Sensors, Image, and Signal Processing for Biomedical Applications
A topical collection in Sensors (ISSN 1424-8220). This collection belongs to the section "Biomedical Sensors".
Viewed by 91622
Share This Topical Collection
Editor
 Dr. Christoph Hintermüller
Dr. Christoph Hintermüller
 Dr. Christoph Hintermüller
Dr. Christoph Hintermüller
E-Mail
Website
Guest Editor
Institute for Biomedical Mechatronics, Johannes Kepler University, 4020 Linz, Austria
Interests: biosignal processing; cardiac electrophysiology; 3D imaging
Topical Collection Information
Recording information from the human body by measuring signals and taking images is important throughout the entire clinical process covering anamnesis, diagnosis, therapy, and treatment. In addition to proper recording, preprocessing and pre-analyzing signals and information from the patient fusion of quantitative data and qualitative information also play an important role. The field of biomedical imaging and signal processing has been and still is open to new developments in other disciplines and fields such as physics and chemistry, independent of how remote these may first appear—this is highlighted in the example of Kinect and another kind of devices originally developed for gaming rather than imaging.
This issue puts the focus on recent developments in the fields of biomedical, medical, and clinical image and signal processing. These include new sensing methods, approaches to analyzing the recorded images and signals, data fusion methods, and algorithms to obtain new and additional insights, and how they help to improve clinical processes and free clinicians and doctors to spend more time in direct contact with their patients rather than interpreting the recorded data and signals.
Dr. Christoph Hintermüller
Guest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Sensors is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2600 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- sensing principles
- sensors
- image and signal processing
- clinical applications
- data fusion
- image and signal analysis
- image and signal classification
Published Papers (18 papers)
Open AccessArticle
Evaluation of Diffuse Reflectance Spectroscopy Vegetal Phantoms for Human Pigmented Skin Lesions
by
Sonia Buendia-Aviles, Margarita Cunill-Rodríguez, José A. Delgado-Atencio, Enrique González-Gutiérrez, José L. Arce-Diego and Félix Fanjul-Vélez
Cited by 2 | Viewed by 2675
Abstract
Pigmented skin lesions have increased considerably worldwide in the last years, with melanoma being responsible for 75% of deaths and low survival rates. The development and refining of more efficient non-invasive optical techniques such as diffuse reflectance spectroscopy (DRS) is crucial for the
[...] Read more.
Pigmented skin lesions have increased considerably worldwide in the last years, with melanoma being responsible for 75% of deaths and low survival rates. The development and refining of more efficient non-invasive optical techniques such as diffuse reflectance spectroscopy (DRS) is crucial for the diagnosis of melanoma skin cancer. The development of novel diagnostic approaches requires a sufficient number of test samples. Hence, the similarities between banana brown spots (BBSs) and human skin pigmented lesions (HSPLs) could be exploited by employing the former as an optical phantom for validating these techniques. This work analyses the potential similarity of BBSs to HSPLs of volunteers with different skin phototypes by means of several characteristics, such as symmetry, color RGB tonality, and principal component analysis (PCA) of spectra. The findings demonstrate a notable resemblance between the attributes concerning spectrum, area, and color of HSPLs and BBSs at specific ripening stages. Furthermore, the spectral similarity is increased when a fiber-optic probe with a shorter distance (240 µm) between the source fiber and the detector fiber is utilized, in comparison to a probe with a greater distance (2500 µm) for this parameter. A Monte Carlo simulation of sampling volume was used to clarify spectral similarities.
Full article
►▼
Show Figures
Open AccessReview
Development of a Sexological Ontology
by
Dariusz S. Radomski, Zuzanna Oscik, Ewa Dmoch-Gajzlerska and Anna Szczotka
Viewed by 3777
Abstract
This study aimed to show what role biomedical engineering can play in sexual health. A new concept of sexological ontology, an essential tool for building evidence-based models of sexual health, is proposed. This ontology should be based on properly validated mathematical models of
[...] Read more.
