Abstract
Entropy algorithms are widely applied in signal analysis to quantify the irregularity of data. In the realm of two-dimensional data, their two-dimensional forms play a crucial role in analyzing images. Previous works have demonstrated the effectiveness of one-dimensional increment entropy in detecting abrupt changes in signals. Leveraging these advantages, we introduce a novel concept, two-dimensional increment entropy (IncrEn2D), tailored for analyzing image textures. In our proposed method, increments are translated into two-letter words, encoding both the size (magnitude) and direction (sign) of the increments calculated from an image. We validate the effectiveness of this new entropy measure by applying it to MIX2D(p) processes and synthetic textures. Experimental validation spans diverse datasets, including the Kylberg dataset for real textures and medical images featuring colon cancer characteristics. To further validate our results, we employ a support vector machine model, utilizing multiscale entropy values as feature inputs. A comparative analysis with well-known bidimensional sample entropy (SampEn2D) and bidimensional dispersion entropy (DispEn2D) reveals that IncrEn2D achieves an average classification accuracy surpassing that of other methods. In summary, IncrEn2D emerges as an innovative and potent tool for image analysis and texture characterization, offering superior performance compared to existing bidimensional entropy measures.
1. Introduction
Entropy, as defined in information theory by Shannon in 1949, quantifies the level of uncertainty present in data [1]. It measures the amount of information contained within a variable. Extracting information from a variable is of significant interest to scientists across various fields (see, e.g., [2,3,4]). In the context of information theory, several entropy measures, e.g., approximate entropy (ApEn) [5], sample entropy (SampEn) [6], permutation entropy (PermEn) [7], distribution entropy (DistrEn) [8], dispersion entropy (DispEn) [9], and increment entropy (IncrEn) [10] have been developed to extract meaningful information from unidimensional signals. These entropy measures allow scientists to analyze and understand the underlying patterns, complexity, and dynamics present in the data [11]. However, new one-dimensional entropy measures are still being proposed to overcome the drawbacks of existing ones.
ApEn [5] assesses pattern regularity by measuring the likelihood that similar patterns of a certain length will remain similar when compared to other subsequences. However, ApEn has some drawbacks. It tends to exhibit a bias toward regularity because of self-matching, lacks relative consistency, and is sensitive to data length. SampEn [6] addresses these issues by excluding self-matches and demonstrates relative consistency. It is less dependent on data length. However, for very short data, SampEn may still lead to unreliable or undefined values. PermEn was also developed as a complexity measure based on ordinal data to quantify the complexity of the given data [7]. Following that, Liu and Wang [12] proposed fine-graining PermEn. By fine-graining the partition, this modified version of PermEn responds more quickly to abrupt changes in amplitude. However, PermEn only considers the order of values in a time series, disregarding changes in magnitude between the elements. Even in the modified version of PermEn, when the data have equal-value elements, it either ignores them or treats them all as one symbol. Besides this, other entropy measures for complexity analysis were introduced like distribution entropy [8] and dispersion entropy [9], but none of them take into account the changes in magnitude between adjacent elements.
To address these limitations, IncrEn was proposed by Liu et al. [10]. IncrEn ranks the magnitudes of the variations between adjacent elements based on a precision factor and the standard deviation of the data. This quantifies the changes in magnitude between adjacent elements and also considers the order of the elements. IncrEn captures the local dynamics and changes between consecutive data points, making it effective for detecting abrupt changes or irregular patterns in the data. Compared to SampEn and PermEn, IncrEn is more adept at capturing and characterizing the structural information present in time series data [13]. IncrEn addresses the issue of undefined values that can occur in SampEn calculations. IncrEn does not suffer from the equal-value problem encountered in some other entropy measures. Moreover, it demonstrates robustness to noise, making it more suitable for analyzing noisy or complex data. Liu et al. [10] compared IncrEn with SampEn and PermEn, investigating the impact of Gaussian noise. They found that IncrEn displayed greater sensitivity to hidden changes in time series compared to PermEn, which treated analogous patterns as identical. Furthermore, both IncrEn and PermEn exhibited good invariance when applied to Gaussian noise sequences, whereas the SampEn results fluctuated significantly [10]. Furthermore, IncrEn showed better performance in seizure detection from real epileptic EEG signals [10]. It also performed well in detecting fault vibration signals [10].
Entropy measures for graph signals have recently been designed [14,15]. Some two-dimensional (2D) entropy algorithms have also been proposed recently to estimate the irregularity of textures or images (see, e.g., [16,17,18,19,20]). Three-dimensional measures have been published too [21]. The two-dimensional entropy measures have been applied to solve many texture problems in multiple domains [22]. Although this research area is still recent, the results obtained with the bidimensional entropy measures are very promising in various domains (see, e.g., [23,24]). The 2D measures can be used to study texture and analyze the irregular structures of images in a similar way to that implemented for signals [25]. Until now, few of the bidimensional entropy measures have been implemented, and many tasks are still to be explored in this way. Moreover, multiscale entropy measures have also emerged as a significant tool in the domain of image analysis, providing a sophisticated means to assess intricate patterns within images. These measures allow us to explore the complex nature of images by quantifying their information across multiple scales.
Recently, multivariate multiscale increment entropy (MMIE) was introduced by Wang et al. [13]. In [26], they proposed the multiscale IncrEn (MIE) and showed that MIE aligns better with the complexity loss theory of disease and aging in various physiological signals at different scales compared to the popular multiscale SampEn (MSE) method and refined composite multiscale dispersion entropy (RCMDE). MIE exhibits superior discrimination ability for physiological conditions affecting time series complexity, and its robustness to parameters outperforms MSE and RCMDE. Additionally, MIE is the most efficient in terms of time consumption, followed by RCMDE and MSE [26].
