Abstract
Root system architecture (RSA) is required for the acquisition of water and mineral nutrients from the soil. One of the essential nutrients, nitrate (NO3−), is sensed and transported by nitrate transporters NRT1.1 and NRT2.1 in the plants. Nitrate transporter 1.1 (NRT1.1) is a dual-affinity nitrate transporter phosphorylated at the T101 residue by calcineurin B-like interacting protein kinase (CIPKs); it also regulates the expression of other key nitrate assimilatory genes. The differential phosphorylation (phosphorylation and dephosphorylation) strategies and underlying Ca2+ signaling mechanism of NRT1.1 stimulate lateral root growth by activating the auxin transport activity and Ca2+-ANR1 signaling at the plasma membrane and the endosomes, respectively. NO3− additionally functions as a signal molecule that forms a signaling system, which consists of a vast array of transcription factors that control root system architecture that either stimulate or inhibit lateral and primary root development in response to localized and high nitrate (NO3−), respectively. This review elucidates the so-far identified nitrate transporters, nitrate sensing, signal transduction, and the key roles of nitrate transporters and its downstream transcriptional regulatory network in the primary and lateral root development in Arabidopsis thaliana under stress conditions.
1. Introduction
Nitrogen significantly influences plant growth and development. Plants adopt numerous strategies to modulate the uptake capacity of their roots to cope with spatial and temporal fluctuations in N availability [1]. In plants, the root architecture adjusts to these environmental fluctuations [2,3] and synchronizes the NO3− supply and demand inside the plants by the coordination of the systemic signal required to deal with root NO3− acquisition [4].
The regulatory pattern of root NO3− uptake simplifies the root transport system in two ways; The first is the rapid uptake after the NO3− provision, which requires de novo protein synthesis [5,6], and the other is the root NO3− efflux, strongly upregulated by N deficiency or low availability and downregulated by high nitrate supply [7,8]. An important hypothesis arising from the recently identified dissimilar NO3− influx and efflux and the low- and high-affinity NO3− transporters has revealed that several diverse carrier proteins are involved in the root nitrate (NO3−) transport system. Studies on Arabidopsis thaliana suggest that at least two transporters, NRT1.1 and NRT2.1, are involved in NO3− sensing [9].
NRT1.1 activates four signaling mechanisms [10]. Firstly, the primary nitrate response (PNR) [11], the long-term response of NRT1.1. Secondly, this then acts as feedback repression of NRT2.1 under a high NO3− supply [12]. Thirdly, the promotion of lateral root (LR) branching by NRT1.1 in response to NO3−, inhibiting the emergence of LR primordia at low NO3− availability [13], and finally, the induction of genes at high NO3− conditions [10]. After nitrate uptake via NRT1s and NRT2s, the part of the NO3− influx into the cell is reduced and thereby assimilated as amino acid through a series of enzymes such as nitrate reductase (NR), nitrite reductase (NiR), glutamine synthesis (GS), and glutamate synthase (GOGAT). These NO3−-mediated developmental processes are governed by a complex network of kinases and phosphatase [14], influencing the primary nitrate response (PNR) [15]. Further studies on sensitive Ca2+ biosensors have revealed that NO3− treatment upgrades Ca2+ concentration in the cytoplasm and nucleus of the protoplast from the mesophyll cells in the tip, pericycle, and stele of the intact roots. In-gel kinase assays have demonstrated that the activity of protein kinases (CPKs) is stimulated by NO3− treatment in protoplasts. Previous studies on protoplasts have distinguished subgroup III of the CPKs as regulators of NO3− responsive genes [16]. This further confirms the function of NO3− as an important signal that regulates gene expression, plant growth, and development [17].
The contribution of the nitrate transporter signaling pathway in the regulation and patterning of root system architecture (RSA) is momentous. This review discusses the significant milestones in the early response signaling and phosphorylation status of NO3− in Arabidopsis root, with greater emphasis on the signal transduction pathways that shape the architecture of the root in response to altered NO3− supply.
2. The Nitrate Signaling Mechanism in RSA
2.1. Nitrate as Early Response Sensing
The molecular identification and the functional characterization of the genes encoding the NO3− transporters in plants began in the mid-1990s and is still an active field of research [18]. The molecular mechanism of NO3− signaling transduction has been discovered in Arabidopsis. Nitrate transporter1/peptide transporter family (NRT1/NPF), nitrate transporter 2 (NRT2), chloride channel (CLC), and slowly activating anion channel (SLAC/SLAH) are the four nitrate transporter families that have been characterized in Arabidopsis [19].
Nitrate transporter 1.1 (NRT1.1), also called CHL1/NPF6.3, belongs to the NRT1/PTR family (NPF) [20]. As a dual-affinity nitrate transporter, NRT1.1 functions in both low and high nitrate affinity states [21,22], subsequently controlling root architecture by acting as a potential nitrate sensor [23,24] and triggering nitrate-dependent changes in gene expression. Moreover, its nitrate uptake function regulates the expression of key nitrate assimilatory genes. Its affinity state changes according to the phosphorylation status of the T101 residue [14,25]. NRT1.1 is capable of triggering independent signaling pathways in response to nitrate in Arabidopsis roots. Different NRT1.1 mutant alleles exhibit distinct responses to nitrate at the transcriptome level as well as the repression of LR development [10]. However, in NRT1.1 (CHL1/NPF6.3), the mutant’s chl1-9 allele, where proline 292 replaces leucine, shows imperfect NO3− affinity but exhibits a biphasic initial NO3− response for NRT2.1 [14]. Different studies have revealed that both chl1-9 and chl1-5 (deletion mutant of NRT1.1) are identical to the long-term suppression of NRT2.1 expression and LR development without nitrate [10].
Both the primary and secondary NO3− responses accomplished by transcriptomic studies indicate that the Affymetrix ATH1 chip has a significant impact on gene expression within 20 min after NO3− treatments. These changes were more apparent in roots than in the shoot, with the root having 1176 affected transcripts and only 183 affected transcripts in the shoot [26]. Another study revealed that after NO3− supply in nitrate-starved conditions, the NO3− transporters NRT1.1, NRT2.1, NRT2.2, and NRT2.4 were stimulated [27].
Hence, an additional sensing system may exist for NO3− influx or efflux with distinctive or overlapping signaling functions related to NRT1.1 [28]. For instance, under low NO3− conditions, the CBL-interacting protein kinases 23 (CIPK23) phosphorylates at threonine residue 101 (T101) by toggling NRT1.1 to a high-affinity nitrate transport system [29]. In the early NO3− response system, Ca2+ is a versatile signaling modulator in various regulatory pathways [30,31]. Ca2+ signaling is associated with NO3− responsive regulatory genes in Arabidopsis roots [16,32]. One should think of how the calcium signal is being triggered. There are some interesting viewpoints about this inquiry, elucidating that NRT1.1 is regulated by CIPK/CBL proteins, which are also themselves being regulated by calcium [14]. However, the mechanism behind the toggling of CPKs in the nucleus in response to nitrate is unknown [15].
2.2. Nitrate and Protein Kinases
The calcineurin B-like protein kinase, CIPK8, is rapidly activated by NO3− and downregulated in chl1-5 mutants. To study the function of CIPK8, the two independent T-DNA insertion mutants (cipk8-1 and cipk8-2) were isolated and a reduction in the cipk8 mutant was apparent via the induction of nitrate-responsive genes NRT1.1, NRT2.1, NIA1, and NiR. This then clearly demonstrated that CIPK8 functions as a positive regulator of the primary NO3− response in the low-affinity system [25]. Another protein kinase complex, CIPK23-CBL1/CBL9 (CIPK, CBL-interacting protein kinase; CBL, calcineurin-B-like protein), has been associated with dual-affinity transition changes of NRT1.1 via phosphorylation [14,33]. Further studies have also shown that FIP1 (factor interacting with poly (A) polymerase 1) adversely regulates the expression of CIPK8 and CIPK23 associated with NO3− signaling. In the fip1 mutant, the increased expression of CIPK23 may affect NO3− uptake and subsequently reduce NO3− content. Molecular genetics suggest that FIP1 and CPSF30-L operate similar NO3− signaling pathways. FIP1-induced NO3− signaling interacts with CPSF30-L and is regulated by CIPK8 and CIPK23 [34,35].
The role of the subgroup III protein kinases (CPKs) CPK10, -30, -32 in NO3− regulated root growth was examined [15]. The NO3−-induced LR primordial density was reduced and LR elongation was significantly hindered in icpk [15], thus associating the inhibition of nitrate–CPK-stimulated genes with transcription, metabolism, and transport activities [15]. The activity of the CPKs can be enriched within 10 min in response to nitrate. These CPKs have been distinguished as the primary regulators that coordinate the essential NO3− response [15] and modulate various essential cell and metabolic functions instantly triggered by NO3− [36,37].
CPK10 and CPK30 have also been shown to be associated with the abscisic acid (ABA) responsiveness of the mesophyll protoplasts, which is a promising avenue of research on the coregulation of NO3− and ABA pathways. Both have been speculated to contribute to the regulation of the root growth and gene expression [37]. For instance, ABI2 (ABA-insensitive 2) phosphatase is a fundamental component of the ABA sensing system [38]. Besides the CIPK23–CBL9 complex functioning in the dual affinity transition changes of NRT1.1, ABI2 and CBL1 also interact with phosphorylated CIPK23, which is recognized as an additional segment of this regulation process. NO3− sensitivity instigates a rapid increase in the cytoplasmic Ca2+ level downstream of NRT1.1 in a PLC-dependent manner [28].
In short, nitrate-mediated CPK signaling phosphorylates transcription factors to regulate the expression of downstream genes that affect nitrogen assimilation, carbon/nitrogen metabolism, and proliferation [15]. However, it is possible that additional NO3− sensors and NRT1.1-independent pathways could be involved in the Ca2+ influx and other signaling measures [28,39] (Figure 1). An increase in Ca2+ initiates a change in the protein phosphorylation status while controlling the movement of the key component of the NO3− signaling pathway. CPK10, 30, and 32 work as regulators of the essential NO3− response, linking the Ca2+ influx with the phosphorylation of the target proteins. CPK activation could also be linked with NRT1.1-dependent pathways [28].
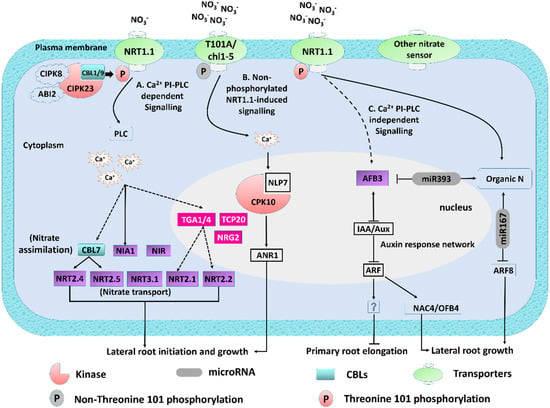
Figure 1.
