Abstract
The efforts to discover HIV therapeutics have continued since the first human immunodeficiency virus (HIV) infected patient was confirmed in the 1980s. Ten years later, the first HIV drug, zidovudine (AZT), targeting HIV reverse transcriptase, was developed. Meanwhile, scientists were enlightened to discover new drugs that target different HIV genes, like integrase, protease, and host receptors. Combination antiretroviral therapy (cART) is the most feasible medical intervention to suppress the virus in people with HIV (PWH) and control the epidemic. ART treatment has made HIV a chronic infection rather than a fatal disease, but ART does not eliminate latent reservoirs of HIV-1 from the host cells; strict and life-long adherence to ART is required for the therapy to be effective in patients. In this review, we first discussed the scientific history of conventional HIV drug discovery since scientists need to develop more and more drugs to solve drug-resistant issues and release the side effects. Then, we summarized the novel research technologies, like gene editing, applied to HIV treatment and their contributions to eliminating HIV as a complementary therapy.
1. Introduction
It has been four decades since the first human immunodeficiency virus (HIV) was identified as the pathogen that caused the disease of acquired immunodeficiency syndrome (AIDS) [,]. Since then, scientists have started working on HIV mechanisms and therapeutics. Until now, over thirty HIV drugs have been approved by the Food and Drug Administration (FDA) []. Based on their molecular mechanism and targets to each step of the viral life cycle, these drugs were classified into six different groups: (1) coreceptor inhibitors (CRIs) and (2) fusion inhibitors (FIs) targeting viral entry; (3) nucleoside reverse transcriptase inhibitors (NRTIs) and (4) non-nucleoside reverse transcriptase inhibitors (NNRTIs) targeting reverse transcription; (5) integrase strand transfer inhibitors (InSTIs) targeting viral integration; and (6) protease inhibitors (PIs) targeting viral maturation [,]. The first generation of drugs is all reverse transcriptase inhibitors, like the first HIV drug, zidovudine (3′-azido-3′-deoxythymidine, or AZT), which was approved by the FDA in 1987 [,]. However, AIDS lethality did not decrease until the advent of antiretroviral therapy (ART) in the middle 1990s, after the protease inhibitors were developed.
In general, ART is the combination of three or more drugs designed against at least two steps in the life cycle of the virus. People with HIV (PWH) should be treated with ART as soon as possible. Usually, the initial ART drugs should contain InSTIs, either first-generation InSTIs like raltegravir (RAL) and elvitegravir (EVG) or the second-generation InSTIs like dolutegravir (DLG) and bictegravir (BIC), to specifically block the viral DNA integrated into the host genome [,]. The clinical trial of ART showed that HIV viral replication was strongly suppressed, and the plasma viral load of PWH was significantly reduced to an undetectable level [,]. There are some clinical studies that demonstrated that no transmissions happened to the partners during condomless intercourse, no matter whether during heterosexual acts or among men who have sex with men (MSM), when the undetectable viral load was maintained for more than six months with ART [,,], which supports the statement that an undetectable viral load equals an untransmittable one (Undetectable = Untransmittable, or U = U) [,]. Eventually, ART development has successfully changed AIDS from a fatal disease into a controlled viral infection with restored immune function [,] and with the mortality rates of PWH close to general mortality rates [,,].
The Department of Health and Human Services Guidelines Panel for the Use of Antiretroviral Agents in Adults and Adolescents with HIV (the Panel) has developed recommendations for the use of statin therapy in people with HIV []. Despite the fact that current ART can durably suppress HIV viremia, the viral load will rebound within several weeks of ART interruption, even in patients with very small HIV reservoirs and minimal ongoing viral transcription []. The lifelong treatment of ART for PWH has brought new challenges, such as side effects, including weight gain, anxiety, irritability, dizziness, insomnia [], and drug resistance [,]. In addition, ART was unable to eradicate the integrated virus in the host cells [,]. The persistence of HIV viral reservoirs in the host cells is always a threat to PWH. Therefore, continuous efforts are demanded to discover novel drugs against different targets with less toxicity and develop alternative therapeutics, such as gene-editing technologies, which may be used to eliminate viral reservoirs in patients and lead to an HIV functional cure.
In this review, we provide an overview of the HIV-1 life cycle and summarize the most updated anti-HIV drugs that the US Food and Drug Administration (FDA) has approved, according to the viral replication life cycle. In addition, we describe the most advanced gene editing tools based on the clustered regularly interspaced short palindromic repeat (CRISPR)/associated nuclease 9 (Cas9) system, which can be a complementary strategy for HIV cure. Finally, the effective in vivo delivery system for programmable elements will also be briefly discussed.
3. CRISPR/Cas9-Based Gene Editing in HIV Treatment
3.1. Gene Editing Targets Host Cell
It has been reported that the 32-base-pair deletion of coreceptor CCR5 renders host cells resistant to R5-tropic HIV-1 variants infection []. In 2008, at the CROI (Conference on Retroviruses and Opportunistic Infections) conference, the Berlin patient was declared the first person “cured of HIV”, who remained free of HIV for more than 13 years without ART after stem cell transplantation from a healthy donor with homozygous CCR5Δ32 mutation. After the Berlin patient, there have been the London patient, the New York patient, the City of Hope patient, and the Düsseldorf patient, who have been cured of HIV from hematopoietic stem cell (HSC) transplantation []. Therefore, intensive work on the disruption of CCR5 expression with gene-editing tools is currently being conducted. The rapidly updated CRISPR/Cas9 technology has boosted the research for curative HIV therapy, such as base editing. Base editing uses programmable DNA-binding proteins to directly convert C⋅G to T⋅A base pairs (cytosine base editing, CBE) [] or convert C⋅G to T⋅A base pairs (adenine base editing, ABE) []. Base editors modify targeted nucleotides with specific base changes in the genome (Figure 1), while CRISPR/Cas9 breaks double-strand and randomly modifies the genomic DNA.