Due to the above-mentioned advantages of IncrEn in comparison with PermEn or SampEn, we introduce two-dimensional increment entropy (IncrEn2D). Inspired by the ability of IncrEn in detecting changes in both the size (magnitude) and direction (sign) of the increments calculated from a signal, we believe that this extension, along with its multiscale variant for images, holds promise for texture pattern analysis. This approach may offer advantages such as better characterization of structural information, improved discrimination ability for diverse physiological conditions, and enhanced robustness to noise and parameter variations.
This new measure was first studied through the analysis of synthetic images. Then, real datasets, including colon cancer images, were processed, and the results were compared with those of other, already existing 2D entropy-based measures.
2. Methods
In the following, the method we propose (IncrEn2D) is described, together with its multiscale version. The datasets processed and the computational steps are also detailed.
2.1. Two-Dimensional Increment Entropy, IncrEn2D
The new entropy measure that we propose here, IncrEn2D, is the extension of 1D IncrEn [10] to its bidimensional form. Its algorithm is defined as follows. Let us consider a grayscale image I represented as a matrix of size , where W is the width (rows) and H, the height (columns). The calculation of IncrEn2D for the given image I consists of the following steps:
- 1.
- First, an increment image V(I) is formed from the original image I. This can be performed in two different ways, as illustrated in Figure 1:
- (a)
- Row-wise increment image. In this case, pixel values are subtracted in adjacent rows so the size of the increment image would be .
- (b)
- Column-wise increment image. In this case, pixel values are subtracted in adjacent columns, and thus, the increment image would be .
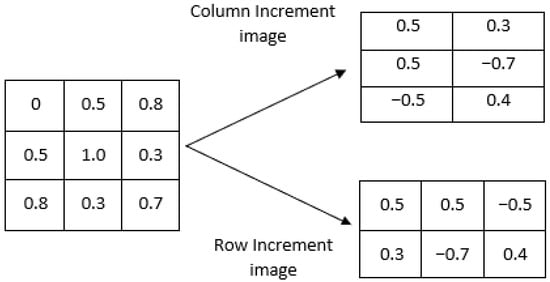 Figure 1. Illustration of the two possible methods for constructing the increment image V(I).
Figure 1. Illustration of the two possible methods for constructing the increment image V(I).
- 2.
- Given a positive integer m, divide the increment image into overlapping blocks of size (see Figure 2, where ). We define Vm(i,j) as the m-length square block in image V(I), where the indices range from row i to and from column j to . The total number of blocks can be computed as for a row-wise increment image and for a column-wise increment image.
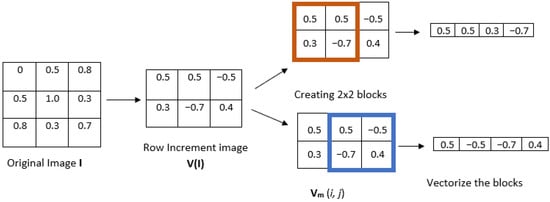 Figure 2. Illustration of steps 1 and 2 of IncrEn2D, using the row-wise method to compute the increment image.
Figure 2. Illustration of steps 1 and 2 of IncrEn2D, using the row-wise method to compute the increment image. - 3.
- Vectorize each block to obtain an increment vector Vm(i,j) and calculate the nth unique word sequence Wn for each Vm(i,j), for a given quantifying resolution R. Each element of the increment vector Vm(i,j) is mapped into word pattern composed of two parts: sign and magnitude . The sign function produces different results depending on the input element. If the input element is greater than 0, the function returns 1. If the input element is equal to 0, the function returns 0. If the input element is less than 0, the function returns −1. The magnitude that represents the extent of difference between these neighboring pixels is determined by the resolution parameter R. Subsequently, every increment vector is transformed into a pattern vector comprising elements (considering both sign and magnitude). To gain a complete understanding of creating a word pattern, let us consider the example of the template vectors illustrated in Figure 3, where the quantifying resolution R is taken as 4. The sign is calculated as = sign(Vm(i,j)), and magnitude is calculated using the following equation:
 Figure 3. Template vectors illustrating the steps to obtain words.
Figure 3. Template vectors illustrating the steps to obtain words. - 4.
- Count the total number of instances of every unique nth word pattern .
- 5.
- Compute the relative frequency of each unique word using the following equation:where is the total number of words for the given increment image.
- 6.
- Calculate the two-dimensional increment entropy, IncrEn2D, of image I using the following equation:
With a specific m and R, each increment vector yields distinct patterns. Thus, IncrEn2D is bounded within . While this upper bound of IncrEn2D reflects theoretical maximum information content, it is important to note that the formulation of IncrEn2D diverges from classical entropy principles. While classical Shannon entropy focuses on individual pixels, IncrEn2D extends this concept by analyzing blocks of pixels and is influenced by a quantification parameter (R). Thus, while it retains elements of Shannon entropy, it introduces a novel approach that considers spatial dependencies within images.
2.2. Multiscale 2D Increment Entropy
One- and two-dimensional entropy measures are commonly employed to quantify the irregularity within signals or images at a specific scale. These methods are adept at identifying repetitive structures and are particularly effective in scenarios of complete randomness, such as white noise. However, their sensitivity to high-frequency components can lead to an oversight of multiple inherent scales in the data [17]. To address this limitation, researchers have introduced multiscale entropy-based approaches. These methods have demonstrated interesting results in the domain of texture classification (see, e.g., [16]). Moreover, they offer the capability to evaluate irregularities across different scales, providing a comprehensive insight into the complexity of a given signal or image. Therefore, we herein propose the multiscale IncrEn2D (MIncrEn2D) to measure the complexity of image textures at different scale factors . The two-step process for calculating MIncrEn2D is described as follows:
- 1.