Summary of early responses in nitrate signaling and assimilation. NO3− signaling pathway switches its affinity via phosphorylation (modified from Undurraga [41]). Nitrate-responsive genes are depicted in light green, transcription factors in purple, and microRNAs in grey. For clarity purposes, the cell nucleus is shown. Phosphatidylinositol-specific (PI-PLC) and Ca2+-dependent pathways. At Low NO3− condition, protein kinases CBL1/9–CIPK23 complex phosphorylates NRT1.1 and changes it into a high-affinity transporter, which activates PLC and results in calcium influx (Ca2+ acts as a second messenger). This cascade mediates changes in the expression of transcription factors (TGA1/4 *) and genes involved in nitrate transport (NRT2.1, NRT2.2, and NRT3.1) and nitrate assimilation (NIA1 and NiR). Nonphosphorylated form of NRT1.1-induced signaling. Nitrate-induced Ca2+-ANR1 signaling that promotes lateral root (LR) initiation is assumed to be a nonphosphorylated form of NRT1.1 signaling after the supply of nitrate in limited-nitrate conditions. (C) PI-PLC and Ca2+-independent pathways. Conversely, AFB3 is regulated by nitrate in a phospholipase C (PLC)- and calcium-independent manner. ABF3 modulates the expression of NAC4 and OBP4 with subsequent effects on root remodeling. Finally, nitrate assimilation produces organic N, which induces miR393 and represses miR167 (grey) and regulates the abundance of AFB3 and ARF8, respectively. * TGA1 and TGA4 are redundant regulatory factors that mediate nitrate responses in Arabidopsis roots. However, the interaction between TGA4 and the PLC–calcium pathway has not been experimentally validated.
3. Nitrate Signaling and Calcium
Previous studies have revealed that nitrate treatments abruptly raise cytoplasmic Ca2+ levels in the roots as well as in the entire seedling [16] (Figure 1). This confirmed that the function of Ca2+ in nitrate signaling originates from early research on corn and barley, where EGTA or LaCL3 alters the expression of NO3−-responsive genes. The potential role of Ca2+ as a second messenger was thus indicated [32,40].
Ca2+ sensor proteins perceive changes in the (Ca2+)cyt and subsequently transduce downstream signaling cascades to stimulate alteration of enzymatic activity, cytoskeleton orientation, phosphorylation, and gene expression [42,43]. This was further confirmed by the pretreatment of seedlings with phospholipase C inhibitors or Ca2+ channel blockers, which severely affected NO3−-responsive gene expression in Arabidopsis, indicating the function of Ca2 as a secondary messenger in NO3− signaling pathways. A model was therefore suggested, where the (Ca2+)cyt level increases by NRT1.1 and phospholipase C activity in response to NO3−, which is required for changes in the prototypical NO3−-responsive gene expression [16]. Taken together, both NRT1.1 and phospholipase activity are mandatory for NO3−- mediated increase in cytoplasmic Ca2+ levels and IP3 (Figure 1) [16].
PLC enzymes are membrane-associated, resulting in the remodeling of lipid membranes by the breakdown of phospholipids and the subsequent production of multiple secondary messengers [16]. In plants, two classes of PLCs exist, and they are distinguished based on their substrate specificity. One is phosphatidylinositol-specific (PI-PLC) and the other is non-specific (NPC). Plant NPCs share homology with bacterial PLCs. NPCs can incline either phosphatidylcholine-specific phospholipase C (PC-PLC), phosphatidylethanolamine (PE-PLC), or phosphatidylserine (PS-PLC). However, PI-PLC is the most considered class of PLC, which hydrolyzes phosphatidylinositol 4, 5-bisphosphate (PIP2) from the plasma membrane to create IP3 and diacylglycerol (DAG) [44]. The nitrate signaling and phosphatidylinositol-specific PI-PLC links were found in Arabidopsis. Nitrate triggers Ca2+ and inositol 1, 4, 5- triphosphate (IP3), which were not witnessed in the plant’s pretreatment with PLC inhibitor U73122. For instance, the NRT1.1 mutants, chl1 and chl9, revealed that this was an NRT1.1-based response. The associated rise in IP3 after NO3− treatment also suggested that the activity of phospholipase C (PLC) was associated with this signaling pathway [16].
In Arabidopsis thaliana, expression analysis of different PI-PLC genes demonstrated that PLC isoforms were differentially expressed in different plant organs [45,46,47] and that the expression of AtPLC1, 2, 3, 4, 5, and 9 were root-specific [48,49].
3.1. Nitrate-Induced Ca2+ and PI-PLC-Dependent Signalling
Phosphatidylinositol-specific phospholipase C (PI-PLC) is the major part of nitrate signaling and transport, modulated by the phosphorylation/dephosphorylating process. Both plasma membrane and tonoplast nitrate transport activity are regulated by phosphorylation [27,29]. In Arabidopsis, Ca2+ has a definite role in plant signal transduction and is also significant for the NO3−-mediated signaling of gene expression. As stated earlier, NO3− treatment rapidly increased the cytoplasmic Ca2+ level in the roots [27,29] (Figure 1) and nitrate is absorbed in the root cell by plasma-membrane-localized nitrate transporter families, NRT1 and NRT2 [22]. NRT1.1/CHL1 is a low-affinity transporter that switches to a high-affinity transport system when NRT1.1 is phosphorylated at the threonine residue 101(T101) by protein kinase CBL1/9-CIPK23 [9]. The protein complex CIPK23–CBL9 (CBL-interacting protein kinase (CIPK); calcineurin-B like protein (CBL)) and CIPK8 have been implicated in the dual-affinity transition changes of NRT1.1 through phosphorylation [33]. More recent studies have revealed that a protein phosphatase 2C (PP2C) family member, ABI2 (ABA-insensitive 2), and the calcium sensor CBL1 were distinguished as supplementary constituents that modulate NRT1.1 transport functions and NRT2.1 expression in root growth NO3− responses [38] (Figure 1).
Hence, the phosphorylation activates a weak upregulation of high-affinity nitrate transporter NRT2.1 [14], and subsequently induces NRT1.1, NRT2.1, NRT2.2, and NRT2.4 under nitrate-starved seedlings after nitrate supply, while upregulating all the nitrate assimilatory genes [27,50]. CPK phosphorylates the NLP TFs, particularly NLP7, which interact with CPK20 in the nucleolus under NO3− availability. Besides NPL7, more TFs, such as TCP20, also contribute to the NO3−-induced transcriptional changes and systemic signaling. In contrast, TGA1/4 controls the genes which participate in the PNR, transport, metabolic, and developmental processes [28].
Under limited-nitrate conditions, the NRT1.1 is, therefore, phosphorylated at the T101 in order to stimulate NRT1.1 association with membrane microdomains at the plasma membrane (PM). When nitrate supply is increased, the nonphosphorylated NRT1.1 shows oligomerization and low structural mobility at the PM, thereby initiating rapid inducible endocytosis. These activities could promote LR growth by switching NRT1.1-auxin transport activity on the PM and stimulating Ca2+-ANR1 signaling from the endosomes (discussed in detail in Section 3.2.1, nonphosphorylated nitrate signaling) [51].
3.2. Differential Phosphorylation State of Nitrate Transporters
NRT2.1 is firmly induced by the nonphosphorylated form of NRT1.1, which transports NO3− at low affinity. After prolonged exposure to NO3− treatment, NRT2.1 is repressed by phosphorylated NRT1.1. The NO3− transport capacity under this condition remains obscure [52].
However, after the point mutation at the plasma membrane, the mode of NRT1.1T101 phosphorylation may be different in both NO3− uptake and signaling. Transgenic plants of T101A, which mimic the NRT1.1/CHL1 dephosphorylation, exhibits only low-affinity NO3− uptake, but can also sense NO3− at high-affinity range, with the high-affinity for NO3− being comparatively less than the wild-type (WT) [14]. These properties propose that WT NRT1.1 and the T101A mutant may have two NO3−-binding sites; high affinity and low affinity. It is worth noting that only the low-affinity binding site of the T101A mutant can be transported over the plasma membrane (PM). Unlike NO3− uptake, NO3− binding to both sites of T101A mutants could trigger the NO3− response. This could justify the reason why the CHL1T101A mutant still exhibits a biphasic primary response [14]. In contrast, T101D- expressing transgenic plants that mimic phosphorylated NRT1.1/CHL1 displayed only high-affinity NO3− uptake activity and are activated only at a high-affinity primary NO3− response. This suggests that T101D can only bind NO3− with a high-affinity uptake system [14]. Subsequently, it could be possible that binding sites with low affinity could be blocked by T101 phosphorylation [14] (Figure 2).
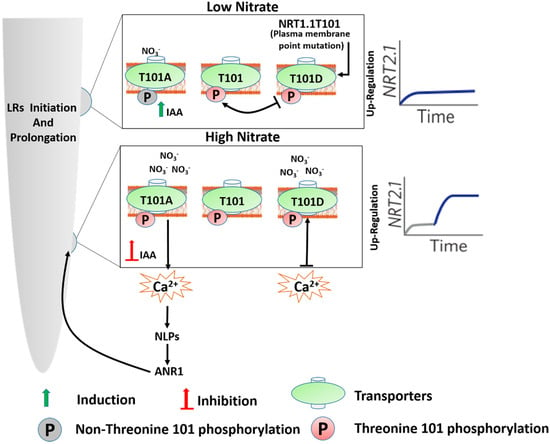
Figure 2.
The schematic diagram describes the differential phosphorylation status of NRT1.1.1T101 at plasma membrane (PM) in the Arabidopsis root, modified by [52]. The layout represents the two binding sites’ low affinity (LA) and high affinity (HA) of T101A. The T101A mutant at the LA binding site follows the NRT1.1-ANR1 signaling pathway upon prolonged exposure to the NO3− under low-nitrate conditions, resulting in LR elongation. This is a nonphosphorylated form of NRT1.1-induced signaling that promotes LRs. In the inserted graph, the grey line represents the weak upregulation of NRT2.1 under low nitrate, and the blue line represents the strong upregulation of NRT2.1 under high nitrate. The graphs on the left and right represent the NRT2.1 induction; see text for more details.
The two NO3− binding sites depicted here (Figure 2) have two adaptations of a single binding site. Taken together, these findings suggest that at the low level of NO3− sensing, T101 phosphorylation keeps the PNR, whereas, for uptake and substrate-binding, T101 phosphorylation may repress the low-affinity NO3− binding and is then required to use the high-affinity transport system [14].
