer understanding of genotype–phenotype relationships and potential adaptive resiliency of endangered loggerheads worldwide and, by extension, other TSD species. We anticipate this new transcriptomic resource integrated with genome-wide surveys of locus-specific polymorphism [19] can inform future conservation efforts that will seek to address growing sex imbalances and survivorship in sea turtle populations experiencing warming climates.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ani11113013/s1, Supplemental file S1 (XLS). Enriched Gene Ontology terms in loggerhead brain and gonadal tissue. Enriched GO term categories are indicated and sorted by increasing p-value and FDR. Supplemental file S2 (PDF). REViGO Gene Ontology treemap. A summary of significantly enriched Gene Ontology terms from loggerhead brain tissue. Supplemental file S3 (PDF). REViGO Gene Ontology treemap. A summary of significantly enriched Gene Ontology terms from loggerhead gonadal tissue. Supplemental file S4 (XLS). Functional annotation and clustering of differentially expressed genes in loggerhead brain and gonadal tissue. Functional annotation terms are shown in ‘annotation’ tabs, and identified clusters are shown in ‘clustering’ tabs in order of decreasing enrichment score. Supplemental file S5 (XLS). Network parameter statistics per gene in loggerhead brain and gonadal tissue. Measures of node degree distributions, centrality, connectivity, and topological coefficients are displayed per gene in brain and gonad networks constructed from significantly differentially expressed genes. Supplemental file S6 (PDF). Visual summary of experimental design. RNA-seq sampling of hatchling transcriptomes per treatment and tissue type (brain, gonad) are summarized. A graphical sequence of sample-specific RNA processing for loggerhead hatchlings experimentally reared at the pivotal temperature for sex determination (29 °C), de novo transcriptome assembly, gene annotation, and differential gene expression analysis is displayed.
Author Contributions
Conceptualization, J.C.C., M.H.G., C.A.H., P.E.A. and A.M.S.; Data curation, J.C.C., N.K., M.M., M.H.G., C.A.H., P.E.A. and A.M.S.; Formal analysis, J.C.C., N.K., M.M., M.H.G., C.A.H., P.E.A. and A.M.S.; Funding acquisition, M.H.G., C.A.H., P.E.A. and A.M.S.; Investigation, J.C.C., N.K., M.M., M.H.G., C.A.H., P.E.A. and A.M.S.; Methodology, J.C.C., N.K., M.M., M.H.G., C.A.H., P.E.A. and A.M.S.; Project administration, M.H.G., C.A.H., P.E.A. and A.M.S.; Resources, J.C.C., M.H.G., C.A.H., P.E.A. and A.M.S.; Software, J.C.C., P.E.A. and A.M.S.; Supervision, M.H.G., C.A.H., P.E.A. and A.M.S.; Validation, J.C.C., M.H.G., C.A.H., P.E.A. and A.M.S.; Visualization, J.C.C., N.K., M.M., P.E.A. and A.M.S.; Writing—original draft, J.C.C., M.H.G., C.A.H., P.E.A. and A.M.S.; Writing—review and editing, J.C.C., M.H.G., C.A.H., P.E.A. and A.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Science Foundation (NSF) REU Site [award DBI-1359301] to [PEA and AMS], Role = student internship training, high-performance computing (HPC) workflow design and implementation, RNA-seq data production and analysis; NSF Research Infrastructure Improvement Established Program to Stimulate Competitive Research (SC EPSCoR) [award #1317771] to [PEA and AMS], Role = HPC infrastructure and distributed data analysis and management; SC EPSCoR Institutional Development Award (IDeA) Network of Biomedical Research Excellence in Bioinformatics [grant #101-11] to [AMS], Role = HPC infrastructure development and distributed data analysis and management; NOAA Research Grant [award #NA10NMF4720035] to [MHG], Role = sampling design, permit-authorized field collection and experimental treatment of turtles; US Department of State Fulbright US Scholar [Award #29480] to [AMS], Role = Results synthesis and manuscript preparation; and grants from the College of Charleston Office of Faculty Research and Development to [AMS], Role = Project design development and RNA-seq data production. The APC was funded by [AMS, PEA, MHG, and CAH].