- Coarse-graining procedure: The non-overlapping moving-average coarse-graining procedure is used in this study to divide the image into multiple scales. In this procedure, a non-overlapping window of size sweeps across the entire image, and the pixels within each window are averaged. These resulting average values shape the coarse-grained images. These images, while not being the subsets of the original, encapsulate information about the entire original image. When considering an image I of dimensions , a coarse-grained image G(τ) at scale factor is expressed mathematically as follows:where , and is the scale factor. For scale factor 1, the coarse-grained image corresponds to the original image. At higher scale factor, the dimensions of the coarse-grained image are reduced by a factor of .
- 2.
- Application of IncrEn2D: In the subsequent step, IncrEn2D is applied individually on each of the coarse-grained images.
2.3. Evaluation Images and Computational Steps
To evaluate the performances of IncrEn2D, MIX2D(p) processes, as well as artificial periodic and synthesized textures, were used. Real datasets were also processed and studied.
2.3.1. Two-Dimensional MIX Process (MIX2D(p))
The creation of MIX2D(p) processes is based on the definition of its unidimensional version, MIX1D(p). MIX2D(p) denotes images where the parameter p corresponds to the level of white noise. As p goes from 0 (representing completely periodic sinusoidal images) to 1 (indicating highly irregular images), the images of size exhibit different degrees of spatial irregularity, described as , , where , and represents a uniform two-dimensional matrix of white noise, spanning from to . In this context, represents a random variable that takes the value 1 with a probability of p and the value 0 with a probability of [25]. Consequently, as the value of p increases, the images become more irregular; see Figure 4.
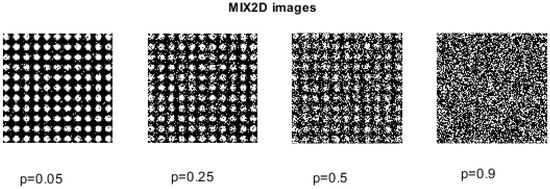
Figure 4.
MIX2D(p) family of images with varying noise levels, .
To assess the effectiveness of IncrEn2D in quantifying images with varying degrees of irregularity and randomness, different MIX2D(p) images were created and used.
2.3.2. Artificial Periodic and Synthesized Texture Images
To assess the behavior of IncrEn2D during the transformation of periodic textures into their synthesized counterparts, four sets of periodic textures and their corresponding synthesized textures were used from [27].
2.3.3. Image with Additive Noise
To examine the influence of two-dimensional white Gaussian noise (WGN2D) and salt-and-pepper noise (SPN2D) on IncrEn2D, we conducted an evaluation using the widely recognized Lena image with size 256 × 256 pixels, as a standard reference. To achieve image normalization, we applied a two-step process. After converting the image to its grayscale form, we subtracted the mean of the image from each pixel value. Next, we divided the resulting values by the standard deviation of the image. Following the normalization of the image within the range of 0 to 1, we systematically introduced various levels of WGN2D with a mean equal to 0 and variance values of 0.01, 0.03, 0.05, and 0.07. The noise was added to almost every pixel of the image. Furthermore, we integrated SPN2D with noise density values of 0.01, 0.05, and 0.09 into the aforementioned normalized reference image. In the case of SPN2D, the noise density, d, determines the amount of noise applied to the image, with d being multiplied by the number of pixels.
2.3.4. Effect of Different Standard Deviation Computations on IncrEn2D
We also explored the influence of the standard deviation computation in Equation (1) on the IncrEn2D results using MIX2D(p) processes. More precisely, we studied IncrEn2D when the standard deviation of the increment block (Vm(i,j)) is used and when the standard deviation of the whole increment image is used in Equation (1).
2.3.5. Effect of Row-Wise and Column-Wise Increment Images
As mentioned above, the two main approaches used to calculate the increment images (the first step of the algorithm) are the row-wise and column-wise increment methods. In this work, we evaluated the influence of these two different ways of computation by creating simulated images with vertical and horizontal stripes (see Figure 5). For the vertical and horizontal stripe pattern images, we set the image dimensions to 256 × 256 pixels, set the stripe width (vertical stripes) and strip height (horizontal stripes), and calculated the number of stripes based on the image width. Then, we generated ten different patterns of each category by creating random binary pattern arrays, where each element represents the color of a stripe (0 for black, 1 for white). We constructed the vertical and horizontal stripe pattern images using these patterns by setting pixel values accordingly. We also carried out an experiment with pure white and black images, where all pixel values were equal to 255 for pure white and 0 for pure black, to evaluate the performance of our proposed method in these extreme cases.
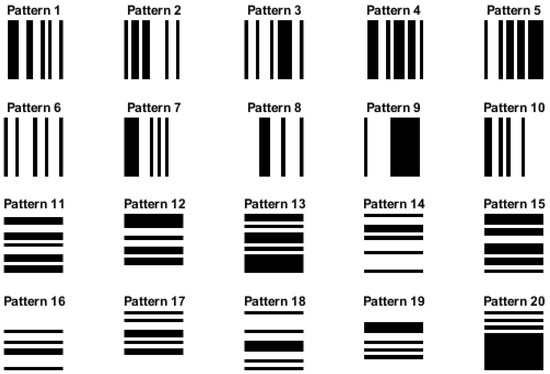
Figure 5.
Simulated images of size 256 × 256 pixels with vertical stripes (patterns 1–10) and horizontal stripes (patterns 11–20).
2.3.6. Kylberg Real Texture Dataset
In this study, a portion of the Kylberg texture dataset was used. The dataset can be accessed publicly from [28]. Specifically, we chose six distinct categories of images. These images represent various fabrics and surfaces: canvas1, cushion1, linsseeds1, sand1, stone1, and seat2 (see Figure 6. Each sample is 576 × 576 pixels in size).

Figure 6.
One image (576 × 576 pixels) of each of the six selected categories from the Kylberg dataset.