This study aimed to show what role biomedical engineering can play in sexual health. A new concept of sexological ontology, an essential tool for building evidence-based models of sexual health, is proposed. This ontology should be based on properly validated mathematical models of sexual reactions identified using reliable measurements of physiological signals. This paper presents a review of the recommended measurement methods. Moreover, a general human sexual reaction model based on dynamic systems built at different levels of time × space × detail is presented, and the actual used modeling approaches are reviewed, referring to the introduced model. Lastly, examples of devices and computer programs designed for sexual therapy are described, indicating the need for legal regulation of their manufacturing, similar to that for other medical devices.
Full article
►▼
Show Figures
Open AccessArticle
Blood Biomarker Detection Using Integrated Microfluidics with Optical Label-Free Biosensor
by
Chiung-Hsi Li, Chen-Yuan Chang, Yan-Ru Chen and Cheng-Sheng Huang
Cited by 3 | Viewed by 3444
Abstract
In this study, we developed an optofluidic chip consisting of a guided-mode resonance (GMR) sensor incorporated into a microfluidic chip to achieve simultaneous blood plasma separation and label-free albumin detection. A sedimentation chamber is integrated into the microfluidic chip to achieve plasma separation
[...] Read more.
In this study, we developed an optofluidic chip consisting of a guided-mode resonance (GMR) sensor incorporated into a microfluidic chip to achieve simultaneous blood plasma separation and label-free albumin detection. A sedimentation chamber is integrated into the microfluidic chip to achieve plasma separation through differences in density. After a blood sample is loaded into the optofluidic chip in two stages with controlled flow rates, the blood cells are kept in the sedimentation chamber, enabling only the plasma to reach the GMR sensor for albumin detection. This GMR sensor, fabricated using plastic replica molding, achieved a bulk sensitivity of 175.66 nm/RIU. With surface-bound antibodies, the GMR sensor exhibited a limit of detection of 0.16 μg/mL for recombinant albumin in buffer solution. Overall, our findings demonstrate the potential of our integrated chip for use in clinical samples for biomarker detection in point-of-care applications.
Full article
►▼
Show Figures
Open AccessArticle
A Novel Online Position Estimation Method and Movement Sonification System: The Soniccup
by
Thomas H. Nown, Madeleine A. Grealy, Ivan Andonovic, Andrew Kerr and Christos Tachtatzis
Viewed by 4151
Abstract
Existing methods to obtain position from inertial sensors typically use a combination of multiple sensors and orientation modeling; thus, obtaining position from a single inertial sensor is highly desirable given the decreased setup time and reduced complexity. The dead reckoning method is commonly
[...] Read more.
Existing methods to obtain position from inertial sensors typically use a combination of multiple sensors and orientation modeling; thus, obtaining position from a single inertial sensor is highly desirable given the decreased setup time and reduced complexity. The dead reckoning method is commonly chosen to obtain position from acceleration; however, when applied to upper limb tracking, the accuracy of position estimates are questionable, which limits feasibility. A new method of obtaining position estimates through the use of zero velocity updates is reported, using a commercial IMU, a push-to-make momentary switch, and a 3D printed object to house the sensors. The generated position estimates can subsequently be converted into sound through sonification to provide audio feedback on reaching movements for rehabilitation applications. An evaluation of the performance of the generated position estimates from a system labeled ‘Soniccup’ is presented through a comparison with the outputs from a Vicon Nexus system. The results indicate that for reaching movements below one second in duration, the Soniccup produces positional estimates with high similarity to the same movements captured through the Vicon system, corresponding to comparable audio output from the two systems. However, future work to improve the performance of longer-duration movements and reduce the system latency to produce real-time audio feedback is required to improve the acceptability of the system.
Full article
►▼
Show Figures
Open AccessArticle
ECG Electrode Localization: 3D DS Camera System for Use in Diverse Clinical Environments
by
Jennifer Bayer, Christoph Hintermüller, Hermann Blessberger and Clemens Steinwender
Cited by 2 | Viewed by 2820
Abstract
Models of the human body representing digital twins of patients have attracted increasing interest in clinical research for the delivery of personalized diagnoses and treatments to patients. For example, noninvasive cardiac imaging models are used to localize the origin of cardiac arrhythmias and
[...] Read more.