3.2.1. Non-Phosphorylating Form of NO3−-Induced Signaling
NRT1.1 contributes to the NO3−-mediated auxin transport, regulates auxin storage, and subsequently influences LR development [53]. The signaling network comprising of Ca2+, Ca2+-protein kinases (CPKs), and NIN-like protein (NLPs) interacts with NO3− via primary transcription to regulate LR growth [23,45]. In addition to this Ca2+-, ARABIDOPSIS NITRATE REGULATED1 (ANR1), a transcription factor functioning downstream of NRT1.1 and NLP7, has been involved in LR elongation under high NO3− (HN) conditions [54]. In a plant developmental network, nitrate-induced Ca2+-ANR1 signaling is a nonphosphorylated form of NRT1.1 signaling, promoting LR growth. NO3− triggers a unique Ca2+-CPKs-NLPs signal, acting as downstream segments of NLP and ANR1, subsequently controlling LR elongation [16] (Figure 1).
NRT1.1 phosphorylation influences cytoplasmic Ca2+ ((Ca2+)cyt) levels in the epidermal cells of the LRs, which was measured by using Fluo-4 dye in various genotypes [52]. In view of the pseudocolor and kymograph pictures of wild-types (WTs), after NO3− stimulation, (Ca2+)cyt signaling was screened at the proposed 60-second period. This was previously depicted by [16]. The researcher found that NO3− explicitly induced Ca2+ signature in the WT but not in chl1-5 mutant seedlings. Under both HN and LN conditions, T101A seedlings exhibited a transient increase in (Ca2+)cyt [51], while T101D seedlings displayed a decrease in [Ca2+]cyt. Concomitantly with [Ca2+]cyt accumulation, HN-stimulated expression of ANR1 in LRs is sensed in T101A, but not in T101D. In the light of these findings, it is suggested that a nonphosphorylated form of NRT1.1 could activate the Ca2+-CPKs-NLPs signaling pathway to induce the expression of ANR1, and subsequently control LR elongation [51]. It was analyzed that intracellular transport of T101A and T101D in LR cells showed that differential phosphorylation of NRT1.1 enhanced the implementation of NRT1.1-stimulated signal transduction in LR growth [51]. Phosphorylated NRT1.1 takes up the sparingly accessible NO3− from the soil at high affinity and induces the NRT2.1 expression to a lower extent compared to the low-affinity state [52] (Figure 2). Under high NO3 conditions, NRT1.1-induced auxin transport is inhibited, and shortly after NO3−-treatment, the dual affinity modes of the NRT1.1 are regulated at Thr-101(T101) phosphorylation [52].
As mentioned earlier, under low NO3− conditions, phosphorylation at T101 stimulates NRT1.1 association with a functional membrane microdomain at PM [51], confirming the NRT1.1-mediated auxin flux, and subsequently repressing their growth by reducing the LRP auxin level. With an increased NO3− level, nonphosphorylated NRT1.1 shows oligomerization and low lateral mobility at the PM and rapid inducible endocytosis. This activity may stimulate LR development by supporting NRT1.1-auxin transport activity on the PM to induce Ca2+-ANR1-signaling from the endosome [51]. Further studies have shown that seedlings of T101A had much higher LR density than that of T101D when grown under low NO3− conditions (0.2 mM), whereas in high NO3− conditions (1 mM), no significant difference was observed in the LR density of the mutants compared to WT plants [51]. These findings confirm that that T101A and more nonphosphorylated WT NRT1.1 promote LR growth in LN by suppressing basipetal auxin transport, and subsequently accumulating auxin in the LR tips [51].
3.3. Nitrate-Induced Ca2+ and PI-PLC-Independent Signaling
Ca2+ and PI-PLC are not affected by the expression of NO3− responsive auxin signaling F-Box3 (AFB3) protein, indicating that beyond Ca2+ and PI-PLC, there is a PI-PLC-independent pathway that controls the regulation of the nitrate-sensitive genes [16,55] (Figure 1). Hence, NRT1.1 toggles within the phosphorylation status of a critical threonine residue from low- to high-affinity states. This residue is amongst the second and third transmembrane helices of NRT1.1 located in the intracellular side [14,25].
In Arabidopsis root, Ca2+ and PI-PLC-independent miR393/AFB3 regulatory modules are recognized as nitrate responsive genes, which assimilate nitrate and auxin signaling [56]. Nitrate induced LRs are dependent on miR167, and its target auxin-responsive factor ARF8 mRNA [57] plays a distinctive role in regulating several genes connected via a network to promote the stimulation of LR initiation and inhibition of elongated roots in response to N [57] (Figure 1). This earlier identified regulatory module, controlled by miR393 microRNA and the AFB3 auxin receptor, stimulates LRs in response to external and internal NO3− applications [51,58]. AFB3 is induced by NO3− and repressed by miR393, whereas nitrate reduction and assimilation produced N metabolites, which induces miR393 [59] (Figure 1). Furthermore, AFB3 coregulates NAC4 and OBP4, and this coregulation is confirmed by using the green fluorescence protein (GFP)-expressing lines after 2 h, in response to nitrate. AFB3, activated in the pericycle, indicated that the AFB3-NAC4-OBF4 complex might build a regulatory module that controls LR growth in a NO3−-dependent manner [56].
Nitrate-stimulated AFB3 induced in the root might be a specific signaling network of Aux/IAA and ARF factors to modulate NAC4 activation and LR growth. The abundant Aux/IAA-ARF modules chronologically generate new LRs and control LR development in Arabidopsis. The lateral root basal meristem (the zone between meristem and elongation) depends on IAA28 and ARF proteins, which include transcription factors ARF5, ARF6, ARF7, ARF8, and ARF19 [13,53]. In plant RSA, the LR initiation and emergence of the AFB3 overexpression line and the afb3 mutant line have emerging roles compared to wild-types and display increased growth of LRs under nitrate-sufficient conditions. Additional findings revealed that the transcription factor NAC4, which functions downstream of AFB3, might be involved in two dependent pathways of RSA regulation [52,58]. Following AFB3, NAC4 acts downstream in the pericycle cell to alter LR density in nitrate treatments [9,51].
Auxin Response Network
Auxin signaling is primarily passed over by transcriptional pathways for morphogenesis and developmental processes, which include TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) proteins, AUXIN/INDLOE-3-ACETIC ACID (AUX/IAA) transcriptional co-regulators and AUXIN RESPONSE FACTOR (ARF) transcription factors [60,61,62]. At low levels of auxin concentration, members of the transcriptional inhibitor family AUXIN/IAA-INDUCIBLE (AUX/IAA) interact with the DNA-binding protein of ARF [56,57], while the ARF proteins function to detect the auxin-response promoter elements (AuxREs) in various auxin-regulated genes to activate or suppress their expression [63,64]. AUX/IAA protein inhibits the ARF function either by passively inhibiting ARF proteins from their target promoters [65] or by binding ARF with the corepressor TOPLESS (TPL) for inactivation of the chromatin and silencing of ARF target genes [56,59,66]. An increase in auxin concentration by an auxin-induced module of the coreceptor complex consists of F-box protein from the TRANSPORT INHIBITOR RESPONSE 1 (TIR1)/AUXIN SIGNALING F-BOX PROTEIN (AFB) family and is an Aux/IAA member [60,67,68]. TIR1/ABFs, a subunit of nuclear S-PHASE KINASE ASSOCIATED PROTEIN 1-CULLIN-F-BOX PROTEIN (SCF)-type E3 ubiquitin-protein ligases (SCFTIR/AFB), stimulate the recognition of substrates. The auxin response is initiated by connecting hormones to the TIR1/AFB receptor. The auxin receptor is part of the SCFTIR1/AFB ubiquitin ligase complex [69,70]. Binding of auxin to its receptor TIR1/AFB activates the information and breakdown of the polyubiquitination of the Aux/IAA inhibitor, which subsequently releases the inhibition of ARF transcription factors, which induce the transcription of auxin-responsive genes [71,72]. This represents the pivot of auxin signaling.
In a simpler form, auxin-initiated AUX/IAA removal relieves ARF inhibition and activates the transcription of primary genes. Remarkably, the auxin response network is enough to reconstitute the AuxRE-dependent activation of reporter genes in yeast [73]. Hence, in Arabidopsis root, a miR393/AFB3 regulatory module is recognized as nitrate-responsive, which assimilates nitrate and auxin signaling to promote root growth [56].
4. The Effects of Nitrate on RSA
4.1. Effects on Primary Root Growth and Development
Generally, the primary root (PR) growth in Arabidopsis is typically found to be relatively insensitive to or even induced by the normal range of NO3− concentration [24,74,75]. It could be inhibited under some culture conditions by moderately high NO3− supply [56]. It was presumed that AFB3 controls LR initiation and PR development by two distinct pathways, of which one is NAC3-dependent, while the other is NAC3-independent [76]. However, studies on the effect of amino acid and peptide on root growth and branching have gained little attention [77]. At a low concentration of glutamate (<50 mM), the PR tip has a unique and differential effect on root architecture, inhibiting PR growth and subsequently stimulating LR growth [78] (Figure 3).
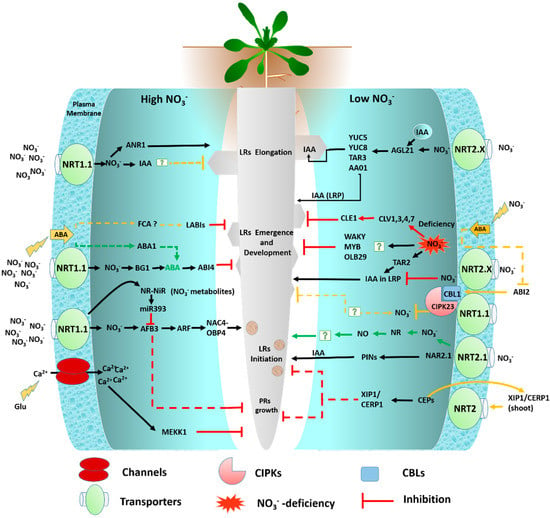
Figure 3.
The schematic diagram presents the multiple pathways regulating the root system architecture (RSA; lateral and primary root) response to the localized and high nitrate conditions in Arabidopsis. Only those pathways discussed in the present review are depicted. The green arrows indicate systemic transport and assimilation, the black arrows indicate positive signaling as a stimulatory effect, red lines indicate negative signaling as an inhibitory effect, the orange lines depict the unknown positive and negative signaling pathways, and dotted lines represent the unconfirmed nitrate-mediated signaling pathways. The low nitrate and severely low nitrate conditions have been reported to have a stimulatory and inhibitory effect on LR development, respectively, while high NO3− supply has an inhibitory effect on LR growth [24] (see text for further information). External NO3 regulates primary root growth in Arabidopsis. The receptor for the external glutamate signal is shown as a glutamate-gated Ca2+ channel because these are known to be activated at root tips [79]. However, its specific role in this signaling pathway is unconfirmed (see text for further information).