2.3.7. Colon Cancer Dataset
Colon adenocarcinoma stands out as the most prevalent forms of colon cancer, accounting for over 95% of all colon cancer instances. It occurs when a specific type of tissue growth, known as adenoma, develops in the large intestine, eventually transforming into cancer [29]. Multiscale increment entropy (MIncrEn2D) was applied on part of the lung and colon cancer histopathological image dataset known as the LC25000 dataset for the validation of the approach. This dataset comprises 25,000 color images showcasing five distinct lung and colon tissue types [30]. For this study, we only considered a portion of the colon cancer part (160 images of cancerous and normal tissue). Some samples of the cancerous and normal tissues from the dataset are presented in Figure 7.
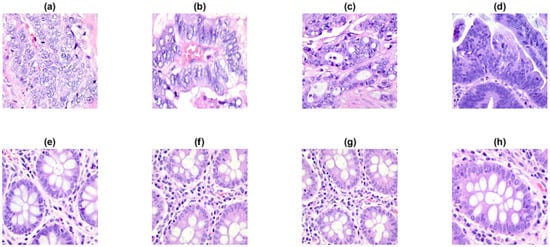
Figure 7.
A few sample images (768 × 768 pixels) from the LC25000 dataset: (a–d) colon cancer tissues and (e–h) normal tissues.
The original images, initially sized at pixels, were preprocessed to ensure uniformity and reduce computational load prior to entropy calculation. During this stage, all images were converted to grayscale images and resized to pixels using bilinear interpolation prior to entropy calculation. Multiscale analysis considered scale factors of up to 4, resulting in the smallest image size of pixels. MIncrEn2D provided entropy values across all scales for both cancerous and normal tissues. This study utilized these entropy values as features for classifying tissues, employing a support vector machine (SVM) with a radial basis kernel function due to its ability to capture intricate relationships. The dataset comprised 160 histopathological images from LC25000, evenly divided between cancerous and normal tissues. Out of these, 70% of the dataset (112 images) were allocated for training, while the remaining 30% (48 images) were reserved for testing. The primary features for classification were entropy values, derived from 4 different scale factors. The SVM classifier was trained on the training dataset and evaluated using the test dataset. This procedure was repeated 5 times and took the average values to ensure consistency in evaluation. To evaluate the classifiers performance, metrics such as the classification accuracy, precision, recall, and F1-score were computed. The results obtained using features given by IncrEn2D were compared with the results given by the now well-known multiscale bidimensional sample entropy (SampEn2D) [25] and multiscale bidimensional dispersion entropy (DispEn2D) [19] measures.
3. Results
This section details the results obtained from the aforementioned images.
3.1. Two-Dimensional MIX Process (MIX2D(p))
We created 256 × 256 pixel MIX2D(p) images by varying the parameters— and p = 0 to 1 with a step size of 0.05—resulting in 21 different p values. Figure 8 illustrates that higher p values correspond to greater IncrEn2D entropy. The entropy increase from to decreases with higher m: for , the range is 1.6435 to 7.6724, while for , it is 0.8280 to 3.2162. Smaller block sizes (e.g., ) capture finer details, while larger sizes encompass broader patterns.
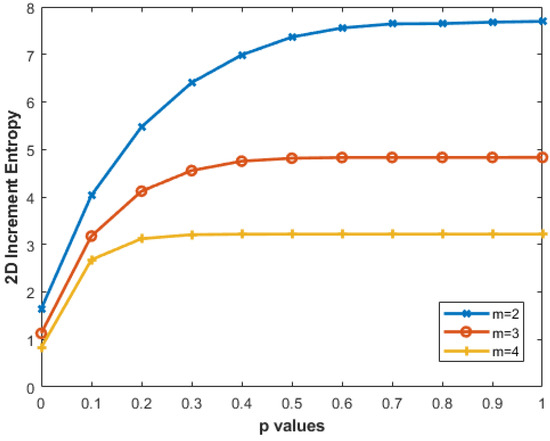
Figure 8.
IncrEn2D for MIX2D(p) images of size 256 × 256 pixels for R=4 and , and 4.
Figure 9 shows that increasing R from 2 to 6 results in a steeper curve, especially notable for . Higher R values provide finer detail, but diminishing differences suggest a saturation point where increased resolution has limited impact on pattern understanding.
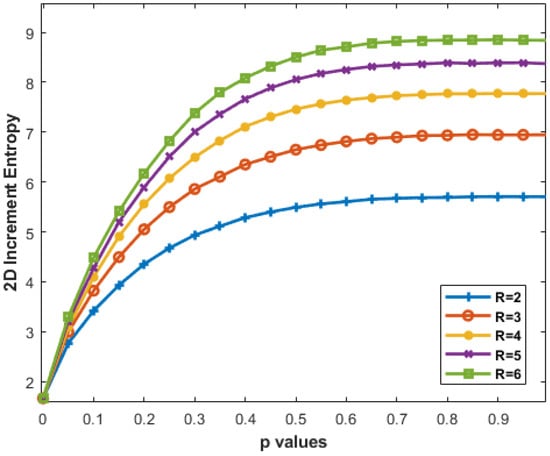
Figure 9.
IncrEn2D for MIX2D(p) images of size 256 × 256 pixels for and varying R values from 2 to 6.
We conducted additional experiments to assess the reliability of our proposed IncrEn2D across varying image sizes, aiming to verify its performance in measuring image irregularity. Specifically, we computed IncrEn2D for MIX2D(p) while progressively increasing image sizes from to pixels, with , , and p varying from 0 to 1. The results, as depicted in Figure 10, consistently demonstrate stable results with changes in image size.