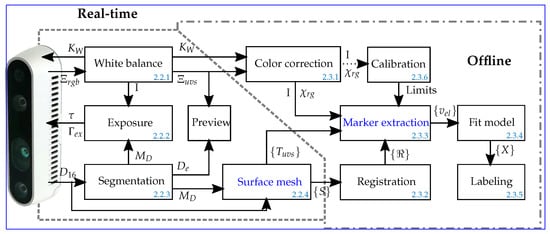
Models of the human body representing digital twins of patients have attracted increasing interest in clinical research for the delivery of personalized diagnoses and treatments to patients. For example, noninvasive cardiac imaging models are used to localize the origin of cardiac arrhythmias and myocardial infarctions. The precise knowledge of a few hundred electrocardiogram (ECG) electrode positions is essential for their diagnostic value. Smaller positional errors are obtained when extracting the sensor positions, along with the anatomical information, for example, from X-ray Computed Tomography (CT) slices. Alternatively, the amount of ionizing radiation the patient is exposed to can be reduced by manually pointing a magnetic digitizer probe one by one to each sensor. An experienced user requires at least 15 min. to perform a precise measurement. Therefore, a 3D depth-sensing camera system was developed that can be operated under adverse lighting conditions and limited space, as encountered in clinical settings. The camera was used to record the positions of 67 electrodes attached to a patient’s chest. These deviate, on average, by 2.0 mm
mm from manually placed markers on the individual 3D views. This demonstrates that the system provides reasonable positional precision even when operated within clinical environments.
Full article
►▼
Show Figures
Open AccessArticle
Specular Reflections Detection and Removal for Endoscopic Images Based on Brightness Classification
by
Chao Nie, Chao Xu, Zhengping Li, Lingling Chu and Yunxue Hu
Cited by 22 | Viewed by 7141
Abstract
Specular Reflections often exist in the endoscopic image, which not only hurts many computer vision algorithms but also seriously interferes with the observation and judgment of the surgeon. The information behind the recovery specular reflection areas is a necessary pre-processing step in medical
[...] Read more.
Specular Reflections often exist in the endoscopic image, which not only hurts many computer vision algorithms but also seriously interferes with the observation and judgment of the surgeon. The information behind the recovery specular reflection areas is a necessary pre-processing step in medical image analysis and application. The existing highlight detection method is usually only suitable for medium-brightness images. The existing highlight removal method is only applicable to images without large specular regions, when dealing with high-resolution medical images with complex texture information, not only does it have a poor recovery effect, but the algorithm operation efficiency is also low. To overcome these limitations, this paper proposes a specular reflection detection and removal method for endoscopic images based on brightness classification. It can effectively detect the specular regions in endoscopic images of different brightness and can improve the operating efficiency of the algorithm while restoring the texture structure information of the high-resolution image. In addition to achieving image brightness classification and enhancing the brightness component of low-brightness images, this method also includes two new steps: In the highlight detection phase, the adaptive threshold function that changes with the brightness of the image is used to detect absolute highlights. During the highlight recovery phase, the priority function of the exemplar-based image inpainting algorithm was modified to ensure reasonable and correct repairs. At the same time, local priority computing and adaptive local search strategies were used to improve algorithm efficiency and reduce error matching. The experimental results show that compared with the other state-of-the-art, our method shows better performance in terms of qualitative and quantitative evaluations, and the algorithm efficiency is greatly improved when processing high-resolution endoscopy images.
Full article
►▼
Show Figures
Open AccessArticle
Comparing the Clinical Viability of Automated Fundus Image Segmentation Methods
by
Gorana Gojić, Veljko B. Petrović, Dinu Dragan, Dušan B. Gajić, Dragiša Mišković, Vladislav Džinić, Zorka Grgić, Jelica Pantelić and Ana Oros
Cited by 2 | Viewed by 2319
Abstract
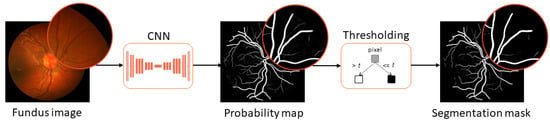
Recent methods for automatic blood vessel segmentation from fundus images have been commonly implemented as convolutional neural networks. While these networks report high values for objective metrics, the clinical viability of recovered segmentation masks remains unexplored. In this paper, we perform a pilot
[...] Read more.