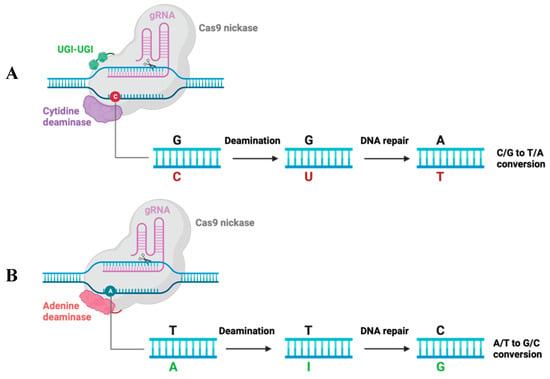
Figure 1.
Mechanism schematic of the cytidine base editor (CBE) and adenine base editor (ABE) systems. (A) CBE contains a Cas9 nickase fused to a deaminase and two UGIs (uracil glycosylase inhibitors). C changed to U under deamination, then changed to T with host DNA repair. As a result, CBE converts the C/G base pair into the T/A base pair. (B) ABE is composed of a Cas9 nickase fused to deaminase that changes A to I, then I to G. As a result, ABE converts the A/T base pair into a G/C base pair.
Xu et al. established an HSC transplantation model with CRISPR/Cas9-conferred CCR5-ablated human CD34+ hematopoietic stem/progenitor cells (HSPCs) []. HSPCs were reconstituted in mice for over 1 year and achieved robust CCR5 disruption, which mediated an HIV-1 resistance effect in vivo. Liu et al. designed two different guide RNA (gRNA) combinations targeting both CXCR4 and CCR5 in a single vector []. The simultaneous genome editing of HIV-1 coreceptors CXCR4 and CCR5 in primary CD4+ T cells with the CRISPR/Cas9 system protects modified cells from X4-tropic or R5-tropic HIV-1 viral infection. Knipping et al. applied base editors to simultaneously disrupt both coreceptors in primary human CD4+ T cells; it prevents transduction with R5-tropic and X4-tropic viral vectors []. Except for targeting coreceptors, Chinnapaiyan et al. used CRISPR/Cas9 to knock down the cellular co-factor cyclin T1, which is crucial for HIV transcription and demonstrated cyclin T1 inhibition-mediated HIV silencing []. Overall, the research on CCR5 gene modification is dominant over other host cellular targets.
3.2. Gene Editing Targets HIV Genome
Although the current HIV-1 treatment of ART and broadly neutralizing antibodies could suppress plasma viral load below the detectable level, ART cannot eliminate the integrated provirus from host cells. Viral load will rebound within a few weeks after ART withdrawal. This challenge can be overcome with gene editing on proviral DNA in CD4+ T cells. Ebina et al. designed gRNA to target HIV-1 long terminal repeat (LTR) in specific locations of the TAR sequence of the R region and NF-κB binding sequence in the U3 region, respectively, and this LTR-targeted CRISPR/Cas9 system can disrupt HIV-1 provirus and active the provirus from the cellular genome []. Kaminski et al. employed a CRISPR/Cas9 editing system to precisely remove the integrated copies of the proviral DNA fragment from latently infected human CD4+ T cells by targeting the highly conserved sequence of U3 LTR region of all isolated HIV-1 strains []. Liao et al. adapted the CRISPR/Cas9 system to disrupt the latently integrated viral genome and provide long-term defense against new viral infections. They screened several potential gRNA targeting sites in the HIV-1 genome, including the structural (gag and env), enzymatic (pol) and accessory genes (vif and rev), as well as LTRs, and they increased the frequency of disruption and activation of the pre-integrated proviral genome by using a multiplexed CRISPR/Cas9 system along with the gRNAs targeting LTR sequences (especially the R region) []. Zhu et al. tested 10 sites in HIV-1 DNA using CRISPR/Cas9 and found that a highly efficient target site is located in the second exon of rev, and it could inactivate provirus efficiently by significantly reducing HIV-1 gene expression and virus production []. In general, among different sites targeted on the viral genome, targeting HIV-1 LTR has a strong impact on viral disruption because LTR serves as a critical element of the viral transcription, especially targeting the LTR-R region, which contains the TAR sequence, which is highly conserved of all HIV-1 subtypes.
In addition, there are combinational targets on both host cells and the viral genome. To improve the efficiency of viral elimination, Dash et al. developed dual CRISPR, which targets both proviral DNA and CCR5 []. There are two CRISPR reagents, including one set designed to target LTR and Gag to activate the HIV-1 LTR-Gag region from latent proviral DNA integrated cells and the other set designed to target host coreceptor CCR5. The viral outgrowth assay (VOA) demonstrated that no progeny virus was recovered in plasma and tissues from CRISPR-treated virus-free mice.