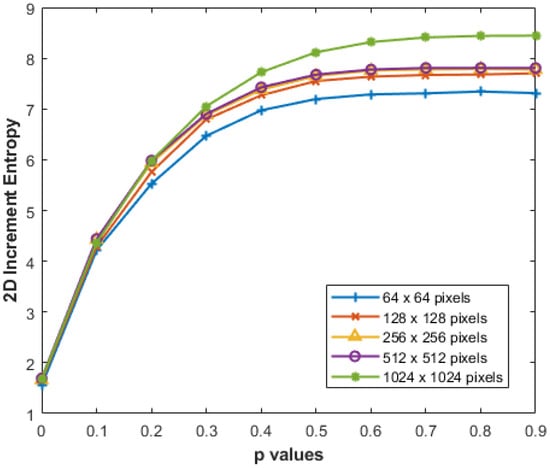
Figure 10.
IncrEn2D for MIX2D(p) images of different sizes for and .
3.2. Artificial Periodic and Synthesized Texture Images
The results obtained with the artificial periodic and synthesized textures are presented in Table 1. We observe that IncrEn2D values are greater for the synthesized textures compared to their corresponding periodic textures.

Table 1.
IncrEn2D for artificial periodic textures and their synthesized textures.
3.3. Image with Additive Noise
Table 2 presents the IncrEn2D values obtained from the Lena image at different levels of WGN2D and SPN2D. The results indicate that when higher variance values are applied to WGN2D, IncrEn2D values also increase. Similarly, a higher noise density in SPN2D leads to higher entropy values.

Table 2.
IncrEn2D values for Lena image on which different levels of 2D white Gaussian noise (WGN2D) and salt-and-pepper noise (SPN2D) were added. Parameters were set as and using the column-wise increment image method and the standard deviation of a block for the IncrEn2D calculation.
3.4. Effect of Different Standard Deviation Computations on IncrEn2D
It is evident from Figure 11 that the results obtained with the standard deviation of the original image I are somewhat misleading: the entropy values for MIX2D(0.9) and MIX2D(1) are lower than those of MIX2D(0.8). The standard deviation of increment block (Vm(i,j)) provides better entropy estimates than the standard deviation of increment image (V(I)). The discrepancy arises from how the standard deviation is computed: the whole increment image considers all pixels, yielding a larger variance, while the increment block focuses on a fraction, providing a more localized measure of variation.
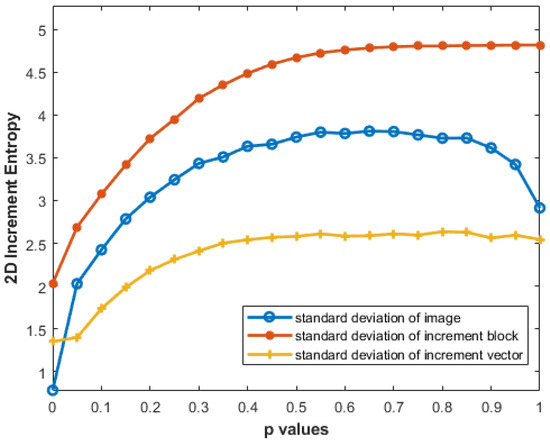
Figure 11.
Effect of different standard deviations on IncrEn2D values with and .
Figure 11 presents the effect of different standard deviation computations on IncrEn2D.
3.5. Effect of Row-Wise and Column-Wise Increment Images
Despite the differences in the calculation, it can be observed, through Figure 12, that the IncrEn2D values obtained with MIX2D(p) from both the row-wise and column-wise approaches are almost the same. This similarity in IncrEn2D values can be attributed to the inherent characteristics of MIX2D(p) images. The results obtained with horizontal and vertical stripe pattern images are presented in Table 3.
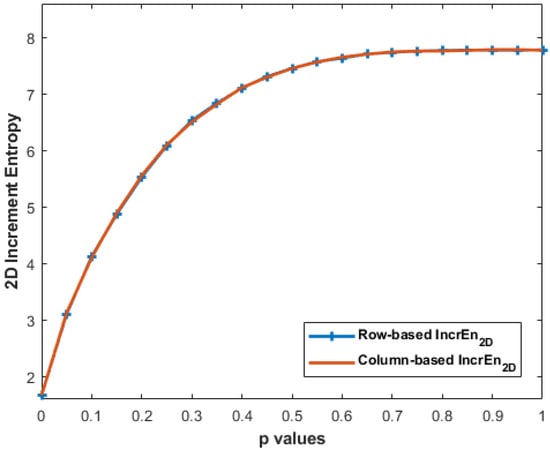
Figure 12.
IncrEn2D values with row-wise and column-wise increment images for MIX2D(p) of size 256 × 256 pixels with and .

Table 3.
IncrEn2D values for vertical and horizontal stripes pattern images. Parameters were set as and .
As anticipated, the experiment with pure white and black images resulted in IncrEn2D values of 0, as these types of images exhibit no irregularities.
3.6. Kylberg Real Texture Dataset
Table 4 presents the IncrEn2D, SampEn2D, and DispEn2D values for the selected Kylberg texture groups. The results indicate distinct entropy values for the six selected groups with almost all entropy measures except that SampEn2D shows minor differences between cushion1 and linseeds1. The same is observed for DispEn2D with sand1 and stone1.

Table 4.
IncrEn2D, SampEn2D, and DispEn2D values for six different groups of texture surfaces from the Kylberg dataset. Parameters were set as follows: for all, for IncrEn2D, of the standard deviation of an image for SampEn2D, and for DispEn2D.
3.7. Colon Cancer Dataset
The anatomical and biological characteristics of colon cancer often exhibit distinct horizontal arrangements along the colon length. For this reason, in the present experimentation, the preference was to employ row-wise increment images. The results obtained with colon cancer data can be seen in Figure 13. We observe lower IncrEn2D values for normal colon tissues and higher IncrEn2D values for cancerous tissues, irrespective of the scale factor used. This relationship between IncrEn2D and tissue types implies its potential as a valuable tool in distinguishing between normal and cancerous colon tissues.
