A Bayesian Model for Paired Data in Genome-Wide Association Studies with Application to Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Single-Marker Analysis
2.1.1. Maximum Likelihood Estimation
2.1.2. Hypothesis Testing
2.2. Bayesian Hierarchical Modeling
2.2.1. Prior Distribution
2.2.2. Joint Posterior Distribution
3. Results
3.1. Simulation Studies
3.2. Real Data Application
3.2.1. Application to Matched-Pair Breast Cancer Data
3.2.2. Single-Marker Analysis
3.2.3. Multi-Marker Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Smith, J.E.; Clark, A.R.; Staggemeier, A.T. A Genetic Approach to Statistical Disclosure Control. In Proceedings of the 11th Annual Conference on Genetic and Evolutionary Computation, Montreal, QC, Canada, 8–12 July 2009; Association for Computing Machinery: New York, NY, USA, 2009. GECCO’09. pp. 1625–1632. [Google Scholar] [CrossRef]
- Stadler, Z.K.; Thom, P.; Robson, M.E.; Weitzel, J.N.; Kauff, N.D.; Hurley, K.E.; Devlin, V.; Gold, B.; Klein, R.J.; Offit, K. Genome-wide association studies of cancer. J. Clin. Oncol. 2010, 28, 4255. [Google Scholar] [CrossRef] [PubMed]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef]
- Kim, H.S.; Minna, J.D.; White, M.A. GWAS Meets TCGA to Illuminate Mechanisms of Cancer Predisposition. Cell 2013, 152, 387–389. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A census of human cancer genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Li, P.; Guo, M.; Wang, C.; Liu, X.; Zou, Q. An overview of SNP interactions in genome-wide association studies. Brief. Funct. Genom. 2014, 14, 143–155. [Google Scholar] [CrossRef]
- Lee, S.; Abecasis, G.R.; Boehnke, M.; Lin, X. Rare-variant association analysis: Study designs and statistical tests. Am. J. Hum. Genet. 2014, 95, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.C.; Lee, S.; Cai, T.; Li, Y.; Boehnke, M.; Lin, X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011, 89, 82–93. [Google Scholar] [CrossRef]