3.3. Clinical Trials of Gene Editing Applied in HIV Treatment
On 8 December 2023, the U.S. FDA approved two milestone treatments, Casgevy and Lyfgenia, for sickle cell disease (SCD) in patients 12 years and older []. Casgevy is the first FDA-approved therapy utilizing CRISPR/Cas9, which greatly inspired the development of HIV treatment in the field of gene therapy. To date, EBT-101 is the only CRISPR/Cas9-based gene therapy for HIV treatment in clinical trials and is now in the status of recruiting (NCT05144386). EBT-101 is comprised of an all-in-one CRISPR/Cas9 system that expresses dual gRNAs targeting viral LTRs and the Gag gene, thereby generating three possible deletions: 5′LTR to Gag, Gag to 3′LTR, and 5′LTR to 3′LTR []. The dual gRNAs excise large sections of proviral DNA (Figure 2), eliminating viral escape and reproduction.
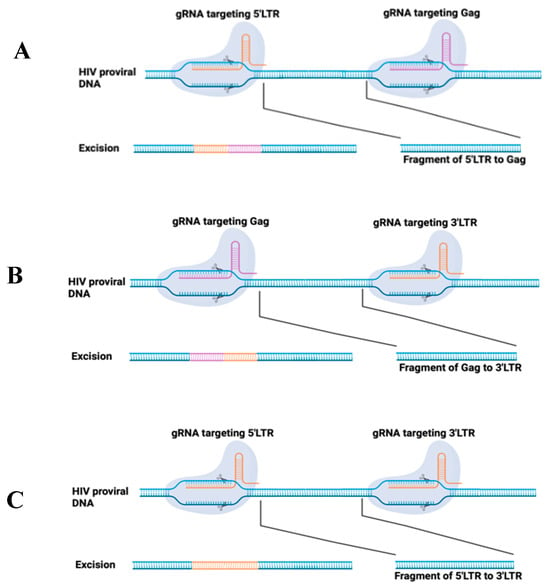
Figure 2.
Schematic of CRISPR/Cas9 system applied for HIV provirus excision. Two gRNAs were designed to target HIV LTR (5′ and 3′) and Gag at specific sites. The induced double-strand breaks are repaired using non-homologous end joining, which will induce the deletion of a fragment from 5′LTR to Gag (A), or Gag to 3′LTR (B), or 5′LTR to 3′LTR (C).
Furthermore, there are zinc finger nucleases (ZFNs) that enable CCR5 gene editing for HIV treatment in clinical trials. A phase I study of autologous T cells genetically modified to target CCR5 using ZFNs was completed (NCT00842634). It demonstrated that the loss of CCR5 protein on the T cells is permanent in the follow-up study up to 6 years. Meanwhile, other phase I (NCT01044654) and phase 1/2 (NCT01252641) clinical trials regarding the dose escalation and single-dose infusion of autologous T cells genetically modified to target CCR5 using ZFNs in HIV-infected patients were completed at almost the same time. Both of the trials proved the safety and feasibility of ZFN-CCR5-modified CD4+ T cells re-infused back to humans as HIV therapy. Further, the phase II clinical trial that is randomly comparing the effect of infusing expanded autologous CD4+ T cells with or without CCR5 modification ex vivo using ZFNs among HIV-infected patients is ongoing (NCT03666871). In addition, there is a phase I pilot study of stem cell gene modification, which is evaluating the feasibility, safety, and engraftment of ZFN-CCR5-modified CD34+ HSPCs in R5 tropic HIV-1 infected patients (NCT02500849).
Although gene editing is a robust tool for HIV treatment, balancing the potential benefits and ethical considerations is important. DNA modification on reproductive embryos is banned in many countries; it is urgently needed to create ethical guidelines and make them transparent. Public engagement is crucial, making the patient group aware of gene editing technology and its pros and cons by involving communication between scientists and patients []. Additionally, the long-term effects and potential unintended edits on the human genome still require extensive study to address the issue. It is essential to avoid off-target effects to ensure maximum benefits and minimum risks in HIV gene therapy [].
4. Conclusions and Future Perspectives
The discovery of anti-HIV drugs is arguably among the most successful achievements for any human disease, considering the number of available anti-HIV agents that have been developed for four decades since the first HIV-1 viral infection case was confirmed in 1981. More than 30 antiretroviral drugs have been approved, and combination therapy has been demonstrated with high efficiency and controllable toxicity for PWH. However, the lifelong treatment of ART and acquired drug resistance are still the key issues in HIV cure. Continuous efforts are demanded to develop new compounds and new drug combinations to achieve therapy success. Although these ART drugs highly suppress the viremia, they are unable to eradicate the integrated viral DNA. With the development of gene-editing tools, such as ZFNs, CRISPR/Cas9, and transcription activator-like effectors (TALENS) etc., more and more research has been conducted on provirus elimination using these new technologies. Especially in recent years, base editing and prime editing, which are derived from the CRISPR/Cas9 system, are much safer compared with classical CRISPR/Cas9 since these two editing methods do not break double-strand targeted DNA. With low off-target effects, these gene modification tools can be applied in various research fields, including anti-HIV therapy. It is convenient and only needs to design different gRNA for different targets. Moreover, it is flexible, and different gRNAs can be combined in one vector for better knock-out efficiency. For example, the current ongoing clinical trial of EBT-101 is a combination of gRNAs targeting HIV-1 LTR and Gag genes.