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the North Carolina State University Institutional Animal Care and Use Committee (IACUC Protocol ID No. 11-103-O) according to the Endangered Species Permit Nos. 11ST53, 12ST53, 13ST50 from the North Carolina Wildlife Resources Commission with federal authority delegated from the United States Fish and Wildlife Service.
Informed Consent Statement
Not applicable.
Data Availability Statement
Original DNA sequence data for this project are openly available in the NCBI Biosample repository for DNA and RNA Resources under the SRA accession PRJNA663187.
Acknowledgments
We thank David Owens (COFC), Michael Arendt and Jeffrey Schwenter (SC Dept. of Natural Resources) for helpful discussions about sea turtle biology; Patricia McClellan-Green, Emily Christiansen, Heather Broadhurst, Stasia Bembenek Bailey for field and lab assistance; Chris Saski (Clemson University) and Bob Wilson (MUSC) for assistance with Illumina cDNA sequencing; COFC OMICS REU student interns David Moore, Katharine King, Franceska Donkor, Connie Truong, and Alyssa Dumas for assistance with preliminary data processing. All aspects of collection, transport, incubation, handling, and euthanasia were authorized by the North Carolina State University (NCSU) Institutional Animal Care and Use Committee (IACUC 11-103-O), the North Carolina Wildlife Resources Commission Sea Turtle Project (Sea Turtle Permits 11ST53, 12ST53, 13ST50), the National Oceanic and Atmospheric Administration (NOAA) Environment Assessment (NA10NMF4720035), and the Cooperative Agreement (Fish and Wildlife Service 14-16-0009-77-906) between the US Fish and Wildlife Service and the NC Wildlife Resources Commission for the conservation of protected species.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| FDR | False discovery rate |
| GO | Gene Ontology |
| TSD | Temperature-dependent sex determination |
References
- Bowen, B.W.; Karl, S.A. Population genetics and phylogeography of sea turtles. Mol. Ecol. 2007, 16, 4886–4907. [Google Scholar] [CrossRef] [PubMed]
- Broderick, A.C.; Coyne, M.S.; Fuller, W.J.; Glen, F.; Godley, B.J. Fidelity and over-wintering of sea turtles. Proc. R. Soc. B. 2007, 274, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.R.; Carthy, R.R.; Ceriani, S.A. Migration routes, foraging behavior, and site fidelity of loggerhead sea turtles (Caretta caretta) satellite tracked from a globally important rookery. Mar. Biol. 2019, 166, 134. [Google Scholar] [CrossRef]
- Siegwalt, F.; Benhamou, S.; Girondot, M.; Jeantet, L.; Martin, J.; Bonola, M.; Lelong, P.; Grand, C.; Chambault, P.; Benhalilou, A.; et al. High fidelity of sea turtles to their foraging grounds revealed by satellite tracking and capture-mark-recapture: New insights for the establishment of key marine conservation areas. Biol. Conserv. 2020, 250, 108742. [Google Scholar] [CrossRef]
- Mrosovsky, N.; Kamel, S.; Rees, A.F.; Margaritoulis, D. Pivotal temperature for loggerhead turtles (Caretta caretta) from KyparissiaBay, Greece. Can. J. Zool. 2002, 80, 2118–2124. [Google Scholar] [CrossRef] [Green Version]
- Lolavar, A.; Wyneken, J. Effects of supplemental watering on loggerhead (Caretta caretta) nests and hatchlings. J. Exp. Mar. Biol. Ecol. 2021, 534, 151476. [Google Scholar] [CrossRef]
- Marco, A.; Abella, E.; Martins, S.; López, O.; Patino-Martinez, J. Female nesting behaviour affects hatchling survival and sex ratio in the loggerhead sea turtle: Implications for conservation programmes. Ethol. Ecol. Evol. 2018, 30, 141–155. [Google Scholar] [CrossRef]