- Pan, W.; Kim, J.; Zhang, Y.; Shen, X.; Wei, P. A powerful and adaptive association test for rare variants. Genetics 2014, 197, 1081–1095. [Google Scholar] [CrossRef]
- Liu, J.Z.; Mcrae, A.F.; Nyholt, D.R.; Medland, S.E.; Wray, N.R.; Brown, K.M.; Hayward, N.K.; Montgomery, G.W.; Visscher, P.M.; Martin, N.G.; et al. A Versatile Gene-Based Test for Genome-wide Association Studies. Am. J. Hum. Genet. 2010, 87, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Macgregor, S. VEGAS2: Software for more flexible gene-based testing. Twin Res. Hum. Genet. 2015, 18, 86–91. [Google Scholar] [CrossRef]
- De Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef]
- Li, M.X.; Gui, H.S.; Kwan, J.S.; Sham, P.C. GATES: A rapid and powerful gene-based association test using extended Simes procedure. Am. J. Hum. Genet. 2011, 88, 283–293. [Google Scholar] [CrossRef]
- Barnett, I.; Mukherjee, R.; Lin, X. The generalized higher criticism for testing SNP-set effects in genetic association studies. J. Am. Stat. Assoc. 2017, 112, 64–76. [Google Scholar] [CrossRef]
- Sun, R.; Lin, X. Genetic variant set-based tests using the generalized Berk–Jones statistic with application to a genome-wide association study of breast cancer. J. Am. Stat. Assoc. 2020, 115, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, S.; Li, Z.; Morrison, A.C.; Boerwinkle, E.; Lin, X. ACAT: A fast and powerful p value combination method for rare-variant analysis in sequencing studies. Am. J. Hum. Genet. 2019, 104, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Cho, J.; Zhao, H. Incorporating biological pathways via a Markov random field model in genome-wide association studies. PLoS Genet. 2011, 7, e1001353. [Google Scholar] [CrossRef][Green Version]