The CRISPR/Cas9 system is indeed a promising tool applied for HIV treatment, but an obstacle is its low delivery efficiency in vivo. The delivery components can be plasmid, ribonucleoprotein (RNP), and mRNA. The delivery vehicle can be viral vector-based delivery, lipid nanoparticle, and microinjection. Using nanoparticles to deliver ART drugs could maintain effective drug concentrations in targeted tissues for HIV treatment []. Adeno-associated virus (AAV) vector is an efficient and widely used delivery agent, and it is the only gene therapy vector that the FDA has approved for human diseases. Viral vector delivery for gene-editing elements also made remarkable progress in clinical HIV treatment. Specifically, EBT-101 adapts AAV9 for CRISPR/Cas9 delivery. Hamann et al. modified the AAV2 capsid protein with a set of novel nanobodies with high affinity for the human CD4 receptor. This CD4 antibody-modified capsid was demonstrated to improve the targeting efficiency of human primary CD4+ T cells in vitro []. It provides a promising strategy for changing AAV tropism to particularly targeting specific cells. The limitation of the AAV vector is its packaging capacity, which is less than 5 kb. The large size of SpCas9 (~4.1 kb) decreases the efficiency of delivery, so it seems impossible to package the base editor and prime editor in AAV. Therefore, Levy et al. came up with a dual AAV system for the delivery of split-base editors []. Another way to solve the issue is to optimize the SpCas9 protein into a truncated variant or use a smaller SaCas9. While there is a long way to go, the combination of antiretroviral drugs and gene-editing technology will lead to promising progress in HIV-1 therapeutics.
Author Contributions
Y.S. took care of conceptualization, writing the original draft, and reviewing and editing. L.W. took care of supervision, resources, conceptualization, writing the original draft, and reviewing and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Barre-Sinoussi, F.; Chermann, J.; Rey, F.; Nugeyre, M.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vezinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 1983, 220, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Gallo, R.C.; Salahuddin, S.Z.; Popovic, M.; Shearer, G.M.; Kaplan, M.; Haynes, B.F.; Palker, T.J.; Redfield, R.; Oleske, J.; Safai, B. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 1984, 224, 500–503. [Google Scholar] [CrossRef]
- Tompa, D.R.; Immanuel, A.; Srikanth, S.; Kadhirvel, S. Trends and strategies to combat viral infections: A review on FDA approved antiviral drugs. Int. J. Biol. Macromol. 2021, 172, 524–541. [Google Scholar] [CrossRef] [PubMed]
- Arts, E.J.; Hazuda, D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2012, 2, a007161. [Google Scholar] [CrossRef] [PubMed]
- Menendez-Arias, L. Molecular basis of human immunodeficiency virus type 1 drug resistance: Overview and recent developments. Antivir. Res. 2013, 98, 93–120. [Google Scholar] [CrossRef] [PubMed]
- Yarchoan, R.; Klecker, R.W.; Weinhold, K.J.; Markham, P.D.; Lyerly, H.K.; Durack, D.T.; Gelmann, E.; Lehrman, S.N.; Blum, R.M.; Barry, D.W. Administration of 3′-azido-3′-deoxythymidine, an inhibitor of HTLV-III/LAV replication, to patients with AIDS or AIDS-related complex. Lancet 1986, 1, 575–580. [Google Scholar] [CrossRef]
- McLeod, G.X.; Hammer, S.M. Zidovudine: Five years later. Ann. Intern Med. 1992, 117, 487–501. [Google Scholar] [PubMed]
- Zhao, A.V.; Crutchley, R.D.; Guduru, R.C.; Ton, K.; Lam, T.; Min, A.C. A clinical review of HIV integrase strand transfer inhibitors (INSTIs) for the prevention and treatment of HIV-1 infection. Retrovirology 2022, 19, 22. [Google Scholar] [CrossRef] [PubMed]
- Mbhele, N.; Chimukangara, B.; Gordon, M. HIV-1 integrase strand transfer inhibitors: A review of current drugs, recent advances and drug resistance. Int. J. Antimicrob. Agents 2021, 57, 106343. [Google Scholar] [CrossRef]
- Lecher, S.L.; Fonjungo, P.; Ellenberger, D.; Toure, C.A.; Alemnji, G.; Bowen, N.; Basiye, F.; Beukes, A.; Carmona, S.; Klerk, M.; et al. HIV Viral Load Monitoring Among Patients Receiving Antiretroviral Therapy—Eight Sub-Saharan Africa Countries, 2013–2018. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 775–778. [Google Scholar] [CrossRef]
- Han, W.M.; Law, M.G.; Egger, M.; Wools-Kaloustian, K.; Moore, R.; McGowan, C.; Kumarasamy, N.; Desmonde, S.; Edmonds, A.; Davies, M.; et al. Global estimates of viral suppression in children and adolescents and adults on antiretroviral therapy adjusted for missing viral load measurements: A multiregional, retrospective cohort study in 31 countries. Lancet HIV 2021, 8, e766–e775. [Google Scholar] [CrossRef]