- Hawkes, L.A.; Broderick, A.C.; Godfrey, M.H.; Godley, B.J. Investigating the potential impacts of climate change on a marine turtle population. Glob. Chang. Biol. 2007, 13, 923–932. [Google Scholar] [CrossRef]
- Dang, W.; Zhang, W.; Du, W.-G. Incubation temperature affects the immune function of hatchling soft-shelled turtles, Pelodiscus sinensis. Sci. Rep. 2015, 5, 10594. [Google Scholar] [CrossRef]
- Fleming, K.A.; Perrault, J.; Stacy, N.; Coppenrath, C.M.; Gainsbury, A.M. Heat, health and hatchlings: Associations of in situ nest temperatures with morphological and physiological characteristics of loggerhead sea turtle hatchlings from Florida. Conserv. Physiol. 2020, 8, coaa046. [Google Scholar] [CrossRef]
- Lu, H.; Jin, J.; Fan, H.; Dang, W. The magnitude of incubation temperature fluctuation affects the immunity of Chinese soft-shelled turtle (Pelodiscus sinensis) hatchlings. Aquac. Res. 2021. [Google Scholar] [CrossRef]
- Fisher, L.R.; Godfrey, M.H.; Owens, D.W. Incubation Temperature Effects on Hatchling Performance in the Loggerhead Sea Turtle (Caretta caretta). PLoS ONE 2014, 9, e114880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janes, D.E.; Organ, C.L.; Fujita, M.K.; Shedlock, A.M.; Edwards, S.V. Genome Evolution in Reptilia, the Sister Group of Mammals. Annu. Rev. Genom. Hum. Genet. 2010, 11, 239–264. [Google Scholar] [CrossRef] [Green Version]
- Radhakrishnan, S.; Literman, R.; Neuwald, J.; Severin, A.; Valenzuela, N. Transcriptomic responses to environmental temperature by turtles with temperature-dependent and genotypic sex determination assessed by RNAseq inform the genetic architecture of embryonic gonadal development. PLoS ONE 2017, 12, e0172044. [Google Scholar] [CrossRef]
- Deveson, I.W.; Holleley, C.E.; Blackburn, J.; Graves JA, M.; Mattick, J.S.; Waters, P.D.; Georges, A. Differential intron retention in Jumonji chromatin modifier genes is implicated in reptile temperature-dependent sex determination. Sci. Adv. 2017, 3, e1700731. [Google Scholar] [CrossRef] [Green Version]
- Yatsu, R.; Miyagawa, S.; Kohno, S.; Parrott, B.B.; Yamaguchi, K.; Ogino, Y.; Miyakawa, H.; Lowers, R.H.; Shigenobu, S.; Guillette, L.J., Jr.; et al. RNA-seq analysis of the gonadal transcriptome during Alligator mississippiensis temperature-dependent sex determination and differentiation. BMC Genom. 2016, 17, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentley, B.P.; Haas, B.J.; Tedeschi, J.; Berry, O. Loggerhead sea turtle embryos (Caretta caretta) regulate expression of stress response and developmental genes when exposed to a biologically realistic heat stress. Mol. Ecol. 2017, 26, 2978–2992. [Google Scholar] [CrossRef] [PubMed]
- Tezak, B.; Sifuentes-Romero, I.; Milton, S.; Wyneken, J. Identifying Sex of Neonate Turtles with Temperature-dependent Sex Determination via Small Blood Samples. Sci. Rep. 2020, 10, 1–8. [Google Scholar]
- Chow, J.C.; Anderson, P.E.; Shedlock, A.M. Sea Turtle Population Genomic Discovery: Global and Locus-Specific Signatures of Polymorphism, Selection, and Adaptive Potential. Genome Biol. Evol. 2019, 11, 2797–2806. [Google Scholar] [CrossRef] [Green Version]
- Godfrey, H.M.; Mrosovsky, N. Pivotal temperature for green sea turtles, Chelonia mydas, nesting in Suriname. Herpetol. J. 2006, 16, 55–61. [Google Scholar]
- Yntema, C.L.; Mrosovsky, N. Sexual Differentiation in Hatchling Loggerheads (Caretta caretta) Incubated at Different Controlled Temperatures. Herpetologica 1980, 36, 33–36. [Google Scholar]
- Harms, C.A.; McClellan-Green, P.; Godfrey, M.H.; Christiansen, E.F.; Broadhurst, H.J.; Godard-Codding, C.A.J. Crude Oil and Dispersant Cause Acute Clinicopathological Abnormalities in Hatchling Loggerhead Sea Turtles (Caretta caretta). Front. Vet. Sci. 2019, 6, 344. [Google Scholar] [CrossRef] [Green Version]