Recent methods for automatic blood vessel segmentation from fundus images have been commonly implemented as convolutional neural networks. While these networks report high values for objective metrics, the clinical viability of recovered segmentation masks remains unexplored. In this paper, we perform a pilot study to assess the clinical viability of automatically generated segmentation masks in the diagnosis of diseases affecting retinal vascularization. Five ophthalmologists with clinical experience were asked to participate in the study. The results demonstrate low classification accuracy, inferring that generated segmentation masks cannot be used as a standalone resource in general clinical practice. The results also hint at possible clinical infeasibility in experimental design. In the follow-up experiment, we evaluate the clinical quality of masks by having ophthalmologists rank generation methods. The ranking is established with high intra-observer consistency, indicating better subjective performance for a subset of tested networks. The study also demonstrates that objective metrics are not correlated with subjective metrics in retinal segmentation tasks for the methods involved, suggesting that objective metrics commonly used in scientific papers to measure the method’s performance are not plausible criteria for choosing clinically robust solutions.
Full article
►▼
Show Figures
Open AccessArticle
Multiple Preprocessing Hybrid Level Set Model for Optic Disc Segmentation in Fundus Images
by
Xiaozhong Xue, Linni Wang, Weiwei Du, Yusuke Fujiwara and Yahui Peng
Cited by 8 | Viewed by 2866
Abstract
The accurate segmentation of the optic disc (OD) in fundus images is a crucial step for the analysis of many retinal diseases. However, because of problems such as vascular occlusion, parapapillary atrophy (PPA), and low contrast, accurate OD segmentation is still a challenging
[...] Read more.
The accurate segmentation of the optic disc (OD) in fundus images is a crucial step for the analysis of many retinal diseases. However, because of problems such as vascular occlusion, parapapillary atrophy (PPA), and low contrast, accurate OD segmentation is still a challenging task. Therefore, this paper proposes a multiple preprocessing hybrid level set model (HLSM) based on area and shape for OD segmentation. The area-based term represents the difference of average pixel values between the inside and outside of a contour, while the shape-based term measures the distance between a prior shape model and the contour. The average intersection over union (IoU) of the proposed method was 0.9275, and the average four-side evaluation (FSE) was 4.6426 on a public dataset with narrow-angle fundus images. The IoU was 0.8179 and the average FSE was 3.5946 on a wide-angle fundus image dataset compiled from a hospital. The results indicate that the proposed multiple preprocessing HLSM is effective in OD segmentation.
Full article
►▼
Show Figures
Open AccessArticle
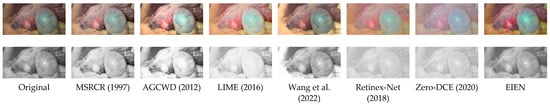
EIEN: Endoscopic Image Enhancement Network Based on Retinex Theory
by
Ziheng An, Chao Xu, Kai Qian, Jubao Han, Wei Tan, Dou Wang and Qianqian Fang
Cited by 13 | Viewed by 4358
Abstract
In recent years, deep convolutional neural network (CNN)-based image enhancement has shown outstanding performance. However, due to the problems of uneven illumination and low contrast existing in endoscopic images, the implementation of medical endoscopic image enhancement using CNN is still an exploratory and
[...] Read more.
In recent years, deep convolutional neural network (CNN)-based image enhancement has shown outstanding performance. However, due to the problems of uneven illumination and low contrast existing in endoscopic images, the implementation of medical endoscopic image enhancement using CNN is still an exploratory and challenging task. An endoscopic image enhancement network (EIEN) based on the Retinex theory is proposed in this paper to solve these problems. The structure consists of three parts: decomposition network, illumination correction network, and reflection component enhancement algorithm. First, the decomposition network model of pre-trained Retinex-Net is retrained on the endoscopic image dataset, and then the images are decomposed into illumination and reflection components by this decomposition network. Second, the illumination components are corrected by the proposed self-attention guided multi-scale pyramid structure. The pyramid structure is used to capture the multi-scale information of the image. The self-attention mechanism is based on the imaging nature of the endoscopic image, and the inverse image of the illumination component is fused with the features of the green and blue channels of the image to be enhanced to generate a weight map that reassigns weights to the spatial dimension of the feature map, to avoid the loss of details in the process of multi-scale feature fusion and image reconstruction by the network. The reflection component enhancement is achieved by sub-channel stretching and weighted fusion, which is used to enhance the vascular information and image contrast. Finally, the enhanced illumination and reflection components are multiplied to obtain the reconstructed image. We compare the results of the proposed method with six other methods on a test set. The experimental results show that EIEN enhances the brightness and contrast of endoscopic images and highlights vascular and tissue information. At the same time, the method in this paper obtained the best results in terms of visual perception and objective evaluation.