To this end, this response is glutamate-specific in Arabidopsis since an ongoing study of the impact of 17 other proteinogenic amino acids on the architecture of the roots found none that could produce its distinctive effect on root architecture [77]. By using a chemical genetic approach, the MEKK1 MAP kinase gene has since been investigated as part of the glutamate signaling pathways in PR tips [80]. MEKK1 functions mainly as a distinctive immune system and its expression was demonstrated to be profoundly receptive to a variety of abiotic factors [81]. Nitrate exhibits a strong signal to stimulate the primary root development by enhancing the activity of the meristem and cytokinin signaling. Cytokinin sensing and biosynthesis mutants showed shorter roots compared with wild-type when subjected to NO3− treatments, especially when NO3− is the primary source [82]. Histological studies of the root tip revealed reduced cell division and elongation in the cytokinin receptor double mutant ahk2/ahk4 (histidine kinase) compared with WT plants under adequate NO3− supply. It is worth noting that as NO3−-mediated restriction in the root growth was observed between 5 and 6 days after planting, the WT plants had the potential to recover from the growth-restricted condition, whereas cytokinin signaling or biosynthesis mutants were most certainly not capable of recovering [82].
In addition, the transcriptomic analysis indicated that genes associated with both cell division and elongation are possibly significant for PR development in response to NO3−, thereby indicating the interaction between nitrate and cytokinin signals in regulating PR development in Arabidopsis [82].
4.2. Effects on Lateral Root Growth and Development
The growth of lateral roots is strongly affected by the concentration of N in the growth environment. For instance, in low NO3− soil, patches of high NO3− have a localized stimulatory impact on LR development, which varies in different plant species [2,74], whereas under high NO3− conditions (with no restricted growth), LR development is repressed [83]. Further studies also revealed that NO3− plays a prominent role in regulating LRs. Generally, low NO3− has a dual effect on the LRs, such as stimulatory as well as inhibitory effects, whereas high NO3− supply only exhibits an inhibitory effect on LR growth and development of LRs [4]. In other words, there are two clear morphological adaptations. Under N-deficient conditions, the LRs are significantly stimulated; however, when exposed to more severe N deficiency, the entire LR length reduces and LR formation disappears [13]. This is initiated by the signaling impact of NO3− itself, rather than downstream metabolites [2].
4.2.1. Stimulatory Effect of Low Nitrate on LR Growth
The low NO3−-stimulated Arabidopsis LR development depends on the role of the auxin biosynthetic gene TAR2 (tryptophan aminotransferase related 2; Figure 3), which is expressed in the pericycle and vasculature of developed roots close to the root tip and is stimulated under low-nitrogen conditions. In WT plants, the low NO3− restored auxin accumulation in the primordial of the nonemerged LRs, with an additional three cell layers and LR emergence. On the other hand, these low N-stimulated auxin accumulation and root developmental responses were disrupted in tar2 null mutants [4,51]. Subsequently, TAR2 is required for restructuring the root architecture in response to low N conditions. Another nitrate responsive gene, BBX16 (bobby sox homolog), belongs to the constans-like zinc finger family. The bbxl16-1 mutant affects lateral root length (LRL) in response to NO3−, with longer LRs by 1 mM KNO3− as low nitrate treatment. The bbx16-1 mutants produce larger LRs under NO3− limitation [48] (Table 1).

Table 1.
Transcription factors of genes associated with nitrogen signaling and nitrogen-associated processes in Arabidopsis thaliana.
When the NO3−-deficient condition becomes severe, the Arabidopsis AGL17-clad MADs-box gene AGL21 is induced by N shortage and auxin to promote LRs in Arabidopsis, whereas agl21 mutants exhibit a reduction in LR elongation in response to low NO3− treatments. Furthermore, the auxin biosynthesis genes YUC5, YUC8, and TAR3 are significantly upregulated in overexpressing (OE) lines and downregulated in agl21 mutants, demonstrating that AGL21 enhances the local auxin activity in the LR primordial, and thus substantially influencing LR growth regulation [77,80,88].
Previous studies about rice have revealed that the AtNRT2.1 homolog OsNAR2.1 knock-out mutant initiates the inhibition of LRs under low NO3− concentration by reducing PIN protein levels in the roots [89]. NRT2.1 positively regulates LRs by influencing the polar transport of auxin under low NO3− conditions. The impact of NRT2.1 on LR growth is possible by a combination of NO3− uptake and signaling. NRT2.1 cannot function independently as a NO3− transporter. Hence, NRT2.1 might act as a key factor in this signaling pathway [4]. It was thus demonstrated that OsNRT2.1 could be involved in the nitrate-dependent pathway of root elongation by regulating auxin transport to the roots under low NO3− conditions [90]. Apart from the aforementioned pathways comprising both transcriptional factors and hormonal signals, nitric oxides (NOs) have been accounted for as a significant NO3−-mediated signal which regulates RSA in plants [79,82]. In rice, NO produced by NR could enhance the inadequate production of N by developing LR initiation under partial NO3− availability [91,92]. To this end, LRs are significantly stimulated by mild NO3− deficiency. Different molecular players are involved in the regulation of different stages of plant growth and development.
4.2.2. Inhibitory Effects of Severely Low Nitrate on LR Growth
Earlier studies have found that the impact of NO3− was related to the ability of the localized NO3− supply to stimulate LR elongation [23,83]. Experimental estimation of using a limited, rather than uniform, NO3− treatment initiates the specific effects of the external NO3− on LR development, and this can be observed under conditions where the systemic effects, due to changes in the N status of the plant, can be limited to a greater extent [2,4]. Under severe N deficiency, both LR formation and length are repressed in plants [93].
A recent investigation [4] featured the vital role of the peptide-receptor signaling module, which comprises N-responsive CLE (CLV3/ENDOSPERM SURROUNDING REGION (ESR))-related peptides and the CLAVATA1 (CLV1) leucine-rich repeat receptor-like kinase regulatory module, in regulating LR growth of Arabidopsis thaliana. CLE1, −3, −4, and −7 are expressed in root pericycle cells of Arabidopsis roots. Under NO3− deficient conditions, overexpression (OE) of CLE genes results in the repression of LR emergence from the PR. This inhibitory action of the CLE peptides also affected LR development required for the feedback function of CLV1 expressed in the phloem of the root companion cells, indicating that the downstream signal is transmitted via phloem for the systemic regulation of RSA [4]. An additional system, downstream of CLV1 feedback, regulates the transcript level of the N-responsive CLE genes in the roots for fine-tuning of the signal amplitude [4,89]. In other words, CLEs-CLV acts as a regulatory module in NO3− signaling pathways, and it also antagonistically controls the growth of LRs under limited N conditions [4,94].
Similarly, one member of the CEP (C-TERMINALLY ENCODED PEPTIDE) gene family has been shown to arrest root growth [95]. The analysis of OE-lines of several CEP genes demonstrates their distinctive function. It was reported that CEPs have an antagonistic effect on LR growth while initiating a delay in PR and LR growth [95].
Another mechanism of the systemic inhibition of LR growth is associated with the inhibition of LRs in response to NO3−. Limited NO3− supply significantly increases abscisic acid (ABA) accumulation, as this ABA accumulation inactivates its coreceptor ABI2 (ABA-insensitive 2) and protein phosphate 2C (PP2C) [96] (Figure 3). The ABI2 then co-interacts with Ca2+-sensor subunit CBL1 and the kinases (CBL1-CIPK23) complexes, with their substrate being NRT1.1/NPF6.3. Hence, under low NO3− conditions, the protein kinase CIPK23 phosphorylates NRT1.1 to sustain movement at low NO3− concentrations [14] (Figure 3). This hypothetical pathway, reconfirmed in recent studies, has revealed that alteration in the ABI2 status promotes the activation of the CBL1–CIPK23 complex, and subsequently reduces root NO3− uptake by inhibiting NRT1.1 transport activity under NO3−-deficient conditions [38]. However, the downstream constituents of this pathway are still unknown. It is thus still unclear whether the antagonist effect of ABA on the LRs, subjected to low N conditions, is a consequence of the disrupted NO3− signaling pathway or physiological function of ABA itself.
Moreover, irrespective of the NO3− activity, NRT2.1 functions as a NO3− sensor or signaling component to inhibit LR initiation under low-NO3 conditions [23,93]. However, their exact underlying mechanism is still unclear. The negative effect of NRT1.1/NRT2.1 on LR growth indicates the distinct systemic pathways under limited NO3− supply [4]. Taken together, NRT1.1/NRT2.1 has a negative role in LR growth and possibly clarifies the inhibitory effect of high NO3− on L development. NRT1.1/NRT2.1 functions negatively and also have an inverse effect on these signaling pathways to control LR growth and development under limited NO3− conditions. The action of each pathway depends on the level of the N deficiency in plants or their specific ecological conditions [4].
4.2.3. Systemic Inhibitory Effect of High External Nitrate on LR Growth
The LRs of Arabidopsis exhibited two different responses to high NO3−. High NO3− (10 mM) conditions decreased the entire root system, whereas, when plants are subjected to low NO3- concentrations (10 µM), the PR part exposed to high NO3− triggered the local induction of LR elongation [2,83]. However, the global inhibitory effect of NO3− appeared to be as a result of prolonged exposure of plants to ample NO3− supply. The LR elongation under this condition was also suppressed in the areas of the root system that were subjected to the state of low NO3− conditions [2,97].
As reported earlier, the AFB3 receptor gene is strongly induced by NO3−, and the LR initiation is specifically diminished in afb3 mutants [59]. Research on the nitrate reductase (NR)-null mutants has revealed that NO3− itself was the main stimulator of AFB3. AFB3 expression feedback is regulated by nitrate-assimilatory products, such as miR393, a micro RNA that targets AFB3 transcript for degradation. This pathway has further confirmed the findings that nitrate (NO3−) induced NAC4 and OBP4 transcription factors, functioning downstream of AFB3. Taken together with the results obtained from nac4 mutants, the afb3 mutant displays an apparent reduction in LR growth in response to NO3− [56]. Similarly to this was the influence of the myb29-1 allele on lateral root length (LRL) when subjected to diverse NO3− conditions, exhibiting shorter lateral root length (LRL) at high NO3− (10 mM KNO3–) treatments [48]. However, the rav2-1 and erf107-1 alleles, which are genotype-dependent, exhibited reduced lateral root length (LRL) when subjected to both 1 and 10 mM KNO3 conditions [48] (Table 1). Recent studies have demonstrated that high-affinity NO3− transporter AtNRT2.1 may be involved in the inhibition of LR initiation at high C: N ratios [98]. Also, the involvement of the ABA affecting LR growth, in response to NO3−, might be connected to the recently identified ABA receptor [98]. Nitrate reductase (NR)-lacking mutants display sensitivity to this systemic inhibitory effect, indicating that NO3− concentration in the tissue of plant cells may function in inhibitory signal induction. Thus, this model defines root branching, as modulated by inhibitory signals via internal N status and external NO3− supply [83].