- Cohen, M.S.; Chen, Y.; McCauley, M.; Gamble, T.; Hosseinipour, M.; Kumarasamy, N.; Hakim, J.; Kuwenda, J.; Grinsztejin, B.; Pilotto, J.; et al. Prevention of HIV-1 infection with early antiretroviral therapy. N. Engl. J. Med. 2011, 365, 493–505. [Google Scholar] [CrossRef]
- Cohen, M.S.; Chen, Y.; McCauley, M.; Gamble, T.; Hosseinipour, M.; Kumarasamy, N.; Hakim, J.; Kuwenda, J.; Grinsztejin, B.; Pilotto, J.; et al. Antiretroviral Therapy for the Prevention of HIV-1 Transmission. N. Engl. J. Med. 2016, 375, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Mujugira, A.; Celum, C.; Coombs, R.W.; Campbell, J.D.; Ndase, P.; Ronald, A.; Were, E.; Bukusi, E.A.; Mugo, N.; Kiarie, J.; et al. HIV Transmission Risk Persists During the First 6 Months of Antiretroviral Therapy. JAIDS J. Acquir. Immune Defic. Syndr. 2016, 72, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Eisinger, R.W.; Dieffenbach, C.W.; Fauci, A.S. HIV Viral Load and Transmissibility of HIV Infection: Undetectable Equals Untransmittable. JAMA 2019, 321, 451–452. [Google Scholar] [CrossRef] [PubMed]
- LeMessurier, J.; Traversy, G.; Varsaneux, O.; Weekes, M.; Avey, M.T.; Niragira, O.; Gervais, R.; Guyatt, G.; Rodin, R. Risk of sexual transmission of human immunodeficiency virus with antiretroviral therapy, suppressed viral load and condom use: A systematic review. CMAJ 2018, 190, E1350–E1360. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Hou, Y.; Zhao, Y.; Dou, Z.; Ma, Y.; Zhang, D.; Wu, Y.; Zhao, D.; Liu, Z.; et al. Immune restoration in HIV-1-infected patients after 12 years of antiretroviral therapy: A real-world observational study. Emerg. Microbes Infect. 2020, 9, 2550–2561. [Google Scholar] [CrossRef]
- Zhang, L.X.; Jiao, Y.M.; Zhang, C.; Song, J.W.; Fab, X.; Xu, R.N.; Huang, H.H.; Zhang, J.Y.; Wang, L.F.; Zhou, C.B.; et al. HIV Reservoir Decay and CD4 Recovery Associated with High CD8 Counts in Immune Restored Patients on Long-Term ART. Front. Immunol. 2020, 11, 1541. [Google Scholar] [CrossRef]
- Edwards, J.K.; Cole, S.R.; Breger, T.L.; Rudolph, J.E.; Filiatreau, L.M.; Buchacz, K.; Humes, E.; Rebeiro, P.F.; D’Souza, G.; Gill, M.J.; et al. Mortality Among Persons Entering HIV Care Compared with the General U. S. Population An Obs. Study Ann. Intern Med. 2021, 174, 1197–1206. [Google Scholar] [CrossRef]
- Fontela, C.; Aguinaga, A.; Moreno-Iribas, C.; Reparaz, J.; Rivero, M.; Gracia, M.; Floristan, Y.; Fresan, U.; Miguel, R.S.; Ezpeleta, C.; et al. Trends and causes of mortality in a population-based cohort of HIV-infected adults in Spain: Comparison with the general population. Sci. Rep. 2020, 10, 8922. [Google Scholar] [CrossRef]
- Reis, K.G.; Desderius, B.; Kingery, J.; Kirabo, A.; Makubi, A.; Myalla, C.; Lee, M.H.; Kapiga, S.; Peck, R.N. Blood pressure, T cells, and mortality in people with HIV in Tanzania during the first 2 years of antiretroviral therapy. J. Clin. Hypertens. (Greenwich) 2020, 22, 1554–1562. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, R.T.; Bedimo, R.; Hoy, J.F.; Landocitz, R.J.; Smith, D.M.; Eaton, E.F.; Lehmann, C.; Springer, S.A.; Sax, P.E.; Thompson, M.A.; et al. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults: 2022 Recommendations of the International Antiviral Society-USA Panel. JAMA 2023, 329, 63–84. [Google Scholar] [CrossRef]
- Pannus, P.; Rutsaert, S.; Wit, S.; Allard, S.D.; Vanham, G.; Cole, B.; Nescoi, C.; Aerts, J.; Spiegelaere, W.; Tsoumanis, A.; et al. Rapid viral rebound after analytical treatment interruption in patients with very small HIV reservoir and minimal on-going viral transcription. J. Int. AIDS Soc. 2020, 23, e25453. [Google Scholar] [CrossRef]
- Kolakowska, A.; Maresca, A.F.; Collins, I.J.; Cailhol, J. Update on Adverse Effects of HIV Integrase Inhibitors. Curr. Treat. Options Infect. Dis. 2019, 11, 372–387. [Google Scholar] [CrossRef]
- Zuo, L.; Liu, K.; Liu, H.; Hu, Y.; Zhang, Z.; Qin, J.; Xu, Q.; Peng, K.; Jin, X.; Wang, J.H.; et al. Trend of HIV-1 drug resistance in China: A systematic review and meta-analysis of data accumulated over 17 years (2001–2017). EClinicalMedicine 2020, 18, 100238, Corrigendum to EClinicalMedicine 2021, 33, 100696. [Google Scholar] [CrossRef] [PubMed]
- Chimukangara, B.; Lessells, R.J.; Rhee, S.Y.; Giandhari, J.; Kharsany, A.B.M.; Naidoo, K.; Lewis, L.; Cawood, C.; Khanyile, D.; Ayalew, K.A.; et al. Trends in Pretreatment HIV-1 Drug Resistance in Antiretroviral Therapy-naive Adults in South Africa, 2000-2016: A Pooled Sequence Analysis. EClinicalMedicine 2019, 9, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, N.; Siebenthal, C.; Vongrad, V.; Turk, T.; Neumann, K.; Beerenwinkel, N.; Bogojeska, J.; Fellay, J.; Roth, V.; Kok, Y.L.; et al. Determinants of HIV-1 reservoir size and long-term dynamics during suppressive ART. Nat. Commun. 2019, 10, 3193. [Google Scholar] [CrossRef]