Full article
►▼
Show Figures
Open AccessArticle
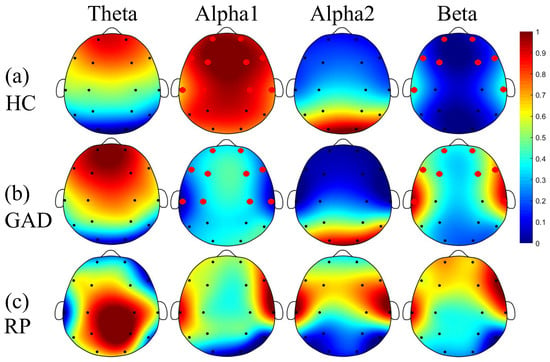
Aberrated Multidimensional EEG Characteristics in Patients with Generalized Anxiety Disorder: A Machine-Learning Based Analysis Framework
by
Zhongxia Shen, Gang Li, Jiaqi Fang, Hongyang Zhong, Jie Wang, Yu Sun and Xinhua Shen
Cited by 42 | Viewed by 7173
Abstract
Although increasing evidences support the notion that psychiatric disorders are associated with abnormal communication between brain regions, scattered studies have investigated brain electrophysiological disconnectivity of patients with generalized anxiety disorder (GAD). To this end, this study intends to develop an analysis framework for
[...] Read more.
Although increasing evidences support the notion that psychiatric disorders are associated with abnormal communication between brain regions, scattered studies have investigated brain electrophysiological disconnectivity of patients with generalized anxiety disorder (GAD). To this end, this study intends to develop an analysis framework for automatic GAD detection through incorporating multidimensional EEG feature extraction and machine learning techniques. Specifically, resting-state EEG signals with a duration of 10 min were obtained from 45 patients with GAD and 36 healthy controls (HC). Then, an analysis framework of multidimensional EEG characteristics (including univariate power spectral density (PSD) and fuzzy entropy (FE), and multivariate functional connectivity (FC), which can decode the EEG information from three different dimensions) were introduced for extracting aberrated multidimensional EEG features via statistical inter-group comparisons. These aberrated features were subsequently fused and fed into three previously validated machine learning methods to evaluate classification performance for automatic patient detection. We showed that patients exhibited a significant increase in beta rhythm and decrease in alpha1 rhythm of PSD, together with the reduced long-range FC between frontal and other brain areas in all frequency bands. Moreover, these aberrated features contributed to a very good classification performance with 97.83 ± 0.40% of accuracy, 97.55 ± 0.31% of sensitivity, 97.78 ± 0.36% of specificity, and 97.95 ± 0.17% of F1. These findings corroborate previous hypothesis of disconnectivity in psychiatric disorders and further shed light on distribution patterns of aberrant spatio-spectral EEG characteristics, which may lead to potential application of automatic diagnosis of GAD.
Full article
►▼
Show Figures
Open AccessArticle
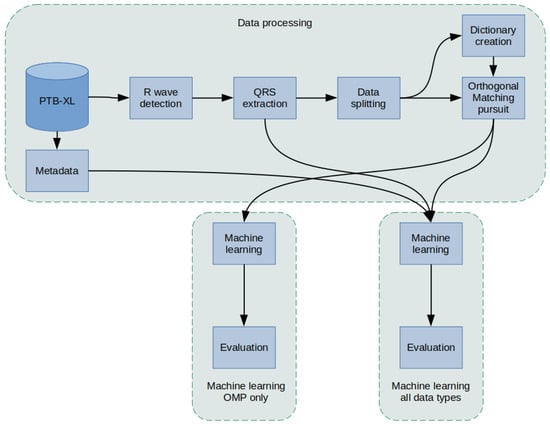
ECG Classification Using Orthogonal Matching Pursuit and Machine Learning
by
Sandra Śmigiel
Cited by 21 | Viewed by 4788
Abstract
Health monitoring and related technologies are a rapidly growing area of research. To date, the electrocardiogram (ECG) remains a popular measurement tool in the evaluation and diagnosis of heart disease. The number of solutions involving ECG signal monitoring systems is growing exponentially in
[...] Read more.