Furthermore, ABA, which is associated with the systematic inhibitory effect of high NO3− on LR growth, might be connected with the recently identified ABA receptor FLOWERING CONTROL LOCUS A (FCA). In addition, root architecture response to the recently identified external L-glutamate conceivably provides a significant tool for studying biological functions of plant glutamate receptors and amino acid signaling [98]. It was also reported that FCA possibly acts as a receptor for ABA. The loss of function mutant fca displays low sensitivity to the inhibitory effect of ABA on LRs, indicating that FCA might be a constituent in signaling transduction pathways associated with high NO3− ABA-mediated inhibition of LRs [4,99,100].
It has been genetically proven that inhibition by ABA and NO3− is mediated by the same signaling mechanism. For instance, the LABI (lateral roots ABA-insensitive) is characterized based on the LR production affinity when exposed to 0.5 µM, which is less sensitive to the high NO3−-induced LR inhibition [4] (Figure 3). The identification of LABI genes could give indepth information about the signaling mechanism underlying this inhibition [98]. Interestingly, all the mutants produced shorter primary roots phenotypes, which indicated that LR development could be intrinsically correlated with PR growth. It was reported that the presence of the PR meristem is required for high NO3− and ABA-induced inhibition; however, this inhibition could be eliminated by the removal of the PRs [4].
Furthermore, root architecture response to glutamate may give an essential experimental framework to study glutamate signaling in plants and to elucidate the possible roles of the glutamate receptor [98]. Recent studies have shown that high NO3− supply (30 mM) stimulated ABA accumulation in the emerging root tips by discharging it from the inactive stores via ER-localized β-GLUCOSIDASE1 (BG1) to regulate root development. This information provides a system for NO3−-induced root development via the regulation of ABA accumulation in the root tips. It was hypothesized that there is a close association between ABA and NO3− signaling to coregulate LR growth [81]. A recent study has also shown that myb29-1 mutants increased the LR length, LR density, and total length under adequate NO3− supply in a genotype-dependent manner [48] (Table 1).
5. Coordinated Regulation of Nitrate and Other Messengers on RSA
Root foraging for NO3− involves both local and systemic signaling. NO3−-auxin-CK regulation could also be a key constituent of N systemic signaling, which coordinates nutritional requirements among various organs at different growth stages [101,102].
5.1. Nitrate-Mediated Auxin Allocation
A systemic regulation that includes the inhibition of auxin translocation from the shoot to root suppresses LR initiation and development and subsequently affects NO3− use efficiency in plants [103]. In such a situation, growing Arabidopsis thaliana on a nitrate medium was observed to have reduced auxin contents in the roots, while increasing the auxin content in the shoots. These findings have demonstrated that high NO3− inhibits the translocation of auxin from the shoots to the roots [78].
In addition, nitric oxide (NO) was found to be a key nitrate-related signal that regulates plant RSA and the signaling cascade of lateral root formation induced by auxin [104]. It can be deduced from the previous observation that a decrease in NO3− provision tends to promote auxin translocation from shoot to root. The high NO3−-inhibited root growth is a consequence of condensed cell elongation, and also probably due to the changes in meristematic length. Higher NO3− supply diminished the IAA concentration in the phloem exudates. The NO3−-induced inhibited root growth was closely associated with the reduction of auxin in the roots, especially in the regions close to the root tips. The regrowth of PRs by external NAA and IAA under high NO3− levels confirms that this inhibitory effect via high NO3− might be partially associated with the reduced IAA level in the roots [42].
However, the effect of NO3− on root growth could be complicated by the fact that high NO3− concentration (50 mM) triggers complete inhibition of LR development [105]. It has been experimentally confirmed that these responses are linked to an auxin transport inhibitor. To this end, the local supply of nitrate reduced the transport of auxin from shoot to root, and this subsequently resulted in decreased root auxin concentration to a level more appropriate for lateral root growth. However, for the stimulation of LRs, a change in the root auxin concentration only is not adequate. Regardless of these models, few ideas concerning the transcriptional gene regulatory system are known [106].
Furthermore, under available nitrate conditions, the auxin level in the root decreased compared to low NO3− conditions, and nitrate application seemed to inhibit auxin transport from shoot to root. In many cases, the external IAA partially lowers the stimulatory effect of localized nitrate. High nitrate supply reduces the IAA concentration in the phloem exudates; thus, suppression of root growth by high nitrate is mainly dependent on the reduction of IAA levels in the roots, specifically in the root tip region. It could be deduced that the inhibitory impact of high nitrate concentration on the restricted root growth may be associated with the decline in auxin content in the roots [42].
The currently accessible information leading to a potential connection between nitrate and auxin accumulation influences the rate of auxin biosynthesis, transport, and allocation of auxin from root to shoot [107].
5.2. Nitrate-Mediated Cytokinin Allocation
Cytokinin (CK) affects intercellular auxin transport by regulating the expression of numerous auxin transport components, and thus balances the auxin distribution to regulate the size of root meristem [108]. Findings have also shown that the NO3−-CK shoot–root dependent system exhibits the NO3− demands of the whole plant, which affects root growth in NO3− rich patches of the soil [109]. Since CK could be widely distributed throughout the entire plant cell, CK-induced root–shoot coordination is a proposed model of systemic signaling for nutritional status [110]. CK activity could be closely associated with NO3− accessibility. Apart from the downstream metabolites of NO3−, NO3− has been known to initiate rapid de novo CK synthesis and accumulation in Arabidopsis roots [111]. The CK biosynthesis occurs in different parts of the plant tissue, where the adenosine phosphate-isopentenyltransferase (IPT) is expressed. IPTs are the primary enzymes that mainly influence the rate of CK biosynthesis, such as the prenylation of adenosine 5′ phosphates and ATP and ADP at the N6- terminal with dimethyl diphosphate (DMAPP) [112].
In Arabidopsis, IPT3 is regulated in a NO3−-dependent manner. The expression of IPT3 with several Arabidopsis response regulators 3, 5, 6 (ARR3, 5, 6) are induced by NO3− during the PNR. Moreover, IPT3 is highly induced in the roots and weakly induced in the shoots in both WT and NR-null mutant plants during the PNR, partially mediated by NRT1.1 [35]. During the PNR, NIA is among the highly inducible genes; thus, NO3− firmly controls CK biosynthesis via activation of IPT3. This indicates that IPT3 is the fundamental determinant of short-term NO3−-dependent CK biosynthesis, specifically in the roots, in response to immediate variation in the soil NO3− [111]. In addition, the type-A ARR genes, including ARR3, 5, and 7, similar to the CK metabolism genes, were found to respond to NO3− but not to NH4+. CYTOKININ RESPONSE FACTORS (CRFs) which are also highly inducible by NO3− [15], are known to be transcriptionally activated by CK and its disruption influences the basal expression of a significant number of CK-regulated genes, including type-A ARRs. CRFs are involved in promoting plant growth and leaf senescence [113]. The close regulation of the CYP735A2 and IPT3 by NO3− could be a major factor shaping NO3−-dependent spatio-temporal CK distribution in plants, and also regulating root system architecture in response to several abiotic stresses [114]. In short, nitrate and two hormonal mediators, CK and its antagonistic partner, auxin, act in synergy to modulate CK biosynthesis for root development.
6. Role of NO3− Transporters in Mitigating Plant Stress
Nitrate transporters are ultimately responsible for the absorption of NO3− from the soil and translocation of NO3− to various aerial parts of the plant [115]. NO3− transporter NRT1.1 acts as a positive growth regulator of vegetative and reproductive organs [116]. Studies have shown that AtNRT1.1/AtNPF6.3/CHL1 might be involved in the tolerance of the plant to proton toxicity; further studies on chl1 mutants, however, have revealed a reduced proton tolerance when compared with WT [117]. Moreover, the accumulation of sodium (Na+) in the plant was found to be defective on npf6.3/nrt1.1 mutants, thus npf6.3/nrt1.1 functions in drought tolerance in the presence of NO3− [118]. The downregulation of NRT1.5 and the upregulation of NRT1.8 were observed in the root of the plant on exposure to cadmium (Cd2+). Thus, increased NO3− accumulation in the root [119] indicates that NRT1.1 and NRG2 function downstream from NRT1.1 to regulate Cd2+ stress and also to stimulate NO3− distribution to the root [119]. ATNPF7.3/ATNRT1.5 is highly expressed in the root and highly inducible by phosphate starvation. The ATNRT1.5 mutant atnrt1.5 exhibits longer PRs, with reduced LR density under Pi-deficient conditions, compared with WT. This is an indication that a reduction in the morphological variation by ethylene synthesis antagonizes CO2 [120].
In addition to the transporters stated earlier, npf6.4 mutants exhibit increased resistance to polyamine [115]. AtNPF2.12/AtNRT2.6 positively regulated seed abortion under NO3−-deficient conditions in Arabidopsis [121]. Moreover, AtNPF2.5 and AtNPF2.3 induced chloride (Cl−) efflux from Arabidopsis roots and subsequently contributed to NO3− translocation [122]. AtNRT1.8/ANPF7.2 tolerates Cd2+ and salt stress. However, its knock-out mutants exhibited sensitivity to abiotic stress [123]. AtNPF3.1 transported ABA and GA (gibberellic acid) in vitro [124]. The interaction between NO3−- and NRT-mediated NO3− uptakes on exposure to Pb in Arabidopsis via NRT-related mutants [125] demonstrates a new strategy for plant tolerance to lead (Pb) contamination [125].
Under low NO3− conditions, an NRT2 member, AtNRT2.1, contributes to iHATS (inducible high-affinity transport system) and plays a crucial role in the RSA, while AtNRT2.4 contributes to plant biomass production. AtNRT2.5 also stimulates mature plants under NO3−-deficient conditions [126]. ATNRT2.6 expression is induced after phytopathogenic bacterium inoculation. Hence, plants with low NRT2.6 expression show lower tolerance to pathogenic attacks [127]. Interestingly, there is a correlation between NRT2.6 expression and reactive oxygen species (ROS) accumulation in response to E. amylovora infection and treatments with the redox-active herbicide methyl viologen. This indicates a probable link between NRT2.6 activity and the production of ROS response to biotic and abiotic stresses [127].
In the chloride channel family (CLC), AtCLC accumulates anions in the vacuole when stomata are open, and also facilitates anion release during stomatal closure in response to stress hormones like abscisic acid (ABA) [128]. In addition to the NO3- transporter, the NO3−-associated transcription factor, phloem-mobile CEPD-like 2 (CEPDL2)-polypeptide contributes to NO3− acquisition, along with CEPD1 and CEPD2, which mediate root N status, and the loss of each of these three proteins severely impair N homeostasis in the plants. A similar study showed that shoots of the CEPDL2/CEPD1/2 genotype characterize a high-affinity NO3− uptake duration in the roots, thereby indicating a systematic regulation of root N acquisition [84]. ANTHOCYANIN PIGMENT1 (PAP1) and its homolog PAP2/MYB90 were strongly stimulated by NO3− [129]. Recent research has demonstrated that three LBDs regulate anthocyanin synthesis via repression of PAP1 and PAP2. MYB and bHLH (basic helix-loop-helix) proteins form complexes with TTG1 (TRANSPARENT TESTA GLABRA1) WD40-repeat protein in Arabidopsis to modulate several other epidermal gene expressions such as anthocyanin regulation, proanthocyanin, and mucilage biosynthesis in the seed coat or trichome and root hair organogenesis [49].