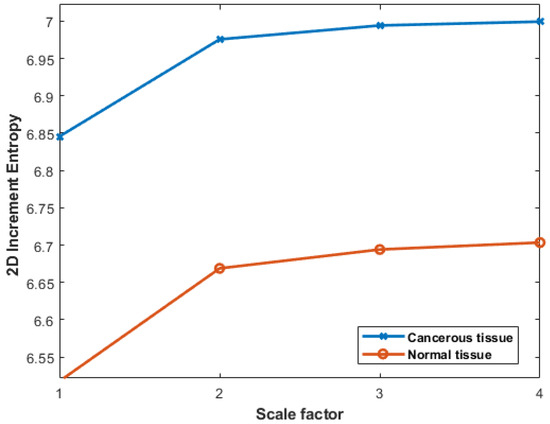
Figure 13.
Multiscale IncrEn2D values with , , and scale factors from 1 to 4 for cancerous and normal tissues.
Moreover, the results based on the SVM model show an average classification accuracy of 77.22%, compared to SampEn2D and DispEn2D, which achieved average classification accuracies of 59.31% and 47.63%, respectively, obtained using the same model (see Table 5).

Table 5.
Classification results obtained from the colon cancer data with the multiscale approaches (from scale factors 1 to 4) for IncrEn2D, SampEn2D, and DispEn2D features.
3.8. Computational Time
In this study, we also conducted a comparative analysis for IncrEn2D, SampEn2D, and DispEn2D in terms of computational time across different image sizes ranging from to pixels. We used MATLAB R2024a on a personal computer equipped with an Intel(R) Core(TM) i7-5600U CPU operating at 2.60 GHz and 8 GB RAM. To achieve this, we utilized images of varying dimensions from the Kylberg dataset. The results are presented in Table 6. We notice that DispEn2D exhibits the shortest computational time, even with larger image sizes. IncrEn2D closely follows, maintaining its computational advantage for small- to moderate-sized images. SampEn2D incurs the highest computational time, particularly with larger image sizes.

Table 6.
Comparison of computational time in seconds to calculate IncrEn2D, SampEn2D, and DispEn2D on one image with different dimensions from the Kylberg dataset. Parameters were set as follows: for all, for IncrEn2D, × standard deviation of image for SampEn2D, and
for DispEn2D.
4. Discussion
The results of this work demonstrate that IncrEn2D offers superior performance in texture analysis across various image sizes and noise conditions, providing insights into its effectiveness for diverse image types and patterns. With the MIX2D(p) images, the behavior of IncrEn2D with respect to the parameter p highlights its sensitivity to entropy changes. Specifically, higher p values correspond to greater IncrEn2D entropy, suggesting that this parameter plays a key role in capturing the complexity of textures. Moreover, the observation that entropy increases from to decrease with higher block sizes m, illustrating the scale-dependent nature of this measure. Smaller block sizes, such as , are more adept at capturing finer details, whereas larger block sizes, such as , are better suited for recognizing broader patterns, aligning with previous findings in multiscale entropy analysis.
The impact of the threshold R is equally important, with higher values yielding finer detail, though the diminishing returns beyond suggest a saturation point. This result is crucial for practical applications, as it implies that increasing R beyond certain values may offer little additional benefit in terms of pattern recognition, potentially optimizing the computational cost for future implementations. Therefore, for the final selection of the R value, one can consider the trade-off between enhanced resolution and computational complexity. Based on the observed results, we can conclude that the optimal parameter set for further experimentation would be and . This combination enables the IncrEn2D to effectively quantify the irregularity of the images under analysis.
Our findings with synthetic and periodic textures reinforce the robustness of IncrEn2D, with synthesized textures consistently exhibiting higher entropy values than their periodic counterparts. This shows the discriminative power of IncrEn2D in distinguishing between periodic and synthesized textures, while also providing a measure of the periodicity within the images. In other words, it provides a reliable method for differentiating between images that possess inherent periodic characteristics and those that have been artificially created or modified. The behavior of IncrEn2D under noise conditions further validates its stability and utility. As the variance of 2D white Gaussian noise (WGN2D) increases, so does the IncrEn2D value, confirming its sensitivity to added randomness in the image. Similarly, salt-and-pepper noise (SPN) at higher densities results in increased entropy values, reflecting its ability to quantify the complexity introduced by these types of distortions. This characteristic can be advantageous in real-world applications where noise is an inevitable factor, such as in medical imaging.
An important aspect of our analysis is the performance of IncrEn2D with different standard deviation measures. The results demonstrate that the standard deviation of the increment vector provides the best differentiation of textures, outperforming both the standard deviation of the image itself and the increment image. This localized approach captures fine-grained entropy changes more effectively. Additionally, the block-based analysis introduces spatial locality, with the standard deviation of the increment block reflecting variations within a specific neighborhood, offering more insight into image structure and patterns. In contrast, the standard deviation of the whole increment image considers variations across the entire image, potentially diluting detailed information. Furthermore, the standard deviation of the increment vector (yellow curve in Figure 11) shows a smoother curve, indicating less sensitivity to localized changes within the image. This could result in a less steep entropy plot, as the measure focuses on overall patterns rather than reacting strongly to localized features.
Our experiments with row-wise and column-wise increment images on MIX2D(p) show a similar curve (see Figure 12). MIX2D(p) images tend to exhibit certain statistical properties, such as local spatial correlation and smooth intensity transitions. These properties result in a relatively balanced distribution of pixel differences in both horizontal and vertical directions. While the row-wise and column-wise increment images may emphasize different aspects of the image structure, their overall impact on IncrEn2D entropy is comparable due to the statistical properties of MIX2D(p) images. The balance between horizontal and vertical variations ensures that the distribution of pixel differences is similar in both cases, leading to similar IncEn2D values.