Health monitoring and related technologies are a rapidly growing area of research. To date, the electrocardiogram (ECG) remains a popular measurement tool in the evaluation and diagnosis of heart disease. The number of solutions involving ECG signal monitoring systems is growing exponentially in the literature. In this article, underestimated Orthogonal Matching Pursuit (OMP) algorithms are used, demonstrating the significant effect of concise representation parameters on improving the performance of the classification process. Cardiovascular disease classification models based on classical Machine Learning classifiers were defined and investigated. The study was undertaken on the recently published PTB-XL database, whose ECG signals were previously subjected to detailed analysis. The classification was realized for class 2, class 5, and class 15 cardiac diseases. A new method of detecting R-waves and, based on them, determining the location of QRS complexes was presented. Novel aggregation methods of ECG signal fragments containing QRS segments, necessary for tests for classical classifiers, were developed. As a result, it was proved that ECG signal subjected to algorithms of R wave detection, QRS complexes extraction, and resampling performs very well in classification using Decision Trees. The reason can be found in structuring the signal due to the actions mentioned above. The implementation of classification issues achieved the highest Accuracy of 90.4% in recognition of 2 classes, as compared to less than 78% for 5 classes and 71% for 15 classes.
Full article
►▼
Show Figures
Open AccessArticle
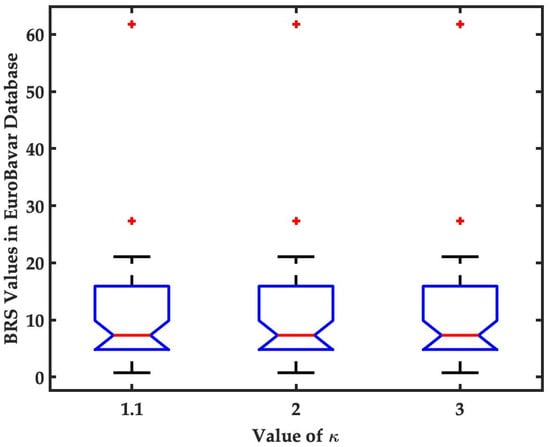
A Novel Method for Baroreflex Sensitivity Estimation Using Modulated Gaussian Filter
by
Tienhsiung Ku, Serge Ismael Zida, Latifa Nabila Harfiya, Yung-Hui Li and Yue-Der Lin
Cited by 1 | Viewed by 3430
Abstract
The evaluation of baroreflex sensitivity (BRS) has proven to be critical for medical applications. The use of
α indices by spectral methods has been the most popular approach to BRS estimation. Recently, an algorithm termed Gaussian average filtering decomposition (GAFD) has been proposed
[...] Read more.
The evaluation of baroreflex sensitivity (BRS) has proven to be critical for medical applications. The use of
α indices by spectral methods has been the most popular approach to BRS estimation. Recently, an algorithm termed Gaussian average filtering decomposition (GAFD) has been proposed to serve the same purpose. GAFD adopts a three-layer tree structure similar to wavelet decomposition but is only constructed by Gaussian windows in different cutoff frequency. Its computation is more efficient than that of conventional spectral methods, and there is no need to specify any parameter. This research presents a novel approach, referred to as modulated Gaussian filter (modGauss) for BRS estimation. It has a more simplified structure than GAFD using only two bandpass filters of dedicated passbands, so that the three-level structure in GAFD is avoided. This strategy makes modGauss more efficient than GAFD in computation, while the advantages of GAFD are preserved. Both GAFD and modGauss are conducted extensively in the time domain, yet can achieve similar results to conventional spectral methods. In computational simulations, the EuroBavar dataset was used to assess the performance of the novel algorithm. The BRS values were calculated by four other methods (three spectral approaches and GAFD) for performance comparison. From a comparison using the Wilcoxon rank sum test, it was found that there was no statistically significant dissimilarity; instead, very good agreement using the intraclass correlation coefficient (ICC) was observed. The modGauss algorithm was also found to be the fastest in computation time and suitable for the long-term estimation of BRS. The novel algorithm, as described in this report, can be applied in medical equipment for real-time estimation of BRS in clinical settings.