- Ziegler, A.; Konig, I.R. A Statistical Approach to Genetic Epidemiology; Wiley-VCH Verlag GmbH & Co. KGaA: Berlin, Germany, 2010. [Google Scholar] [CrossRef]
- Emigh, T.H. A Comparison of Tests for Hardy-Weinberg Equilibrium. Biometrics 1980, 36, 627. [Google Scholar] [CrossRef]
- Minard, M.E.; Kim, L.S.; Price, J.E.; Gallick, G.E. The Role of the Guanine Nucleotide Exchange Factor Tiam1 in Cellular Migration, Invasion, Adhesion and Tumor Progression. Breast Cancer Res. Treat. 2004, 84, 21–32. [Google Scholar] [CrossRef]
- Adam, L.; Vadlamudi, R.K.; McCrea, P.; Kumar, R. Tiam1 Overexpression Potentiates Heregulin-induced Lymphoid Enhancer Factor-1/β-Catenin Nuclear Signaling in Breast Cancer Cells by Modulating the Intercellular Stability. J. Biol. Chem. 2001, 276, 28443–28450. [Google Scholar] [CrossRef] [PubMed]
- Walch, A.; Seidl, S.; Hermannstadter, C.; Rauser, S.; Deplazes, J.; Langer, R.; von Weyhern, C.H.; Sarbia, M.; Busch, R.; Feith, M.; et al. Combined analysis of Rac1, IQGAP1, Tiam1 and E-cadherin expression in gastric cancer. Mod. Pathol. 2008, 21, 544–552. [Google Scholar] [CrossRef]
- Engers, R.; Mueller, M.; Walter, A.; Collard, J.G.; Willers, R.; Gabbert, H.E. Prognostic relevance of Tiam1 protein expression in prostate carcinomas. Br. J. Cancer 2006, 95, 1081–1086. [Google Scholar] [CrossRef]
- Minard, M.E.; Ellis, L.M.; Gallick, G.E. Tiam1 regulates cell adhesion, migration and apoptosis in colon tumor cells. Clin. Exp. Metastasis 2006, 23, 301–313. [Google Scholar] [CrossRef]