The results obtained with horizontal and vertical stripe pattern images (see Figure 5) emphasize the critical significance of considering texture orientation when applying IncrEn2D; see Table 3. The row-wise increment image is able to capture changes within each row, exhibiting sensitivity to horizontal variations and effectively detecting features and patterns extending horizontally across the image. Consequently, the row-wise increment image yields non-zero IncrEn2D values for horizontal pattern images, while registering zero values for vertical pattern images. In contrast, the column-wise increment image is able to capture changes within each column, demonstrating sensitivity to vertical variations and in detecting features and patterns that extend vertically. As a result, the column-wise increment image yields non-zero IncrEn2D values for vertical pattern images and, conversely, reports zero values for horizontal pattern images.
It is important to note that the choice between row-wise and column-wise increment images may depend on the specific characteristics of the image data or the analysis goals. For instance, if the image dataset predominantly contains horizontally oriented features or patterns, the row-wise increment image might provide more relevant information. Conversely, the column-wise increment image may be more informative if the dataset consists of vertically oriented features.
The application of IncrEn2D to the Kylberg dataset revealed distinct entropy values for most texture groups, providing a strong basis for its use in texture classification tasks. The results suggest that IncrEn2D, like SampEn2D and DispEn2D, could serve as a valuable tool for distinguishing between different patterns of fabrics and surfaces. They also highlight the potential ability of the measures for discriminating measures in texture analysis.
Finally, the colon cancer dataset results underscore the superior performance of IncrEn2D in medical image analysis. Achieving an average classification accuracy of 77.22% with the SVM model, IncrEn2D significantly outperformed SampEn2D (59.31%) and DispEn2D (47.63%) under the same conditions. Additionally, IncrEn2D achieves the highest precision and sensitivity, indicating that, when it predicts positive cases (colon cancer), it is more accurate compared to SampEn2D and DispEn2D. Moreover, it is better at capturing actual positive cases. On the other hand, SampEn2D exhibits the highest specificity, suggesting that it performs better at correctly identifying negative cases compared to IncrEn2D and DispEn2D. However, the major problem of SampEn2D is that it can lead to undefined values at high-scale factors. DispEn2D and IncrEn2D do not have this undefined value problem. Moreover, the F1-score indicates that IncrEn2D achieves a better balance between precision and sensitivity.
From a computational perspective, DispEn2D exhibited the shortest computational time across all image sizes, followed closely by IncrEn2D. However, IncrEn2D maintains a favorable balance between computational efficiency and performance, particularly for small- to moderate-sized images. In contrast, SampEn2D incurs significantly higher computational costs, especially with larger images, which may limit its practical applicability in real-time or large-scale applications.
Though IncrEn2D demonstrates effectiveness across different datasets, it is imperative to acknowledge its inherent limitations. Notably, the performance of IncrEn2D may be hindered when dealing with large images (like pixels), where computational efficiency may become a constraint. Additionally, it is pertinent to mention that IncrEn2D currently operates solely on grayscale images, indicating a limitation in its applicability to colored images. These considerations underscore the need for further research to address these constraints and improve the applicability of the proposed method.
5. Conclusions
In this paper, we introduced a novel bidimensional entropy measure, IncrEn2D, and its multiscale form, to, respectively, estimate the irregularity and complexity of an image. These measures take into account both the size (magnitude) and direction (sign) of the increments calculated from an image. We studied their capability as powerful techniques for image analysis. We thoroughly examined the influence of the two parameters, namely the embedding dimension m and the quantification parameter R. We validated the performance of IncrEn2D using MIX2D(p) processes, as well as artificial and synthesized textures. We also investigated the impact of different standard deviations and increment image choices on the IncrEn2D calculation. Furthermore, the choice between row-wise and column-wise increment images should align with the specific image dataset’s predominant orientation or analytical objectives.
We validated IncrEn2D on two real texture datasets: Kylberg and a medical dataset with colon cancer images. For Kylberg, IncrEn2D effectively distinguished fabric patterns with distinct entropy values in designated groups, showing potential comparable to DispEn2D and faster computation than SampEn2D. In biomedical images, IncrEn2D differentiated normal and cancerous colon tissues across scale factors, outperforming multiscale SampEn2D and multiscale DispEn2D in accuracy and precision. This highlights IncrEn2D as a reliable texture descriptor with broad relevance in biomedical applications. Overall, the introduced IncrEn2D emerges as an innovative and possible tool for image analysis and texture characterization.