Full article
►▼
Show Figures
Open AccessArticle
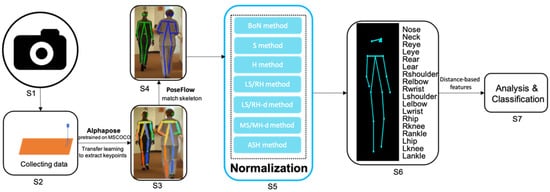
2D Gait Skeleton Data Normalization for Quantitative Assessment of Movement Disorders from Freehand Single Camera Video Recordings
by
Wei Tang, Peter M. A. van Ooijen, Deborah A. Sival and Natasha M. Maurits
Cited by 12 | Viewed by 3695
Abstract
Overlapping phenotypic features between Early Onset Ataxia (EOA) and Developmental Coordination Disorder (DCD) can complicate the clinical distinction of these disorders. Clinical rating scales are a common way to quantify movement disorders but in children these scales also rely on the observer’s assessment
[...] Read more.
Overlapping phenotypic features between Early Onset Ataxia (EOA) and Developmental Coordination Disorder (DCD) can complicate the clinical distinction of these disorders. Clinical rating scales are a common way to quantify movement disorders but in children these scales also rely on the observer’s assessment and interpretation. Despite the introduction of inertial measurement units for objective and more precise evaluation, special hardware is still required, restricting their widespread application. Gait video recordings of movement disorder patients are frequently captured in routine clinical settings, but there is presently no suitable quantitative analysis method for these recordings. Owing to advancements in computer vision technology, deep learning pose estimation techniques may soon be ready for convenient and low-cost clinical usage. This study presents a framework based on 2D video recording in the coronal plane and pose estimation for the quantitative assessment of gait in movement disorders. To allow the calculation of distance-based features, seven different methods to normalize 2D skeleton keypoint data derived from pose estimation using deep neural networks applied to freehand video recording of gait were evaluated. In our experiments, 15 children (five EOA, five DCD and five healthy controls) were asked to walk naturally while being videotaped by a single camera in 1280 × 720 resolution at 25 frames per second. The high likelihood of the prediction of keypoint locations (mean = 0.889, standard deviation = 0.02) demonstrates the potential for distance-based features derived from routine video recordings to assist in the clinical evaluation of movement in EOA and DCD. By comparison of mean absolute angle error and mean variance of distance, the normalization methods using the Euclidean (2D) distance of left shoulder and right hip, or the average distance from left shoulder to right hip and from right shoulder to left hip were found to better perform for deriving distance-based features and further quantitative assessment of movement disorders.
Full article
►▼
Show Figures
Open AccessArticle
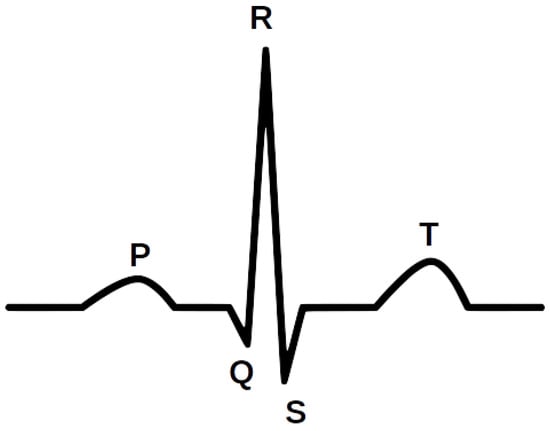
Study of the Few-Shot Learning for ECG Classification Based on the PTB-XL Dataset
by
Krzysztof Pałczyński, Sandra Śmigiel, Damian Ledziński and Sławomir Bujnowski
Cited by 51 | Viewed by 11284
Abstract
The electrocardiogram (ECG) is considered a fundamental of cardiology. The ECG consists of P, QRS, and T waves. Information provided from the signal based on the intervals and amplitudes of these waves is associated with various heart diseases. The first step in isolating
[...] Read more.