7. Conclusions
RSA response of the plant to NO3− accessibility represents a prominent model to study developmental plasticity; however, the underlying mechanism remains highly obscure [130]. One of the most important discoveries in the past few years has been the involvement of NO3− transporters NRT1.1 and NRT2.1 in early response signaling, and their effects on the morphological adaptation of the plant RSA. Despite their roles as transporters and in signaling response, NRT1.1 cannot fully explain the complete mechanism of the NO3− responses observed in plants [43]. However, some findings have supported the previous speculation that NO3− transporters could act as early NO3− sensors [98]. This provides critical insights into understanding the ability to sense NO3− as well as other nutrients [52].
In this review, we have summarized in depth the characterization of the nitrate transporters NRT1.1 and NRT2.1 in Arabidopsis (Figure 1), delivering clues on how NO3− is sensed, taken up, and mobilized, and their modification by phosphorylation at the T101 residue has also been well demonstrated. In addition, the influences of physiological growth on RSA under low and high NO3− conditions, and the underlying molecular players, including TFs and N metabolites, are hypothesized and are associated with the transcriptional control of significant NO3−-responsive genes, which include NIA1, NIA2, NiR, NR, NRT2.1, -2.2, -2.4, -2.5, and NRT3.1. However, the fact is that different TFs, NLP7, TGA1/4, and TCP20, can regulate the expression of the same target gene, NRT2.1 (Figure 1). These TFs co-interact in response to NO3− to regulate root growth.
Despite the development of multiple NO3− signaling pathways regulating RSA and the characterization of primary Ca2+-induced responses elucidated in the present review, many important inquires on how PLCs are implicated in nitrate signaling and the specificity of the protein kinases that switch the different constituents of PLCs are yet to be answered. Moreover, the speculated nonphosphorylated form of the NRT1.1-signaling Ca2+-CPKs-NLPs pathway has received trivial experimental attention. Additionally, PLC- and Ca2+-independent nitrate signaling pathways have another component, as evidenced by AFB3 expression and its downstream TFs, which lead to the possibility that there might be another second messenger involved in nitrate responses.
There are more nitrate regulatory modules in existence, with no clues about their signaling pathways and components; however, they are the fundamental contributors controlling LR development. Hence, functional identification and characterization of the various players associated with this and other NO3− signaling pathways and their possible functions in the root architecture of Arabidopsis is the next step to try and comprehend the NO3− responses that will facilitate crop genetics improvement.
Author Contributions
Conceptualization, M.A. and Z.U.; methodology, M.A. and Z.U.; software, M.A. and Z.U.; validation, O.O.A., L.A., H.L. and Q.W.; formal analysis, M.A.; investigation, Z.U.; resources, F.X.; data curation, M.A. and Z.U.; writing—original draft preparation, M.A. and Z.U; writing—review and editing, O.O.A. and L.A.; supervision, H.L. and Q.W.; project administration, H.L. and Q.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Shandong Provincial Natural Science Foundation, China (ZR2017QC003), Science Foundation for Young Scholars of Tobacco Research Institute of Chinese Academy of Agricultural Sciences (2016B02), the Agricultural Science and Technology Innovation Program (ASTIP-TRIC02) and the Fundamental Research Funds for China Agricultural Academy of Sciences (1610232016005).
Acknowledgments
The authors would like to thank Anelia Marais for the critical reading of the manuscript.
Conflicts of Interest
The authors declared that they have no conflict of interest.
Abbreviations
| NRT1 | Nitrate transporter1 |
| CLC | Chloride Channel |
| CIPK | CBL-Interacting Protein Kinase |
| FIP1 | Factor interacting with poly (A) polymerase 1 |
| CPSF30-L | Cleavage and Polyadenylation Specificity Factor 30-L |
| ABI2 | ABA-insensitive 2 |
| AFB3 | AUXIN RECEPTOR F-BOX3 |
| NAC4 | NAC-domain containing protein 4 |
| OBP4 | OBF Binding Protein 4 |
| TGA1/4 | Targets the activation sequence1/4 |
| EGTA | Ethylene glycol tetraacetic acid |
| CEP | C- TERMINALLY ENCODED PEPTIDE |
| LaCl3 | Lanthanum (III) chloride hydrate |
| PI-PLC | Phosphatidylinositol-specific phospholipase C |
| TCP20 | Teosinte branched1/cycloidea/proliferating cell factor1-20 |
| ANR1 | ARABIDOPSIS NITRATE REGULATED1 |
| ARF | AUXIN RESPONSE FACTOR |
| SCFTIR/AFB | S-PHASE KINASE ASSOCIATED PROTEIN 1-CULLIN-F-BOX PROTEIN (SCF)-type E3 ubiquitin-protein ligases |
| MEKK1 | Mitogen-Activated Protein Kinase Kinase 1 |
| BBX | Bobby sox homolog |
| ROXY | Floral glutaredoxins |
| CEPD | Phloem-specific peptides |
| ERFs | Ethylene-responsive element binding factors |
| RAV2 | Regulator of V-ATPase in vacuolar membrane protein 2 |
References
- Nacry, P.; Bouguyon, E.; Gojon, A. Nitrogen acquisition by roots: Physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 2013, 370, 1–29. [Google Scholar] [CrossRef]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS-box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Gojon, A.; Nacry, P.; Davidian, J.C. Root uptake regulation: A central process for NPS homeostasis in plants. Curr. Opin. Plant Biol. 2009, 12, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.H.; Yu, J.Q.; Hu, D.G. Nitrate: A crucial signal during lateral roots development. Front. Plant Sci. 2017, 8, 485. [Google Scholar] [CrossRef]
- Jackson, W.A.; Flesher, D.; Hageman, R.H. Nitrate uptake by dark-grown corn seedlings. Plant Physiol. 1973, 51, 120–127. [Google Scholar] [CrossRef]
- Hole, D.J.; Emran, A.M.; Fares, Y.; Drew, M.C. Induction of nitrate transport in maize roots, and kinetics of influx, measured with nitrogen-13. Plant Physiol. 1990, 93, 642–647. [Google Scholar] [CrossRef]
- Lee, R.B. Control of net uptake of nutrients by regulation of influx in barley plants recovering from nutrient deficiency. Ann. Bot. 1993, 72, 223–230. [Google Scholar] [CrossRef]
- Imsande, J.; Touraine, B. N demand and the regulation of nitrate uptake. Plant Physiol. 1994, 105, 3–7. [Google Scholar] [CrossRef]
- Liu, K.H.; Tsay, Y.F. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 2003, 22, 1005–1013. [Google Scholar] [CrossRef]
- Bouguyon, E.; Brun, F.; Meynard, D.; Kubeš, M.; Pervent, M.; Leran, S.; Lacombe, B.; Krouk, G.; Guiderdoni, E.; Zažímalová, E.; et al. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 2015, 1, 15015. [Google Scholar] [CrossRef]
- Muños, S.; Cazettes, C.; Fizames, C.; Gaymard, F.; Tillard, P.; Lepetit, M.; Lejay, L.; Gojon, A. Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1 W inside a box sign. Plant Cell 2004, 16, 2433–2447. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Tillard, P.; Gojon, A. Regulation of the high-affinity NO3- uptake system by NRT1.1-mediated NO3- - demand signalling in Arabidopsis. Plant Physiol. 2006, 142, 1075–1086. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef]
- Ho, C.-H.; Lin, S.-H.; Hu, H.-C.; Tsay, Y.-F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef]
- Riveras, E.; Alvarez, J.M.; Vidal, E.A.; Oses, C.; Vega, A.; Gutiérrez, R.A. The calcium ion is a second messenger in the nitrate signalling pathway of Arabidopsis. Plant Physiol. 2015, 169, 1397–1404. [Google Scholar] [CrossRef]
- Krouk, G.; Crawford, N.M.; Coruzzi, G.M.; Tsay, Y.-F. Nitrate signalling: Adaptation to fluctuating environments. Curr. Opin. Plant Biol. 2010, 13, 266–273. [Google Scholar] [CrossRef]
- Dreyer, I.; Horeau, C.; Lemaillet, G.; Zimmermann, S.; Bush, D.R.; Rodríguez-Navarro, A.; Schachtman, D.P.; Spalding, E.P.; Sentenac, H.; Gaber, R.F. Identification and characterization of plant transporters. J. Exp. Bot. 1999, 50, 1073–1087. [Google Scholar] [CrossRef]
- Krapp, A. Plant nitrogen assimilation and its regulation: A complex puzzle with missing pieces. Curr. Opin. Plant Biol. 2015, 25, 115–122. [Google Scholar] [CrossRef]
- Medici, A.; Krouk, G. The Primary Nitrate Response: A multifaceted signalling pathway. J. Exp. Bot. 2014, 65, 5567–5576. [Google Scholar] [CrossRef]
- Wang, R.; Liu, D.; Crawford, N.M. The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. USA 1998, 95, 15134–15139. [Google Scholar] [CrossRef]
- Tsay, Y.F.; Schroeder, J.I.; Feldmann, K.A.; Crawford, N.M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 1993, 72, 705–713. [Google Scholar] [CrossRef]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1.1 transporter participates in the signalling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 2006, 103, 19206–19211. [Google Scholar] [CrossRef]
- Walch-Liu, P.; Forde, B.G. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J. 2008, 54, 820–828. [Google Scholar] [CrossRef]
- Hu, H.-C.; Wang, Y.-Y.; Tsay, Y.-F. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009, 57, 264–278. [Google Scholar] [CrossRef]
- Wang, R.; Okamoto, M.; Xing, X.; Crawford, N.M. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003, 132, 556–567. [Google Scholar] [CrossRef]
- Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar]
- Armijo, G.; Gutiérrez, R.A. Emerging players in the nitrate signalling pathway. Mol. Plant 2017, 10, 1019–1022. [Google Scholar] [CrossRef]
- O’Brien, J.A.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutiérrez, R.A.A. Nitrate Transport, Sensing, and Responses in Plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Cold calcium signalling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 1996, 8, 489–503. [Google Scholar]