- Bandera, A.; Gori, A.; Clerici, M.; Sironi, M. Phylogenies in ART: HIV reservoirs, HIV latency and drug resistance. Curr. Opin. Pharmacol. 2019, 48, 24–32. [Google Scholar] [CrossRef]
- Bour, S.; Geleziunas, R.; Wainberg, M.A. The human immunodeficiency virus type 1 (HIV-1) CD4 receptor and its central role in promotion of HIV-1 infection. Microbiol. Rev. 1995, 59, 63–93. [Google Scholar] [CrossRef]
- Feng, Y.; Broder, C.C.; Kennedy, P.E.; Berger, E.A. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 1996, 272, 872–877. [Google Scholar] [CrossRef]
- Berger, E.A.; Murphy, P.M.; Farber, J.M. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 1999, 17, 657–700. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C.; Fass, D.; Berger, J.M.; Kim, P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 1997, 89, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Telesnitsky, A.; Goff, S. Reverse Transcriptase and the Generation of Retroviral DNA. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Lusic, M.; Siliciano, R.F. Nuclear landscape of HIV-1 infection and integration. Nat. Rev. Microbiol. 2017, 15, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.E.; Saad, J.S. The Interplay between HIV-1 Gag Binding to the Plasma Membrane and Env Incorporation. Viruses 2020, 12, 548. [Google Scholar] [CrossRef] [PubMed]
- Rein, A. RNA Packaging in HIV. Trends Microbiol. 2019, 27, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Checkley, M.A.; Luttge, B.G.; Freed, E.O. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J. Mol. Biol. 2011, 410, 582–608. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Ganser-Pornillos, B.K. Maturation of retroviruses. Curr. Opin. Virol. 2019, 36, 47–55. [Google Scholar] [CrossRef]
- Xiao, T.; Cai, Y.; Chen, B. HIV-1 Entry and Membrane Fusion Inhibitors. Viruses 2021, 13, 735. [Google Scholar] [CrossRef]
- Fletcher, C.V. Enfuvirtide, a new drug for HIV infection. Lancet 2003, 361, 1577–1578. [Google Scholar] [CrossRef]
- He, Y. Synthesized peptide inhibitors of HIV-1 gp41-dependent membrane fusion. Curr. Pharm. Des. 2013, 19, 1800–1809. [Google Scholar] [CrossRef]
- Patel, I.H.; Zhang, X.; Nieforth, K.; Salgo, M.; Buss, N. Pharmacokinetics, pharmacodynamics and drug interaction potential of enfuvirtide. Clin. Pharmacokinet. 2005, 44, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Lalezari, J.P.; Henry, K.; O’Hearn, M.; Montaner, J.; Piliero, P.J.; Trottier, B.; Walmsley, S.; Cohen, C.; Kuritzkes, D.R.; Eron, J.J., Jr.; et al. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 2003, 348, 2175–2185. [Google Scholar] [CrossRef] [PubMed]
- Dorr, P.; Westby, M.; Dobbs, S.; Griffin, P.; Irvine, B.; Macartney, M.; Mori, J.; Rickett, G.; Smith-Burchnell, C.; Napier, C.; et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 2005, 49, 4721–4732. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Zhu, Y.; Li, J.; Chen, Z.; Han, G.W.; Kufareva, I.; Li, T.; Ma, L.; Fenalti, G.; Li, J.; et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 2013, 341, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Tiraboschi, J.M.; Niubo, J.; Curto, J.; Podzamczer, D. Maraviroc concentrations in cerebrospinal fluid in HIV-infected patients. JAIDS J. Acquir. Immune Defic. Syndr. 2010, 55, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Tiraboschi, J.M.; Niubo, J.; Curto, J.; Podzamczer, D. Maraviroc concentrations in seminal plasma in HIV-infected patients. JAIDS J. Acquir. Immune Defic. Syndr. 2010, 55, e35–e36. [Google Scholar] [CrossRef] [PubMed]
- Dumond, J.B.; Patterson, K.B.; Pecha, A.L.; Werner, R.E.; Andrews, E.; Damle, B.; Tressler, R.; Worsley, J.; Kashuba, A.D.M. Maraviroc concentrates in the cervicovaginal fluid and vaginal tissue of HIV-negative women. JAIDS J. Acquir. Immune Defic. Syndr. 2009, 51, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Fatkenheuer, G.; Nelson, M.; Lazzarin, A.; Konourina, I.; Hoepelman, A.I.M.; Lampiris, H.; Hirschel, B.; Tebas, P.; Raffi, F.; Trottier, B.; et al. Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. N. Engl. J. Med. 2008, 359, 1442–1455. [Google Scholar] [CrossRef] [PubMed]
- Beccari, M.V.; Beccari, M.V.; Mogle, B.T.; Sidman, E.F.; Mastro, K.A.; Asiago-Reddy, E.; Kufel, W.D. Ibalizumab, a Novel Monoclonal Antibody for the Management of Multidrug-Resistant HIV-1 Infection. Antimicrob. Agents Chemother. 2019, 63, e00110-19. [Google Scholar] [CrossRef]