- Ding, Y.; Chen, B.; Wang, S.; Zhao, L.; Chen, J.; Ding, Y.; Chen, L.; Luo, R. Overexpression of Tiam1 in hepatocellular carcinomas predicts poor prognosis of HCC patients. Int. J. Cancer 2009, 124, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, S.T.; Tsai, M.H.; Chen, C.L.; Lee, J.X.; Jao, T.M.; Yu, S.L.; Yen, S.J.; Yang, Y.C. NDST4 Is a Novel Candidate Tumor Suppressor Gene at Chromosome 4q26 and Its Genetic Loss Predicts Adverse Prognosis in Colorectal Cancer. PLoS ONE 2013, 8, e67040. [Google Scholar] [CrossRef]
- Meurs, E.F.; Galabru, J.; Barber, G.N.; Katze, M.G.; Hovanessian, A.G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 1993, 90, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Shir, A.; Levitzki, A. Inhibition of glioma growth by tumor-specific activation of double-stranded RNA–dependent protein kinase PKR. Nat. Biotechnol. 2002, 20, 895–900. [Google Scholar] [CrossRef]
- Kim, T.H.; Cho, S.G. Kisspeptin inhibits cancer growth and metastasis via activation of EIF2AK2. Mol. Med. Rep. 2017, 16, 7585–7590. [Google Scholar] [CrossRef]
- Kim, S.H.; Forman, A.P.; Mathews, M.B.; Gunnery, S. Human breast cancer cells contain elevated levels and activity of the protein kinase, PKR. Oncogene 2000, 19, 3086–3094. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Kunkeaw, N.; Lee, Y.S. Protein kinase R and its cellular regulators in cancer: An active player or a surveillant? WIREs RNA 2019, 11, e1558. [Google Scholar] [CrossRef]
- Garcia, M.A.; Gil, J.; Ventoso, I.; Guerra, S.; Domingo, E.; Rivas, C.; Esteban, M. Impact of Protein Kinase PKR in Cell Biology: From Antiviral to Antiproliferative Action. Microbiol. Mol. Biol. Rev. 2006, 70, 1032–1060. [Google Scholar] [CrossRef]
- Schmit, K.; Michiels, C. TMEM Proteins in Cancer: A Review. Front. Pharmacol. 2018, 9, 1345. [Google Scholar] [CrossRef]