Author Contributions
Conceptualization, M.A., M.S.H. and A.H.-H.; methodology, M.A. and A.H.-H.; software, M.A., H.A. and A.H.-H.; validation, M.S.H., R.A., H.A. and A.H.-H.; resources, M.S.H., R.A., H.A. and A.H.-H.; writing—original draft preparation, M.A.; writing—review and editing, M.S.H., R.A., H.A. and A.H.-H.; supervision, M.S.H., R.A. and A.H.-H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The synthetic data generated and used in this study are available upon request. The real datasets analyzed in this study are publicly available.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Shannon, C.E.; Weaver, W. A Mathematical Model of Communication; University of Illinois Press: Urbana, IL, USA, 1949; Volume 11, pp. 11–20. [Google Scholar] [CrossRef]
- Boaretto, B.R.R.; Budzinski, R.C.; Rossi, K.L.; Masoller, C.; Macau, E.E.N. Spatial permutation entropy distinguishes resting brain states. Chaos Solitons Fractals 2023, 171, 113453. [Google Scholar] [CrossRef]
- Deng, Z.; Li, T.; Deng, D.; Liu, K.; Zhang, P.; Zhang, S.; Luo, Z. Feature selection for label distribution learning using dual-similarity based neighborhood fuzzy entropy. Inf. Sci. 2022, 615, 385–404. [Google Scholar] [CrossRef]
- Xu, W.; Jiang, L.; Li, C. Improving data and model quality in crowdsourcing using cross-entropy-based noise correction. Inf. Sci. 2021, 546, 803–814. [Google Scholar] [CrossRef]
- Pincus, S.M. Approximate entropy as a measure of system complexity. Proc. Natl. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J.-Physiol.-Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef]
- Bandt, C.; Pompe, B. Permutation entropy: A natural complexity measure for time series. Phys. Rev. Lett. 2002, 88, 174102. [Google Scholar] [CrossRef]
- Li, P.; Liu, C.; Li, K.; Zheng, D.; Liu, C.; Hou, Y. Assessing the complexity of short-term heartbeat interval series by distribution entropy. Med Biol. Eng. Comput. 2015, 53, 77–87. [Google Scholar] [CrossRef]
- Rostaghi, M.; Azami, H. Dispersion entropy: A measure for time-series analysis. IEEE Signal Process. Lett. 2016, 23, 610–614. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, A.; Xu, N.; Xue, J. Increment entropy as a measure of complexity for time series. Entropy 2016, 18, 22. [Google Scholar] [CrossRef]
- Li, Y.; Tang, B.; Jiao, S.; Zhou, Y. Optimized multivariate multiscale slope entropy for nonlinear dynamic analysis of mechanical signals. Chaos Solitons Fractals 2024, 179, 114436. [Google Scholar] [CrossRef]
- Xiao-Feng, L.; Yue, W. Fine-grained permutation entropy as a measure of natural complexity for time series. Chin. Phys. B 2009, 18, 2690. [Google Scholar] [CrossRef]
- Wang, D.; Jin, N. Multivariate multiscale increment entropy: A complexity measure for detecting flow pattern transition in multiphase flows. Nonlinear Dyn. 2020, 100, 3853–3865. [Google Scholar] [CrossRef]
- Fabila-Carrasco, J.S.; Tan, C.; Escudero, J. Permutation entropy for graph signals. IEEE Trans. Signal Inf. Process. Netw. 2022, 8, 288–300. [Google Scholar] [CrossRef]
- Fabila-Carrasco, J.S.; Tan, C.; Escudero, J. Dispersion entropy for graph signals. Chaos Solitons Fractals 2023, 175, 113977. [Google Scholar] [CrossRef]
- Morel, C.; Humeau-Heurtier, A. Multiscale permutation entropy for two-dimensional patterns. Pattern Recognit. Lett. 2021, 150, 139–146. [Google Scholar] [CrossRef]
- Hilal, M.; Berthin, C.; Martin, L.; Azami, H.; Humeau-Heurtier, A. Bidimensional multiscale fuzzy entropy and its application to pseudoxanthoma elasticum. IEEE Trans. Biomed. Eng. 2019, 67, 2015–2022. [Google Scholar] [CrossRef]
- Gaudêncio, A.S.; Hilal, M.; Cardoso, J.M.; Humeau-Heurtier, A.; Vaz, P.G. Texture analysis using two-dimensional permutation entropy and amplitude-aware permutation entropy. Pattern Recognit. Lett. 2022, 159, 150–156. [Google Scholar] [CrossRef]
- Azami, H.; da Silva, L.E.V.; Omoto, A.C.M.; Humeau-Heurtier, A. Two-dimensional dispersion entropy: An information-theoretic method for irregularity analysis of images. Signal Process. Image Commun. 2019, 75, 178–187. [Google Scholar] [CrossRef]
- Espinosa, R.; Bailón, R.; Laguna, P. Two-dimensional EspEn: A new approach to analyze image texture by irregularity. Entropy 2021, 23, 1261. [Google Scholar] [CrossRef]
- Zheng, J.; Ying, W.; Tong, J.; Li, Y. Multiscale three-dimensional Holo–Hilbert spectral entropy: A novel complexity-based early fault feature representation method for rotating machinery. Nonlinear Dyn. 2023, 111, 10309–10330. [Google Scholar] [CrossRef]
- Ghalati, M.K.; Nunes, A.; Ferreira, H.; Serranho, P.; Bernardes, R. Texture analysis and its applications in biomedical imaging: A survey. IEEE Rev. Biomed. Eng. 2021, 15, 222–246. [Google Scholar] [CrossRef]
- Versaci, M.; Morabito, F.C. Image edge detection: A new approach based on fuzzy entropy and fuzzy divergence. Int. J. Fuzzy Syst. 2021, 23, 918–936. [Google Scholar] [CrossRef]
- Espinosa, M.; Ricardo, A. EspEn Graph for the Spatial Analysis of Entropy in Images. Entropy 2023, 25, 159. [Google Scholar] [CrossRef]
- Silva, L.E.V.; Senra Filho, A.C.S.; Fazan, V.P.S.; Felipe, J.C.; Junior, L.M. Two-dimensional sample entropy: Assessing image texture through irregularity. Biomed. Phys. Eng. Express 2016, 2, 045002. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Pang, W.; Jiang, A. Multiscale increment entropy: An approach for quantifying the physiological complexity of biomedical time series. Inf. Sci. 2022, 586, 279–293. [Google Scholar] [CrossRef]
- Simoncelli, E. Texture Synthesis: Misc Textures. 2023. Available online: https://graphics.stanford.edu/projects/texture/demo/synthesis_eero.html (accessed on 15 July 2023).
- Kylberg, G. The Kylberg Texture Dataset v. 1.0. External Report (Blue Series) 35, Centre for Image Analysis; Swedish University of Agricultural Sciences and Uppsala University: Uppsala, Sweden, 2011. [Google Scholar]
- Masud, M.; Sikder, N.; Nahid, A.; Bairagi, A.K.; AlZain, M.A. A machine learning approach to diagnosing lung and colon cancer using a deep learning-based classification framework. Sensors 2021, 21, 748. [Google Scholar] [CrossRef]
- Borkowski, A.A.; Bui, M.M.; Thomas, L.B.; Wilson, C.P.; DeLand, L.A.; Mastorides, S.M. Lung and colon cancer histopathological image dataset (lc25000). arXiv 2019, arXiv:1912.12142. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).