- Bembenek-Bailey, S.A.; Niemuth, J.N.; McClellan-Green, P.D.; Godfrey, M.H.; Harms, C.A.; Gracz, H.; Stoskopf, M.K. NMR Metabolomic Analysis of Skeletal Muscle, Heart, and Liver of Hatchling Loggerhead Sea Turtles (Caretta caretta) Experimentally Exposed to Crude Oil and/or Corexit. Metabolites 2019, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Bembenek Bailey, S.A.; Niemuth, J.N.; McClellan-Green, P.D.; Godfrey, M.H.; Harms, C.A.; Stoskopf, M.K. 1H-NMR metabolomic study of whole blood from hatchling loggerhead sea turtles (Caretta caretta) exposed to crude oil and/or Corexit. R. Soc. Open Sci. 2017, 4, 171433. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High -throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Smuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Shaffer, H.B.; Minx, P.; Warren, D.E.; Shedlock, A.M.; Thomson, R.C.; Valenzuela, N.; Abramyan, J.; Amemiya, C.T.; Badenhorst, D.; Biggar, K.K.; et al. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 2013, 14, R28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Pascual-Anaya, J.; Zadissa, A.; Li, W.; Niimura, Y.; Huang, Z.; Li, C.; White, S.; Xiong, Z.; Fang, D.; et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat. Genet. 2013, 45, 701–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.X.; Teng, J.; Zhao, Y.; Li, N.; Wang, H.; Ji, X.S. Gonad Transcriptome Analysis of High-Temperature-Treated Females and High-Temperature-Induced Sex-Reversed Neomales in Nile Tilapia. Int. J. Mol. Sci. 2018, 19, 689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, G.; Li, S.; Zhao, Q.; Chu, J.; Zhou, B.; Fan, S.; Shi, F.; Wei, X.; Hu, X.; Zheng, X.; et al. Transcriptomic and metabolomic insights into the variety of sperm storage in oviduct of egg layers. Poult. Sci. 2021, 100, 101087. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Buemio, A.; Chu, R.; Vafaee, M.; Crews, D. Epigenetic Control of Gonadal Aromatase (cyp19a1) in Temperature-Dependent Sex Determination of Red-Eared Slider Turtles. PLoS ONE 2013, 8, e63599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhen, T.; Metzger, K.; Schroeder, A.; Woodward, R. Expression of Putative Sex-Determining Genes during the Thermosensitive Period of Gonad Development in the Snapping Turtle, Chelydra serpentina. Sex. Dev. 2007, 1, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, C.; Ramsey, M.; Queen, J.; Crews, D. Expression ofSox9,Mis, andDmrt1 in the gonad of a species with temperature-dependent sex determination. Dev. Dyn. 2007, 236, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Hernández, V.; Marmolejo-Valencia, A.; Merchant-Larios, H. Exogenous estradiol alters gonadal growth and timing ofmtemperature sex determination in gonads of sea turtle. Dev. Biol. 2015, 408, 79–89. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, A.L.; Metzger, K.J.; Miller, A.; Rhen, T. A Novel Candidate Gene for Temperature-Dependent Sex Determination in the Common Snapping Turtle. Genetics 2016, 203, 557–571. [Google Scholar] [CrossRef] [Green Version]
- Mohammed, H.; D’Santos, C.; Serandour, A.A.; Ali, R.; Brown, G.D.; Atkins, A.; Palacio, O.R.; Holmes, K.-A.; Theodorou, V.; Robinson, J.L.; et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2021, 3, 342–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luangpraseuth-Prosper, A.; Lesueur, E.; Jouneau, L.; Pailhoux, E.; Cotinot, C.; Mandon-Pépin, B. TOPAZ1, a germ cell specific factor, is essential for male meiotic progression. Dev. Biol. 2015, 406, 158–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arango, N.A.; Li, L.; Dabir, D.; Nicolau, F.; Pieretti-Vanmarcke, R.; Koehler, C.; McCarrey, J.R.; Lu, N.; Donahoe, P.K. Meiosis I arrest abnormalities lead to severe oligozoospermia in meiosis 1 arresting protein (M1ap)-deficient mice. Biol. Reprod. 2013, 88, 76. [Google Scholar] [CrossRef]