- Wang, X.; Pankratz, V.S.; Fredericksen, Z.; Tarrell, R.; Karaus, M.; McGuffog, L.; Pharaoh, P.D.; Ponder, B.A.; Dunning, A.M.; Peock, S.; et al. Common variants associated with breast cancer in genome-wide association studies are modifiers of breast cancer risk in BRCA1 and BRCA2 mutation carriers. Hum. Mol. Genet. 2010, 19, 2886–2897. [Google Scholar] [CrossRef]
- Giovannone, A.J.; Winterstein, C.; Bhattaram, P.; Reales, E.; Low, S.H.; Baggs, J.E.; Xu, M.; Lalli, M.A.; Hogenesch, J.B.; Weimbs, T. Soluble syntaxin 3 functions as a transcriptional regulator. J. Biol. Chem. 2018, 293, 5478–5491. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Han, L.; Ma, J.; Yang, C.; Su, R.; He, J. STX3 represses the stability of the tumor suppressor PTEN to activate the PI3K-Akt-mTOR signaling and promotes the growth of breast cancer cells. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2018, 1864, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Shi, R.; Xiang, W.; Kang, X.; Tang, B.; Li, C.; Gao, L.; Zhang, X.; Zhang, L.; Dai, R.; et al. The Agpat4/LPA axis in colorectal cancer cells regulates antitumor responses via p38/p65 signaling in macrophages. Signal Transduct. Target. Ther. 2020, 5, 24. [Google Scholar] [CrossRef]
- Launonen, V.; Stenback, F.; Puistola, U.; Bloigu, R.; Huusko, P.; Kytola, S.; Kauppila, A.; Winqvist, R. Chromosome 11q22.3-q25 LOH in Ovarian Cancer: Association with a More Aggressive Disease Course and Involved Subregions. Gynecol. Oncol. 1998, 71, 299–304. [Google Scholar] [CrossRef]
- Gentile, M.; Wiman, A.; Thorstenson, S.; Loman, N.; Borg, A.; Wingren, S. Deletion mapping of chromosome segment 11q24-q25, exhibiting extensive allelic loss in early onset breast cancer. Int. J. Cancer 2001, 92, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wu, Y.; Meng, Q.; Xia, Z. Elevated microRNA-25 inhibits cell apoptosis in lung cancer by targeting RGS3. Vitr. Cell. Dev. Biol.-Anim. 2015, 52, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Esparza, A.; Bartoschek, M.; Gialeli, C.; Okroj, M.; Owen, S.; Jirstrom, K.; Orimo, A.; Jiang, W.G.; Pietras, K.; Blom, A.M. Complement inhibitor CSMD1 acts as tumor suppressor in human breast cancer. Oncotarget 2016, 7, 76920–76933. [Google Scholar] [CrossRef] [PubMed]