- Ebert, D.H.; Greenberg, M.E. Activity-dependent neuronal signalling and autism spectrum disorder. Nature 2013, 493, 327–337. [Google Scholar] [CrossRef]
- Sakakibara, H.; Kobayashi, K.; Deji, A.; Sugiyama, T. Partial characterization of the signalling pathway for the nitrate-dependent expression of genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant Cell Physiol. 1997, 38, 837–843. [Google Scholar] [CrossRef]
- Noguero, M.; Lacombe, B. Transporters involved in root nitrate uptake and sensing by Arabidopsis. Front. Plant Sci. 2016, 7, 1–7. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, W.; Li, Z.; Li, Z.; Bi, Y.; Crawford, N.M.; Wang, Y. FIP1 plays an important role in nitrate signalling and regulates CIPK8 and CIPK23 expression in Arabidopsis. Front. Plant Sci. 2018, 9, 1–14. [Google Scholar]
- Wang, R.; Tischner, R.; Gutiérrez, R.A.; Hoffman, M.; Xing, X.; Chen, M.; Coruzzi, G.; Crawford, N.M.; Wang, R.; Tischner, R.; et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004, 136, 2512–2522. [Google Scholar] [CrossRef]
- Krouk, G.; Mirowski, P.; LeCun, Y.; Shasha, D.E.; Coruzzi, G.M. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 2010, 11, R123. [Google Scholar] [CrossRef]
- Ristova, D.; Carré, C.; Pervent, M.; Medici, A.; Kim, G.J.; Scalia, D.; Ruffel, S.; Birnbaum, K.D.; Lacombe, B.; Busch, W.; et al. Combinatorial interaction network of transcriptomic and phenotypic responses to nitrogen and hormones in the Arabidopsis thaliana root. Sci. Signal. 2016, 9, 1–11. [Google Scholar] [CrossRef]
- Léran, S.; Edel, K.H.; Pervent, M.; Hashimoto, K.; Corratgé-Faillie, C.; Offenborn, J.N.; Tillard, P.; Gojon, A.; Kudla, J.; Lacombe, B. Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal. 2015, 8, ra43. [Google Scholar] [CrossRef]
- Sueyoshi, K.; Mitsuyama, T.; Sugimoto, T.; Kleinhofs, A.; Warner, R.L.; Oji, Y. Effects of inhibitors for signalling components on the expression of the genes for nitrate reductase and nitrite reductase in excised barley leaves. Soil Sci. Plant Nutr. 1999, 45, 1015–1019. [Google Scholar] [CrossRef]
- Sanders, D.; Dodd, A.N. The language of calcium signalling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar]
- Undurraga, S.F.; Ibarra-Henríquez, C.; Fredes, I.; Álvarez, J.M.; Gutiérrez, R.A. Nitrate signalling and early responses in Arabidopsis roots. J. Exp. Bot. 2017, 68, 2541–2551. [Google Scholar] [CrossRef]
- Tian, Q.; Chen, F.; Liu, J.; Zhang, F.; Mi, G. Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J. Plant Physiol. 2008, 165, 942–951. [Google Scholar] [CrossRef]
- Wang, R.; Xing, X.; Wang, Y.; Tran, A.; Crawford, N.M. A Genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 2009, 151, 472–478. [Google Scholar] [CrossRef]
- Rupwate, S.D.; Rajasekharan, R. Plant phosphoinositide-specific phospholipase C: An insight. Plant Signal. Behav. 2012, 7, 11–14. [Google Scholar] [CrossRef]
- Helling, D.; Possart, A.; Cottier, S.; Klahre, U.; Kost, B. Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell 2006, 18, 3519–3534. [Google Scholar] [CrossRef]
- Pokotylo, I.; Kolesnikov, Y.; Kravets, V.; Zachowski, A.; Ruelland, E. Plant phosphoinositide-dependent phospholipases C: Variations around a canonical theme. Biochimie 2014, 96, 144–157. [Google Scholar] [CrossRef]
- Tasma, I.M.; Brendel, V.; Whitham, S.A.; Bhattacharyya, M.K. Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol. Biochem. 2008, 46, 627–637. [Google Scholar] [CrossRef]
- Gaudinier, A.; Rodriguez-Medina, J.; Zhang, L.; Olson, A.; Liseron-Monfils, C.; Bågman, A.M.; Foret, J.; Abbitt, S.; Tang, M.; Li, B.; et al. Transcriptional regulation of nitrogen-associated metabolism and growth. Nature 2018, 563, 259–264. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef]
- Hashimoto, K.; Kudla, J. Calcium decoding mechanisms in plants. Biochimie 2011, 93, 2054–2059. [Google Scholar] [CrossRef]
- Zhang, X.; Cui, Y.; Yu, M.; Su, B.; Gong, W.; Baluška, F.; Komis, G.; Šamaj, J.; Shan, X.; Lin, J. Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signalling in lateral root growth. Plant Physiol. 2019, 181, 480–498. [Google Scholar] [CrossRef] [PubMed]
- Giehl, R.F.H.; Von Wirén, N. Nitrate signalling: Functions of a nitrate transceptor. Nat. Plants 2015, 1, 15021. [Google Scholar] [CrossRef]
- Krouk, G. Nitrate signalling: Calcium bridges the nitrate gap. Nat. Plants 2017, 3, 1–2. [Google Scholar] [CrossRef]
- Gan, Y.; Bernreiter, A.; Filleur, S.; Abram, B.; Forde, B.G. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 2012, 53, 1003–1016. [Google Scholar] [CrossRef]
- Vidal, E.A.; Álvarez, J.M.; Gutiérrez, R.A. Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Signal. Behav. 2014, 9, 1–5. [Google Scholar] [CrossRef]
- Vidal, E.A.; Moyano, T.C.; Riveras, E.; Contreras-López, O.; Gutiérrez, R.A. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Natl. Acad. Sci. USA 2013, 110, 12840–12845. [Google Scholar] [CrossRef]
- Gifford, M.L.; Dean, A.; Gutierrez, R.A.; Coruzzi, G.M.; Birnbaum, K.D. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 2008, 105, 803–808. [Google Scholar] [CrossRef]
- Li, Z.; Xu, Z.; He, G.; Yang, G.; Chen, M. The voltage-dependent anion channel 1 (AtVDAC1) negatively regulates plant cold responses during germination and seedling development in Arabidopsis and interacts with calcium sensor CBL1. Int. J. Mol. Sci. 2013, 14, 701–713. [Google Scholar] [CrossRef]
- Vidal, E.A.; Araus, V.; Lu, C.; Parry, G.; Green, P.J.; Coruzzi, G.M.; Gutiérrez, R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482. [Google Scholar] [CrossRef]
- Kato, H.; Nishihama, R.; Weijers, D.; Kohchi, T. Evolution of nuclear auxin signalling: Lessons from genetic studies with basal land plants. J. Exp. Bot. 2018, 69, 291–301. [Google Scholar] [CrossRef]
- Ma, W.; Li, J.; Qu, B.; He, X.; Zhao, X.; Li, B.; Fu, X.; Tong, Y. Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 2014, 78, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Heologis, A.T.T. Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 11786–11791. [Google Scholar]
- Vernoux, T.; Brunoud, G.; Farcot, E.; Morin, V.; Van Den Daele, H.; Legrand, J.; Oliva, M.; Das, P.; Larrieu, A.; Wells, D.; et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011, 7, 508. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Author’s personal copy auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Farcot, E.; Lavedrine, C.; Vernoux, T. A modular analysis of the auxin signalling network. PLoS ONE 2015, 10, 1–26. [Google Scholar] [CrossRef]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef]
- De Rybel, B.; Vassileva, V.; Parizot, B.; Demeulenaere, M.; Grunewald, W.; Audenaert, D.; Van Campenhout, J.; Overvoorde, P.; Jansen, L.; Vanneste, S.; et al. Article A novel aux/IAA28 signalling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 2010, 20, 1697–1706. [Google Scholar] [CrossRef]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef]
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef]
- Calderón Villalobos, L.I.A.; Lee, S.; De Oliveira, C.; Ivetac, A.; Brandt, W.; Armitage, L.; Sheard, L.B.; Tan, X.; Parry, G.; Mao, H.; et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 2012, 8, 477–485. [Google Scholar] [CrossRef] [PubMed]
- dos Santos Maraschin, F.; Memelink, J.; Offringa, R. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 2009, 59, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Pierre-Jerome, E.; Jang, S.S.; Havens, K.A.; Nemhauser, J.L.; Klavins, E. Recapitulation of the forward nuclear auxin response pathway in yeast. Proc. Natl. Acad. Sci. USA 2014, 111, 9407–9412. [Google Scholar] [CrossRef]
- Tian, H.; De Smet, I.; Ding, Z. Shaping a root system: Regulating lateral versus primary root growth. Trends Plant Sci. 2014, 19, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Drew, M.C.; Saker, L.R. Nutrient supply and the growth of the seminal root system in barley: II. Localized, compensatory increases in lateral root growth and rates op nitrate uptake when nitrate supply is restricted to only part of the root system. J. Exp. Bot. 1975, 26, 79–90. [Google Scholar] [CrossRef]
- Forde, B.G. Nitrogen signalling pathways shaping root system architecture: An update. Curr. Opin. Plant Biol. 2014, 21, 30–36. [Google Scholar] [CrossRef]
- Forde, B.G. Glutamate signalling in roots. J. Exp. Bot. 2014, 65, 779–787. [Google Scholar] [CrossRef]
- Walch-Liu, P.; Liu, L.H.; Remans, T.; Tester, M.; Forde, B.G. Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 1045–1057. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, H.; Hamera, S.; Chen, X.; Fang, R. MiR444a has multiple functions in the rice nitrate-signalling pathway. Plant J. 2014, 78, 44–55. [Google Scholar] [CrossRef]
- Forde, B.G.; Cutler, S.R.; Zaman, N.; Krysan, P.J. Glutamate signalling via a MEKK1 kinase-dependent pathway induces changes in Arabidopsis root architecture. Plant J. 2013, 75, 1–10. [Google Scholar] [CrossRef]
- Shinozaki, K.; Mizoguchi, T.; Irie, K.; Hirayama, T.; Hayashida, N.; Yamaguchi-Shinozaki, K.; Matsumoto, K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1996, 93, 765–769. [Google Scholar]
- Naulin, P.A.; Armijo, G.; Vega, A.; Tamayo, K.P.; Gras, D.E.; de la Cruz, J.; Gutiérrez, R.A. nitrate induction of primary root growth requires cytokinin signalling in Arabidopsis thaliana. Plant Cell Physiol. 2019, 1–28. [Google Scholar]
- Zhang, H.; Jennings, A.; Barlow, P.W.; Forde, B.G. Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 1999, 96, 6529–6534. [Google Scholar] [CrossRef]
- Ota, R.; Ohkubo, Y.; Yamashita, Y.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Patterson, K.; Walters, L.A.; Cooper, A.M.; Olvera, J.G.; Rosas, M.A.; Rasmusson, A.G.; Escobar, M.A. Nitrate-regulated glutaredoxins control Arabidopsis primary root growth. Plant Physiol. 2016, 170, 989–999. [Google Scholar] [CrossRef]