- Ge, C.; Ye, J.; Weber, C.; Sun, W.; Zhang, H.; Zhou, Y.; Cai, C.; Qian, G.; Capel, B. The histone demethylase KDM6B regulates temperature-dependent sex determination in a turtle species. Science 2018, 360, 645–648. [Google Scholar] [CrossRef] [Green Version]
- Weber, C.; Zhou, Y.; Lee, J.G.; Looger, L.L.; Qian, G.; Ge, C.; Capel, B. Temperature-dependent sex determination is mediated by pSTAT3 repression of Kdm6b. Science 2020, 368, 303–306. [Google Scholar] [CrossRef]
- Purnell, B.A. How egg temperature sets sex. Science 2020, 368, 278. [Google Scholar]
- Boyle, M.C.; Fitzsimmons, N.N.; Limpus, C.J.; Kelez, S.; Velez-Zuazo, X.; Waycott, M. Evidence for transoceanic migrations by loggerhead sea turtles in the southern Pacific Ocean. Proc. R. Soc. B Biol. Sci. 2009, 276, 1993–1999. [Google Scholar] [CrossRef] [Green Version]
- Bowen, B.W.; Kamezaki, N.; Limpus, C.J.; Hughes, G.R.; Meylan, A.B.; Avise, J.C. Global Phylogeography of the Loggerhead Turtle (Caretta caretta) as Indicated by Mitochondrial DNA Haplotypes. Evolution 1994, 48, 1820. [Google Scholar]
- Lohmann, K.J.; Putman, N.F.; Lohmann, C.M.F. Geomagnetic imprinting: A unifying hypothesis of long-distance natal homing in salmon and sea turtles. Proc. Natl. Acad. Sci. USA 2008, 105, 19096–19101. [Google Scholar] [CrossRef] [Green Version]
- Shamblin, B.M.; Bolten, A.; Abreu-Grobois, F.A.; Bjorndal, K.; Cardona, L.; Carreras, C.; Clusa, M.; Monzón-Argüello, C.; Nairn, C.J.; Nielsen, J.T.; et al. Geographic Patterns of Genetic Variation in a Broadly Distributed Marine Vertebrate: New Insights into Loggerhead Turtle Stock Structure from Expanded Mitochondrial DNA Sequences. PLoS ONE 2014, 9, e85956. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.A.; Lau, J.C.S.; Hogben, H.J.; Biskup, T.; Kattnig, D.R.; Hore, P.J. Alternative radical pairs for cryptochrome-based magnetoreception. J. R. Soc. Interface 2014, 11, 20131063. [Google Scholar] [CrossRef] [Green Version]
- Boss, J.; Liedvogel, M.; Lundberg, M.; Olsson, P.; Reischke, N.; Naurin, S.; Åkesson, S.; Hasselquist, D.; Wright, A.; Grahn, M.; et al. Gene expression in the brain of a migratory songbird during breeding and migration. Mov. Ecol. 2016, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- Mueller, J.C.; Pulido, F.; Kempenaers, B. Identification of a gene associated with avian migratory behaviour. Proc. R. Soc. B Biol. Sci. 2011, 278, 2848–2856. [Google Scholar] [CrossRef] [PubMed]
- Toews, D.P.L.; Taylor, S.A.; Streby, H.M.; Kramer, G.R.; Lovette, I.J. Selection on VPS13A linked to migration in a songbird. Proc. Natl. Acad. Sci. USA 2019, 116, 18272–18274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouw, L.G.; Reading, N.S.; Jenson, S.D.; Lim, M.S.; Elenitoba-Johnson, K.S.J. Expression of the Rho-family GTPase gene RHOF in lymphocyte subsets and malignant lymphomas. Br. J. Haematol. 2005, 129, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Tokue, M.; Ikami, K.; Mizuno, S.; Takagi, C.; Miyagi, A.; Takada, R.; Noda, C.; Kitadate, Y.; Hara, K.; Mizuguchi, H.; et al. SHISA6 Confers Resistance to Differentiation-Promoting Wnt/β-Catenin Signaling in Mouse Spermatogenic Stem Cells. Stem Cell Rep. 2017, 8, 561–575. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).