- Kamal, M.; Shaaban, A.M.; Zhang, L.; Walker, C.; Gray, S.; Thakker, N.; Toomes, C.; Speirs, V.; Bell, S.M. Loss of CSMD1 expression is associated with high tumour grade and poor survival in invasive ductal breast carcinoma. Breast Cancer Res. Treat. 2009, 121, 555–563. [Google Scholar] [CrossRef]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef]
- Conte, N.; Delaval, B.; Ginestier, C.; Ferrand, A.; Isnardon, D.; Larroque, C.; Prigent, C.; Séraphin, B.; Jacquemier, J.; Birnbaum, D. TACC1-chTOG-Aurora A protein complex in breast cancer. Oncogene 2003, 22, 8102–8116. [Google Scholar] [CrossRef]
- Ma, C.; Quesnelle, K.M.; Sparano, A.; Rao, S.; Park, M.S.; Cohen, M.A.; Wang, Y.; Samanta, M.; Kumar, M.S.; Aziz, M.U.; et al. Characterization CSMD1 in a large set of primary lung, head and neck, breast and skin cancer tissues. Cancer Biol. Ther. 2009, 8, 907–916. [Google Scholar] [CrossRef]
- Toyooka, K.O.; Toyooka, S.; Virmani, A.K.; Sathyanarayana, U.G.; Euhus, D.M.; Gilcrease, M.; Minna, J.D.; Gazdar, A.F. Loss of expression and aberrant methylation of the CDH13 (H-cadherin) gene in breast and lung carcinomas. Cancer Res. 2001, 61, 4556–4560. [Google Scholar]
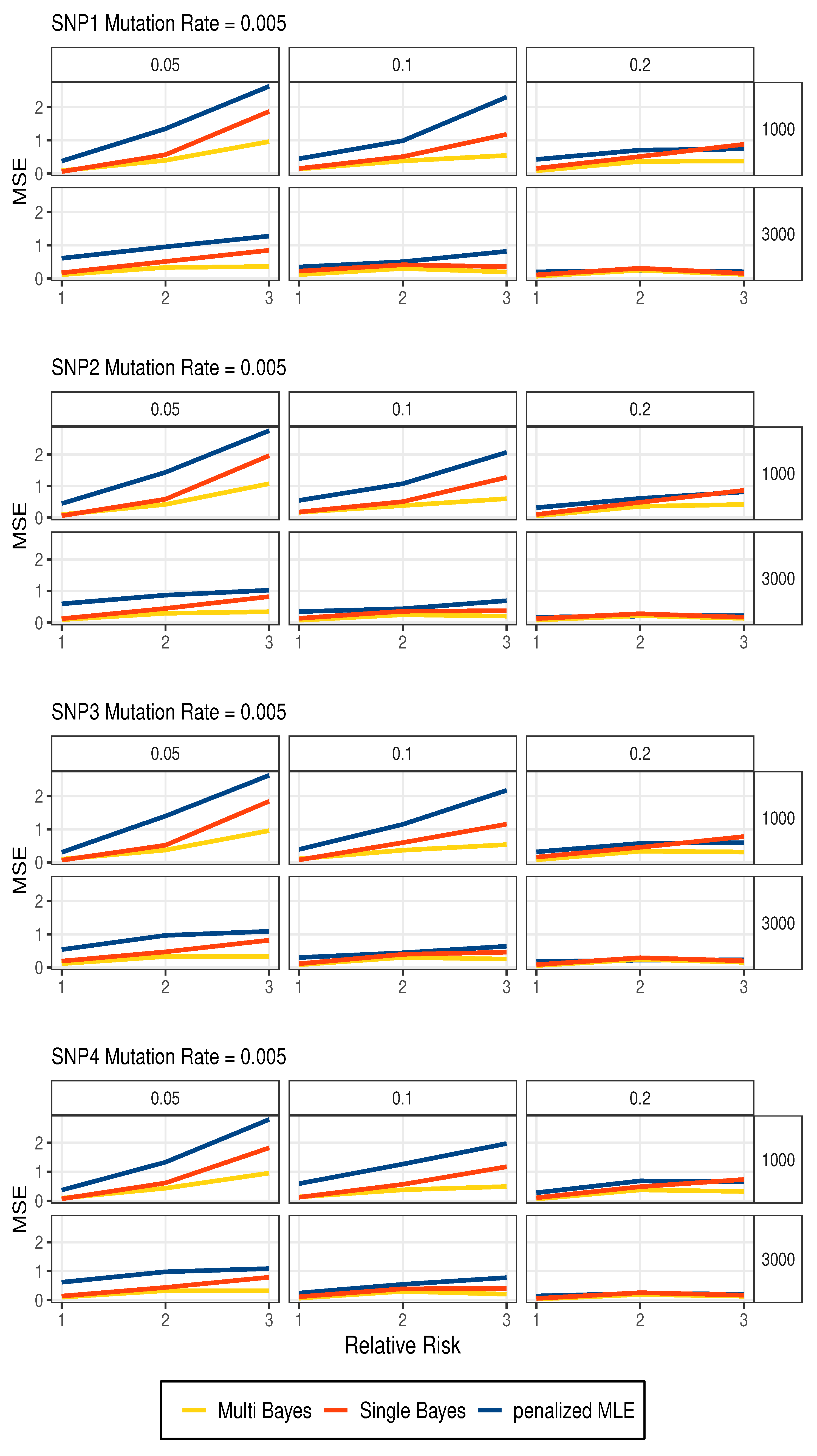

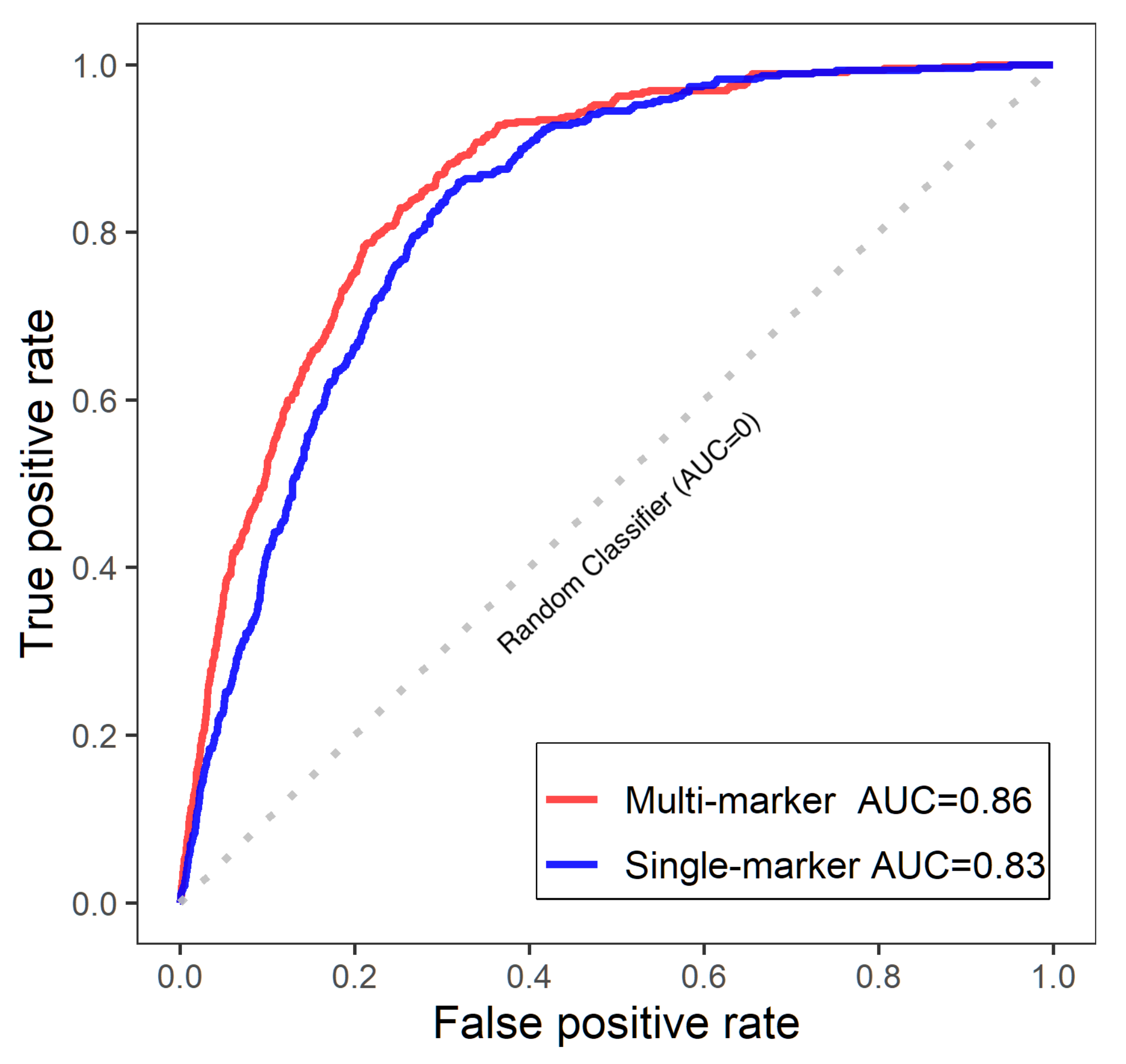
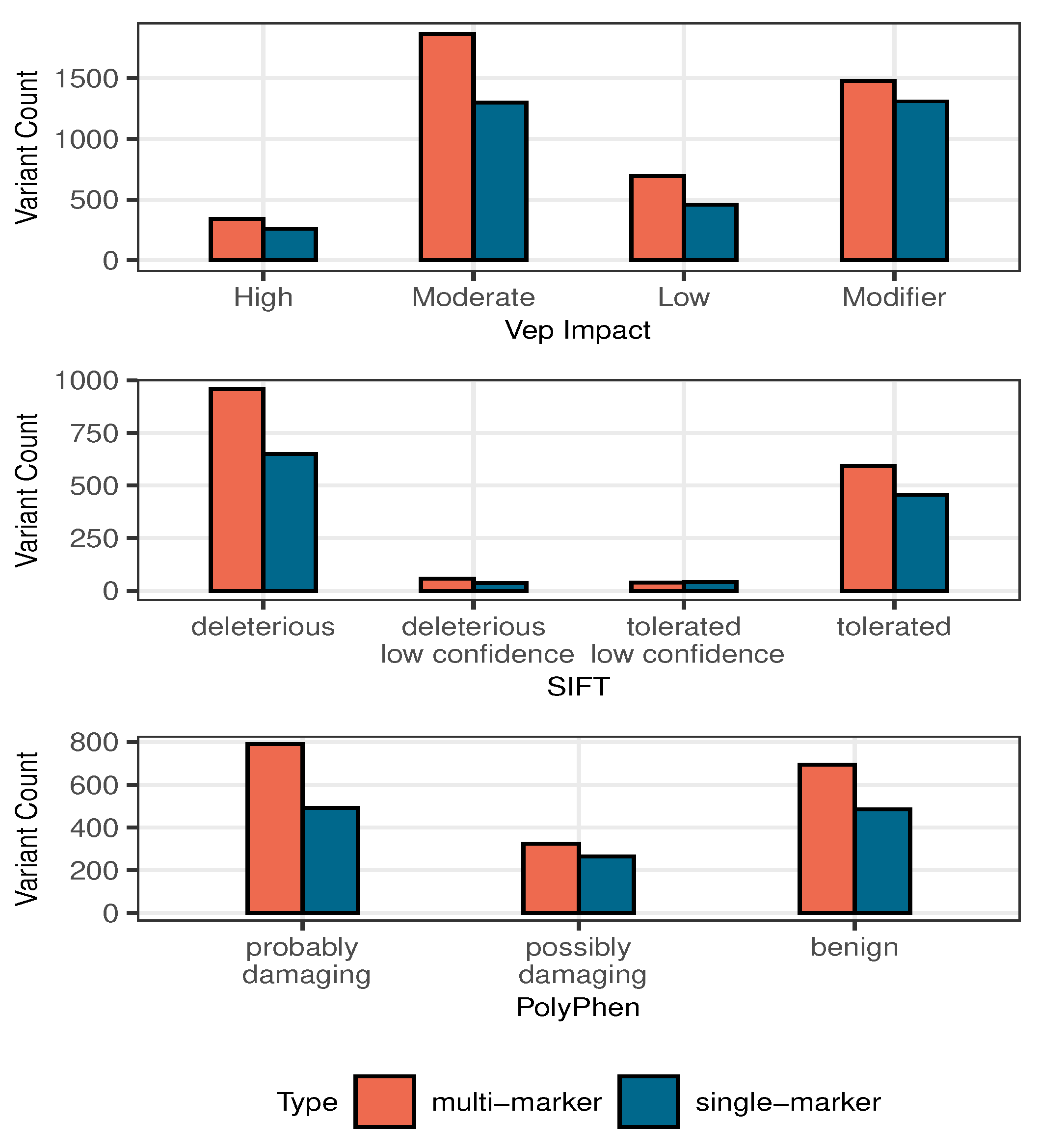
| n | SNP | Allele | Multi Bayes | Single Bayes | Penalized MLE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq | = 1 | = 2 | = 3 | = 1 | = 2 | = 3 | = 1 | = 2 | = 3 | ||
| 1000 | SNP1 | 0.05 | 0.14 | 0.31 | 0.64 | 0.24 | 0.32 | 0.45 | 0.26 | 0.13 | 0.09 |