- Pace, C.S.; Fordyce, M.W.; Franco, D.; Kao, C.Y.; Seaman, M.S.; Ho, D.D. Anti-CD4 monoclonal antibody ibalizumab exhibits breadth and potency against HIV-1, with natural resistance mediated by the loss of a V5 glycan in envelope. JAIDS J. Acquir. Immune Defic. Syndr. 2013, 62, 1–9. [Google Scholar] [CrossRef]
- Wang, L.; Yukselten, Y.; Nuwagaba, J.; Sutton, R.E. JAK/STAT signaling pathway affects CCR5 expression in human CD4+ T cells. Sci. Adv. 2024, 10, eadl0368. [Google Scholar] [CrossRef]
- Sluis-Cremer, N.; Arion, D.; Parniak, M.A. Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Cell. Mol. Life Sci. 2000, 57, 1408–1422. [Google Scholar] [CrossRef] [PubMed]
- Kohlstaedt, L.A.; Wang, J.; Friedman, J.M.; Rice, P.A.; Steitz, T.A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 1992, 256, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Tronchet, J.M.; Seman, M. Nonnucleoside inhibitors of HIV-1 reverse transcriptase: From the biology of reverse transcription to molecular design. Curr. Top. Med. Chem. 2003, 3, 1496–1511. [Google Scholar] [CrossRef]
- De Clercq, E. The role of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Antivir. Res. 1998, 38, 153–179. [Google Scholar] [CrossRef] [PubMed]
- Bertagnolio, S.; Hermans, L.; Jordan, M.R.; Avila-Rios, S.; Iwuji, C.; Derache, A.; Delaporte, E.; Wensing, A.; Aves, T.; Borhan, A.S.M.; et al. Clinical Impact of Pretreatment Human Immunodeficiency Virus Drug Resistance in People Initiating Nonnucleoside Reverse Transcriptase Inhibitor-Containing Antiretroviral Therapy: A Systematic Review and Meta-analysis. J. Infect. Dis. 2021, 224, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.; Goldstein, F.; Reichmuth, M.L.; Kouyos, R.D.; Wandeler, G.; Egger, M.; Riou, J. Acquired HIV drug resistance mutations on first-line antiretroviral therapy in Southern Africa: Systematic review and Bayesian evidence synthesis. J. Clin. Epidemiol. 2022, 148, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Feng, D.; Sun, Y.; Fang, Z.; Wei, F.; De Clercq, E.; Pannecouque, C.; Liu, X.; Zhan, P. Structure-Based Bioisosterism Yields HIV-1 NNRTIs with Improved Drug-Resistance Profiles and Favorable Pharmacokinetic Properties. J. Med. Chem. 2020, 63, 4837–4848. [Google Scholar] [CrossRef]
- Kohl, N.E.; Emini, E.A.; Schleif, W.A.; Davis, L.J.; Heimbach, J.C.; Dixon, R.A.; Scolnick, E.M.; Sigal, I.S. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 1988, 85, 4686–4690. [Google Scholar] [CrossRef]
- Noble, S.; Faulds, D. Saquinavir. A review of its pharmacology and clinical potential in the management of HIV infection. Drugs 1996, 52, 93–112. [Google Scholar] [CrossRef]
- Bozzette, S.A.; Ake, C.F.; Tam, H.K.; Chang, S.W.; Louis, T.A. Cardiovascular and cerebrovascular events in patients treated for human immunodeficiency virus infection. N. Engl. J. Med. 2003, 348, 702–710. [Google Scholar] [CrossRef]
- Alvi, R.M.; Neilan, A.M.; Tariq, N.; Awadalla, M.; Afshar, M.; Banerji, D.; Rokicki, A.; Mulligan, C.; Triant, V.A.; Zanni, M.V.; et al. Protease Inhibitors and Cardiovascular Outcomes in Patients with HIV and Heart Failure. J. Am. Coll. Cardiol. 2018, 72, 518–530. [Google Scholar] [CrossRef]
- Sham, H.L.; Zhao, C.; Li, L.; Betebenner, D.A.; Saldivar, A.; Vasavanonda, S.; Kempf, D.; Plattner, J.J.; Norbeck, D.W. Novel lopinavir analogues incorporating non-Aromatic P-1 side chains--synthesis and structure--activity relationships. Bioorg. Med. Chem. Lett. 2002, 12, 3101–3103. [Google Scholar] [CrossRef] [PubMed]
- Callebaut, C.; Stray, K.; Tsai, L.; Williams, M.; Yang, Z.Y.; Cannizzaro, C.; Leavitt, S.A.; Liu, X.; Wang, K.; Murray, B.P.; et al. In vitro characterization of GS-8374, a novel phosphonate-containing inhibitor of HIV-1 protease with a favorable resistance profile. Antimicrob. Agents Chemother. 2011, 55, 1366–1376. [Google Scholar] [CrossRef] [PubMed]
- Hruz, P.W.; Yan, Q.; Tsai, L.; Koster, J.; Xu, L.; Cihlar, T.; Callebaut, C. GS-8374, a novel HIV protease inhibitor, does not alter glucose homeostasis in cultured adipocytes or in a healthy-rodent model system. Antimicrob. Agents Chemother. 2011, 55, 1377–1382. [Google Scholar] [CrossRef][Green Version]
- LaFemina, R.L.; Schneider, C.L.; Robbins, H.L.; Callahan, P.L.; LeGrow, K.; Roth, E.; Schleif, W.A.; Emini, E.A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 1992, 66, 7414–7419. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.L.; Felock, P.J.; Hastings, J.C.; Blau, C.U.; Hazuda, D.J. The role of manganese in promoting multimerization and assembly of human immunodeficiency virus type 1 integrase as a catalytically active complex on immobilized long terminal repeat substrates. J. Virol. 1996, 70, 1424–1432. [Google Scholar] [CrossRef]
