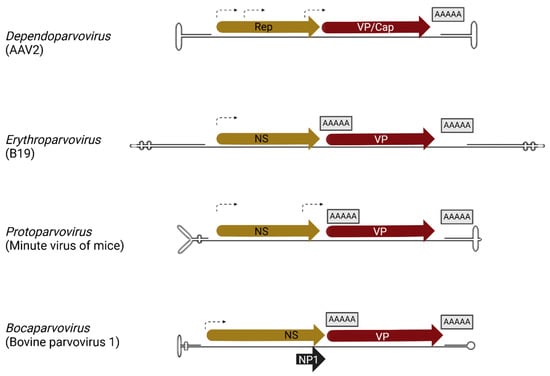
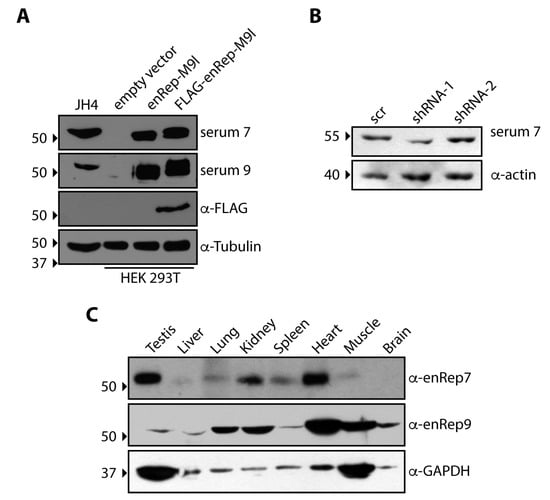
Parvoviridae
A topical collection in Viruses (ISSN 1999-4915). This collection belongs to the section "Human Virology and Viral Diseases".
Viewed by 6578Editor
Interests: parvovirus B19; virus-cell interactions; viral infections; recombinant viruses; virological diagnosis; antiviral strategies
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Viruses in the Parvoviridae family constitute a most diverse group and an ever-intriguing field of research. While all share an ssDNA genome and small dimensions, they can differ widely in structure, genome organization and expression, virus–cell interactions, host range, and impacts on hosts. Within the family, some viruses are human and veterinary pathogens; other viruses have long been studied and engineered as tools for oncolytic therapy, or as sophisticated gene delivery vectors, and can now display their wide and expanding applicative potential. Exploring the biological diversity within the family, the inherent complexity in these apparently simple viruses, and expanding the translational potential, is an ongoing endeavour and commitment for the scientific community.
This Topical Collection on ‘Parvoviridae’ continues the series of collections dedicated to viruses belonging to the Parvoviridae family. The aim is to compile contributions in the field of parvovirus research, encouraging new insights and research on unresolved issues, as well as new approaches exploiting systemic methodologies. Evolution, structural biology, viral replication, virus–host interaction, pathogenesis and immunity, gene therapy, and viral oncotherapy are all topics of relevance to the community involved in parvovirus research and of interest to wider audiences, studies of which may be contributed to this collection.
Prof. Dr. Giorgio Gallinella
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Viruses is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2600 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- Parvoviridae
- Parvovirus evolution
- Parvovirus structure
- Parvovirus genetics
- Parvovirus–host interactions
- Parvovirus pathogenesis and immunity
- Parvovirus oncolytic therapy
- Parvovirus viral vectors
- antiviral strategies
Related Special Issues
- New Insights into Parvovirus Research in Viruses (21 articles)
- Advances in Parvovirus Research 2020 in Viruses (15 articles)
- Advances in Parvovirus Research 2022 in Viruses (18 articles)
- Advances in Parvovirus Research 2024 in Viruses (11 articles)