| 0.10 | 0.14 | 0.38 | 0.82 | 0.23 | 0.36 | 0.59 | 0.18 | 0.10 | 0.21 | ||
| 0.20 | 0.07 | 0.39 | 0.91 | 0.18 | 0.34 | 0.65 | 0.18 | 0.09 | 0.55 | ||
| SNP2 | 0.05 | 0.14 | 0.31 | 0.62 | 0.23 | 0.31 | 0.44 | 0.30 | 0.16 | 0.12 | |
| 0.10 | 0.15 | 0.39 | 0.81 | 0.25 | 0.38 | 0.57 | 0.22 | 0.13 | 0.18 | ||
| 0.20 | 0.07 | 0.40 | 0.91 | 0.15 | 0.35 | 0.67 | 0.18 | 0.10 | 0.55 | ||
| SNP3 | 0.05 | 0.14 | 0.32 | 0.64 | 0.23 | 0.34 | 0.47 | 0.26 | 0.12 | 0.14 | |
| 0.10 | 0.14 | 0.36 | 0.82 | 0.21 | 0.32 | 0.59 | 0.20 | 0.11 | 0.20 | ||
| 0.20 | 0.07 | 0.41 | 0.92 | 0.18 | 0.37 | 0.67 | 0.15 | 0.09 | 0.53 | ||
| SNP4 | 0.05 | 0.14 | 0.31 | 0.64 | 0.24 | 0.31 | 0.47 | 0.19 | 0.12 | 0.11 | |
| 0.10 | 0.14 | 0.38 | 0.83 | 0.23 | 0.35 | 0.59 | 0.21 | 0.13 | 0.15 | ||
| 0.20 | 0.07 | 0.44 | 0.92 | 0.15 | 0.42 | 0.70 | 0.19 | 0.15 | 0.58 | ||
| 3000 | SNP1 | 0.05 | 0.12 | 0.45 | 0.91 | 0.22 | 0.38 | 0.68 | 0.24 | 0.04 | 0.17 |
| 0.10 | 0.08 | 0.55 | 0.98 | 0.19 | 0.44 | 0.84 | 0.15 | 0.17 | 0.80 | ||
| 0.20 | 0.05 | 0.62 | 0.99 | 0.12 | 0.47 | 0.91 | 0.11 | 0.49 | 0.99 | ||
| SNP2 | 0.05 | 0.11 | 0.44 | 0.91 | 0.20 | 0.38 | 0.67 | 0.18 | 0.03 | 0.14 | |
| 0.10 | 0.06 | 0.55 | 0.98 | 0.14 | 0.42 | 0.84 | 0.16 | 0.13 | 0.82 | ||
| 0.20 | 0.05 | 0.62 | 0.99 | 0.13 | 0.46 | 0.90 | 0.18 | 0.42 | 1.00 | ||
| SNP3 | 0.05 | 0.12 | 0.44 | 0.91 | 0.23 | 0.40 | 0.68 | 0.12 | 0.05 | 0.16 | |
| 0.10 | 0.06 | 0.55 | 0.97 | 0.12 | 0.44 | 0.82 | 0.16 | 0.12 | 0.83 | ||
| 0.20 | 0.04 | 0.64 | 0.99 | 0.10 | 0.51 | 0.90 | 0.09 | 0.50 | 0.99 | ||
| SNP4 | 0.05 | 0.11 | 0.46 | 0.91 | 0.19 | 0.42 | 0.69 | 0.18 | 0.02 | 0.12 | |
| 0.10 | 0.06 | 0.55 | 0.98 | 0.14 | 0.44 | 0.82 | 0.15 | 0.15 | 0.77 | ||
| 0.20 | 0.03 | 0.62 | 0.99 | 0.09 | 0.45 | 0.90 | 0.18 | 0.48 | 1.00 | ||