- Engelman, A.; Mizuuchi, K.; Craigie, R. HIV-1 DNA integration: Mechanism of viral DNA cleavage and DNA strand transfer. Cell 1991, 67, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Grobler, J.A.; Stillmock, K.; Hu, B.; Witmer, M.; Felock, P.; Espeseth, A.S.; Wolfer, A.; Egbertson, M.; Bourgeois, M.; Melamed, J.; et al. Diketo acid inhibitor mechanism and HIV-1 integrase: Implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. USA 2002, 99, 6661–6666. [Google Scholar] [CrossRef]
- Scarsi, K.K.; Havens, J.P.; Podany, A.T.; Avedissian, S.N.; Fletcher, C.V. HIV-1 Integrase Inhibitors: A Comparative Review of Efficacy and Safety. Drugs 2020, 80, 1649–1676. [Google Scholar] [CrossRef]
- Allers, K.; Schneider, T. CCR5Delta32 mutation and HIV infection: Basis for curative HIV therapy. Curr. Opin. Virol. 2015, 14, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Payra, S.; Manjhi, P.K.; Singh, S.; Kumar, R.; Singh, S.K.; Kumar, A.; Maharshi, V. HIV cure: Are we going to make history? HIV Med. 2024, 25, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yang, H.; Gao, Y.; Chen, Z.; Xie, L.; Liu, Y.; Liu, Y.; Wang, X.; Li, H.; Lai, W.; et al. CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo. Mol. Ther. 2017, 25, 1782–1789. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, S.; Jin, X.; Wang, Q.; Yang, K.; Li, C.; Xiao, Q.; Hou, P.; Liu, S.; Wu, S.; et al. Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4(+) T cells from HIV-1 infection. Cell Biosci. 2017, 7, 47. [Google Scholar] [CrossRef]
- Knipping, F.; Newby, G.A.; Eide, C.R.; McElroy, A.N.; Nielsen, S.C.; Smith, K.; Fang, Y.; Cornu, T.I.; Costa, C.; Gutierrez-Guerrero, A.; et al. Disruption of HIV-1 co-receptors CCR5 and CXCR4 in primary human T cells and hematopoietic stem and progenitor cells using base editing. Mol. Ther. 2022, 30, 130–144. [Google Scholar] [CrossRef]
- Chinnapaiyan, S.; Santiago, M.J.; Panda, K.; Rahman, M.S.; Alluin, J.; Rossi, J.; Unwalla, H.J. A conditional RNA Pol II mono-promoter drives HIV-inducible, CRISPR-mediated cyclin T1 suppression and HIV inhibition. Mol. Ther. Nucleic Acids 2023, 32, 553–565. [Google Scholar] [CrossRef]
- Ebina, H.; Misawa, N.; Kanemura, Y.; Koyanagi, Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013, 3, 2510. [Google Scholar] [CrossRef]
- Kaminski, R.; Chen, Y.; Fischer, T.; Tedaldi, E.; Napoli, A.; Zhang, Y.; Karn, J.; Hu, W.; Khalili, K. Elimination of HIV-1 Genomes from Human T-lymphoid Cells by CRISPR/Cas9 Gene Editing. Sci. Rep. 2016, 6, 22555. [Google Scholar] [CrossRef]
- Liao, H.K.; Gu, Y.; Diaz, A.; Marlett, J.; Takahashi, Y.; Li, M.; Suzuku, K.; Xu, R.; Hishida, T.; Chang, C.J.; et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015, 6, 6413. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Lei, R.; Le Duff, Y.; Li, J.; Guo, F.; Wainberg, M.A.; Liang, C. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology 2015, 12, 22. [Google Scholar] [CrossRef]
- Dash, P.K.; Chen, C.; Kaminski, R.; Su, H.; Mancuso, P.; Sillman, B.; Zhang, C.; Liao, S.; Sravanam, S.; Liu, H.; et al. CRISPR editing of CCR5 and HIV-1 facilitates viral elimination in antiretroviral drug-suppressed virus-infected humanized mice. Proc. Natl. Acad. Sci. USA 2023, 120, e2217887120. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-gene-therapies-treat-patients-sickle-cell-disease (accessed on 8 December 2023).
- Burdo, T.H.; Chen, C.; Kaminski, R.; Sariyer, I.K.; Mancuso, P.; Donadoni, M.; Smith, M.D.; Saryier, R.; Caocci, M.; Liao, S.; et al. Preclinical safety and biodistribution of CRISPR targeting SIV in non-human primates. Gene Ther. 2024, 31, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Ghosh, S.; Raghunath, M.; Bhaskar, R.; Sinha, J.K. Balancing potential benefits and ethical considerations of gene editing. Lancet 2023, 401, 2109–2110. [Google Scholar] [CrossRef] [PubMed]
- Uddin, F.; Rudin, C.M.; Sen, T. CRISPR Gene Therapy: Applications, Limitations, and Implications for the Future. Front. Oncol. 2020, 10, 1387. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Kraft, J.C.; Yu, D.; Ho, R.J.Y. Recent developments of nanotherapeutics for targeted and long-acting, combination HIV chemotherapy. Eur. J. Pharm. Biopharm. 2019, 138, 75–91. [Google Scholar] [CrossRef] [PubMed]
- Hamann, M.V.; Beschorner, N.; Vu, X.; Hauber, I.; Lange, U.; Traenkle, B.; Kaiser, P.D.; Foth, D.; Schneider, C.; Buning, H.; et al. Improved targeting of human CD4+ T cells by nanobody-modified AAV2 gene therapy vectors. PLoS ONE 2021, 16, e0261269. [Google Scholar] [CrossRef]
- Levy, J.M.; Yeh, W.H.; Pendse, N.; Davis, J.R.; Hennessey, E.; Butcher, R.; Koblan, L.W.; Comander, J.; Liu, Q.; Liu, D. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 2020, 4, 97–110. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