- Ohta, M.; Matsui, K.; Hiratsu, K.; Shinshi, H.; Ohme-Takagi, M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 2001, 13, 1959–1968. [Google Scholar] [CrossRef]
- Bielecka, M.; Watanabe, M.; Morcuende, R.; Scheible, W.R.; Hawkesford, M.J.; Hesse, H.; Hoefgen, R. Transcriptome and metabolome analysis of plant sulfate starvation and resupply provides novel information on transcriptional regulation of metabolism associated with sulfur, nitrogen, and phosphorus nutritional responses in Arabidopsis. Front. Plant Sci. 2015, 5, 1–18. [Google Scholar] [CrossRef]
- Yu, L.-H.; Miao, Z.-Q.; Qi, G.-F.; Wu, J.; Cai, X.-T.; Mao, J.-L. MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 2014, 7, 1653–1669. [Google Scholar] [CrossRef]
- Huang, S.; Chen, S.; Liang, Z.; Zhang, C.; Yan, M.; Chen, J.; Xu, G.; Fan, X.; Zhang, Y. Knockdown of the partner protein OsNAR2.1 for high-affinity nitrate transport represses lateral root formation in a nitrate-dependent manner. Sci. Rep. 2015, 5, 15–18. [Google Scholar] [CrossRef]
- Naz, M.; Luo, B.; Guo, X.; Li, B.; Chen, J.; Fan, X. Overexpression of nitrate transporter osnrt2.1 enhances nitrate-dependent root elongation. Genes (Basel) 2019, 10, 290. [Google Scholar] [CrossRef]
- Meng, Z.B.; Chen, L.Q.; Suo, D.; Li, G.X.; Tang, C.X.; Zheng, S.J. Nitric oxide is the shared signalling molecule in phosphorus- and iron-deficiency-induced formation of cluster roots in white lupin (Lupinus albus). Ann. Bot. 2012, 1055–1064. [Google Scholar] [CrossRef]
- Sun, H.; Li, J.; Song, W.; Tao, J.; Huang, S.; Chen, S.; Hou, M.; Xu, G.; Zhang, Y. Nitric oxide generated by nitrate reductase increases nitrogen uptake capacity by inducing lateral root formation and inorganic nitrogen uptake under partial nitrate nutrition in rice. J. Exp. Bot. 2015, 66, 2449–2459. [Google Scholar] [CrossRef]
- Gruber, B.D.; Giehl, R.F.H.; Friedel, S.; von Wirén, N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Araya, T.; Miyamoto, M.; Wibowo, J.; Suzuki, A.; Kojima, S.; Tsuchiya, Y.N.; Sawa, S.; Fukuda, H.; Von Wirén, N.; Takahashi, H. CLE-CLAVATA1 peptide-receptor signalling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 2029–2034. [Google Scholar] [CrossRef] [PubMed]
- Delay, C.; Imin, N.; Djordjevic, M.A. CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J. Exp. Bot. 2013, 64, 5383–5394. [Google Scholar] [CrossRef] [PubMed]
- Joshi-Saha, A.; Valon, C.; Leung, J. A brand new START: Abscisic acid perception and transduction in the guard cell. Sci. Signal. 2011, 4, re4. [Google Scholar] [CrossRef]
- Malamy, J.E.; Ryan, K.S. Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 2001, 127, 899–909. [Google Scholar] [CrossRef]
- Zhang, H.; Rong, H.; Pilbeam, D. Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 2329–2338. [Google Scholar] [CrossRef]
- Little, D.Y.; Rao, H.; Oliva, S.; Daniel-Vedele, F.; Krapp, A.; Malamy, J.E. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. USA 2005, 102, 13693–13698. [Google Scholar] [CrossRef]
- Razem, F.A.; El-Kereamy, A.; Abrams, S.R.; Hill, R.D. The RNA-binding protein FCA is an abscisic acid receptor. Nature 2006, 439, 290–294. [Google Scholar] [CrossRef]
- Mounier, E.; Pervent, M.; Ljung, K.; Gojon, A.; Nacry, P. Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ. 2014, 37, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Bellegarde, F.; Gojon, A.; Martin, A. Signals and players in the transcriptional regulation of root responses by local and systemic N signalling in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 2553–2565. [Google Scholar] [CrossRef]
- Puig, J.; Pauluzzi, G.; Guiderdoni, E.; Gantet, P. Regulation of shoot and root development through mutual signalling. Mol. Plant 2012, 5, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.W.; Yang, J.L.; Qin, C.; Jin, C.W.; Mo, J.H.; Ye, T.; Chen, W.W.; Yang, J.L.; Qin, C.; Jin, C.W.; et al. Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron. Plant Physiol. 2016, 154, 810–819. [Google Scholar]
- Signora, L.; De Smet, I.; Foyer, C.H.; Zhang, H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 2001, 28, 655–662. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, F.; Zhang, F.; Mi, G. Auxin transport from shoot to root is involved in the response of lateral root growth to localized supply of nitrate in maize. Plant Sci. 2005, 169, 894–900. [Google Scholar] [CrossRef]
- Caba, J.M.; Centeno, M.L.; Fernández, B.; Gresshoff, P.M.; Ligero, F. Inoculation and nitrate alter phytohormone levels in soybean roots: Differences between a supernodulating mutant and the wild type. Planta 2000, 211, 98–104. [Google Scholar] [CrossRef]
- Růzǐčka, K.; Šimášková, M.; Duclercq, J.; Petrášek, J.; Zažímalová, E.; Simon, S.; Friml, J.; Van Montagu, M.C.E.; Benková, E. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 2009, 106, 4284–4289. [Google Scholar] [CrossRef]
- Ruffel, S.; Krouk, G.; Ristova, D.; Shasha, D.; Birnbaum, K.D.; Coruzzi, G.M. Nitrogen economics of root foraging: Transitive closure of the nitrate-cytokinin relay and distinct systemic signalling for N supply vs. demand. Proc. Natl. Acad. Sci. USA 2011, 108, 18524–18529. [Google Scholar] [CrossRef]
- Sakakibara, H. CYTOKININS: Activity, Biosynthesis, and Translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef]
- Takei, K.; Ueda, N.; Aoki, K.; Kuromori, T.; Hirayama, T.; Shinozaki, K.; Yamaya, T.; Sakakibara, H. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 2004, 45, 1053–1062. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinin Biosynthesis and Regulation. Vitam. Horm. 2005, 72, 271–287. [Google Scholar] [PubMed]
- Raines, T.; Shanks, C.; Cheng, C.Y.; McPherson, D.; Argueso, C.T.; Kim, H.J.; Franco-Zorrilla, J.M.; Lõpez-Vidriero, I.; Solano, R.; Vaňková, R.; et al. The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 2016, 85, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Ramireddy, E.; Chang, L.; Schmülling, T. Cytokinin as a mediator for regulating root system architecture in response to environmental cues. Plant Signal. Behav. 2014, 9, e27771. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.B.; Meng, S.; Gong, J.M. The expected and unexpected roles of nitrate transporters in plant abiotic stress resistance and their regulation. Int. J. Mol. Sci. 2018, 19, 1–15. [Google Scholar] [CrossRef]
- Guo, F.; Wang, R.; Chen, M.; Crawford, N.M. The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 2001, 13, 1761–1777. [Google Scholar] [CrossRef]
- Fang, X.Z.; Tian, W.H.; Liu, X.X.; Lin, X.Y.; Jin, C.W.; Zheng, S.J. Alleviation of proton toxicity by nitrate uptake specifically depends on nitrate transporter 1.1 in Arabidopsis. New Phytol. 2016, 211, 149–158. [Google Scholar] [CrossRef]
- Álvarez-Aragón, R.; Rodríguez-Navarro, A. Nitrate-dependent shoot sodium accumulation and osmotic functions of sodium in Arabidopsis under saline conditions. Plant J. 2017, 91, 208–219. [Google Scholar] [CrossRef]
- Jian, S.; Luo, J.; Liao, Q.; Liu, Q.; Guan, C.; Zhang, Z. NRT1.1 regulates nitrate allocation and cadmium tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Cui, Y.N.; Li, X.T.; Yuan, J.Z.; Wang, F.Z.; Wang, S.M.; Ma, Q. Nitrate transporter NPF7.3/NRT1.5 plays an essential role in regulating phosphate deficiency responses in Arabidopsis. Biochem. Biophys. Res. Commun. 2019, 508, 314–319. [Google Scholar] [CrossRef]
- Almagro, A.; Shan, H.L.; Yi, F.T. Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 2008, 20, 3289–3299. [Google Scholar] [CrossRef] [PubMed]
- Taochy, C.; Gaillard, I.; Ipotesi, E.; Oomen, R.; Leonhardt, N.; Zimmermann, S.; Peltier, J.B.; Szponarski, W.; Simonneau, T.; Sentenac, H.; et al. The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. Plant J. 2015, 83, 466–479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-B.; Yi, H.Y.; Gong, J.M. The Arabidopsis Ethylene/Jasmonic acid-NRT signalling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell 2014, 26, 3984–3998. [Google Scholar] [CrossRef] [PubMed]
- Tal, I.; Zhang, Y.; Jørgensen, M.E.; Pisanty, O.; Barbosa, I.C.R.; Zourelidou, M.; Regnault, T.; Crocoll, C.; Erik Olsen, C.; Weinstain, R.; et al. The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 2016, 7, 11486. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Fang, X.Z.; Dai, Y.J.; Zhu, Y.X.; Chen, H.S.; Lin, X.Y.; Jin, C.W. Nitrate transporter 1.1 alleviates lead toxicity in Arabidopsis by preventing rhizosphere acidification. J. Exp. Bot. 2019, 70, 6363–6374. [Google Scholar] [CrossRef]
- Kiba, T.; Krapp, A. Plant nitrogen acquisition under low availability: Regulation of uptake and root architecture. Plant Cell Physiol. 2016, 57, 707–714. [Google Scholar] [CrossRef]
- Dechorgnat, J.; Patrit, O.; Krapp, A.; Fagard, M.; Daniel-Vedele, F. Characterization of the Nrt2.6 gene in Arabidopsis thaliana: A link with plant response to biotic and abiotic stress. PLoS ONE 2012, 7, e42491. [Google Scholar] [CrossRef]
- Wege, S.; De Angeli, A.; Droillard, M.J.; Kroniewicz, L.; Merlot, S.; Cornu, D.; Gambale, F.; Martinoia, E.; Barbier-Brygoo, H.; Thomine, S.; et al. Phosphorylation of the vacuolar anion exchanger AtCLCa is required for the stomatal response to abscisic acid. Sci. Signal. 2014, 7, 1–11. [Google Scholar] [CrossRef]
- Morcuende, R.; Bari, R.; Gibon, Y.; Zheng, W.; Pant, B.D.; Bläsing, O.; Usadel, B.; Czechowski, T.; Udvardi, M.K.; Stitt, M.; et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007, 30, 85–112. [Google Scholar] [CrossRef]
- Xu, P.; Cai, W. Nitrate-responsive OBP4-XTH9 regulatory module controls lateral root development in Arabidopsis thaliana. PLoS Genet. 2019, 15, e1008465. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).