| Summary | Min | Q1 | Median | Mean | Q3 | Max |
|---|---|---|---|---|---|---|
| SNP Counts | 1 | 3 | 8 | 42 | 39 | 1242 |
| Gene Length | 21 | 16,215 | 47,282 | 159,292 | 166,206 |
| Summary | Min | Q1 | Median | Mean | Q3 | Max |
|---|---|---|---|---|---|---|
| SNP counts | 2 | 3 | 5 | 6 | 8 | 40 |
| Gene Name | Chr | Gene Status |
|---|---|---|
| IL7 | chr8 | 0.975 |
| TIAM1 | chr21 | 0.972 |
| CKAP2L | chr2 | 0.961 |
| TTC28 | chr22 | 0.956 |
| NDST4 | chr4 | 0.952 |
| EIF2AK2 | chr2 | 0.952 |
| CACNB4 | chr2 | 0.95 |
| PARD3B | chr2 | 0.946 |
| TMEM117 | chr12 | 0.944 |
| ATP6V0D1 | chr16 | 0.943 |
| Gene Name | Chr | Gene Status |
|---|---|---|
| LINC00383 | chr13 | 0.999 |
| KIRREL3 | chr11 | 0.999 |
| STX3 | chr11 | 0.999 |
| AGPAT4 | chr6 | 0.999 |
| SYCE1 | chr10 | 0.997 |
| RCBTB1 | chr13 | 0.997 |
| PKNOX2 | chr11 | 0.997 |
| RGS3 | chr9 | 0.997 |
| GCSH | chr16 | 0.997 |
| CSMD1 | chr8 | 0.996 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bu, Y.; Chen, M.; Xuan, Z.; Wang, X. A Bayesian Model for Paired Data in Genome-Wide Association Studies with Application to Breast Cancer. Entropy 2025, 27, 1077. https://doi.org/10.3390/e27101077
Bu Y, Chen M, Xuan Z, Wang X. A Bayesian Model for Paired Data in Genome-Wide Association Studies with Application to Breast Cancer. Entropy. 2025; 27(10):1077. https://doi.org/10.3390/e27101077
Chicago/Turabian StyleBu, Yashi, Min Chen, Zhenyu Xuan, and Xinlei Wang. 2025. "A Bayesian Model for Paired Data in Genome-Wide Association Studies with Application to Breast Cancer" Entropy 27, no. 10: 1077. https://doi.org/10.3390/e27101077
APA StyleBu, Y., Chen, M., Xuan, Z., & Wang, X. (2025). A Bayesian Model for Paired Data in Genome-Wide Association Studies with Application to Breast Cancer. Entropy, 27(10), 1077. https://doi.org/10.3390/e27101077