The electrocardiogram (ECG) is considered a fundamental of cardiology. The ECG consists of P, QRS, and T waves. Information provided from the signal based on the intervals and amplitudes of these waves is associated with various heart diseases. The first step in isolating the features of an ECG begins with the accurate detection of the R-peaks in the QRS complex. The database was based on the PTB-XL database, and the signals from Lead I–XII were analyzed. This research focuses on determining the Few-Shot Learning (FSL) applicability for ECG signal proximity-based classification. The study was conducted by training Deep Convolutional Neural Networks to recognize 2, 5, and 20 different heart disease classes. The results of the FSL network were compared with the evaluation score of the neural network performing softmax-based classification. The neural network proposed for this task interprets a set of QRS complexes extracted from ECG signals. The FSL network proved to have higher accuracy in classifying healthy/sick patients ranging from 93.2% to 89.2% than the softmax-based classification network, which achieved 90.5–89.2% accuracy. The proposed network also achieved better results in classifying five different disease classes than softmax-based counterparts with an accuracy of 80.2–77.9% as opposed to 77.1% to 75.1%. In addition, the method of R-peaks labeling and QRS complexes extraction has been implemented. This procedure converts a 12-lead signal into a set of R waves by using the detection algorithms and the k-mean algorithm.
Full article
►▼
Show Figures
Open AccessArticle
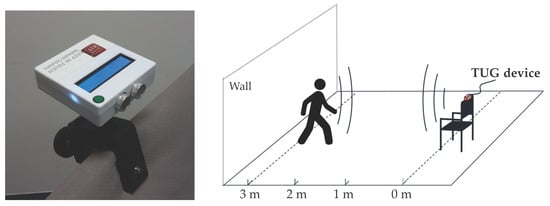
Quantification of the Link between Timed Up-and-Go Test Subtasks and Contractile Muscle Properties
by
Andreas Ziegl, Dieter Hayn, Peter Kastner, Ester Fabiani, Boštjan Šimunič, Kerstin Löffler, Lisa Weidinger, Bianca Brix, Nandu Goswami and Schreier Günter
Cited by 8 | Viewed by 3723
Abstract
Frailty and falls are a major public health problem in older adults. Muscle weakness of the lower and upper extremities are risk factors for any, as well as recurrent falls including injuries and fractures. While the Timed Up-and-Go (TUG) test is often used
[...] Read more.
Frailty and falls are a major public health problem in older adults. Muscle weakness of the lower and upper extremities are risk factors for any, as well as recurrent falls including injuries and fractures. While the Timed Up-and-Go (TUG) test is often used to identify frail members and fallers, tensiomyography (TMG) can be used as a non-invasive tool to assess the function of skeletal muscles. In a clinical study, we evaluated the correlation between the TMG parameters of the skeletal muscle contraction of 23 elderly participants (22 f, age 86.74 ± 7.88) and distance-based TUG test subtask times. TUG tests were recorded with an ultrasonic-based device. The sit-up and walking phases were significantly correlated to the contraction and delay time of the muscle vastus medialis (
ρ = 0.55–0.80,
p < 0.01). In addition, the delay time of the muscles vastus medialis (
ρ = 0.45,
p = 0.03) and gastrocnemius medialis (
ρ = −0.44,
p = 0.04) correlated to the sit-down phase. The maximal radial displacements of the biceps femoris showed significant correlations with the walk-forward times (
ρ = −0.47,
p = 0.021) and back (
ρ = −0.43,
p = 0.04). The association of TUG subtasks to muscle contractile parameters, therefore, could be utilized as a measure to improve the monitoring of elderly people’s physical ability in general and during rehabilitation after a fall in particular. TUG test subtask measurements may be used as a proxy to monitor muscle properties in rehabilitation after long hospital stays and injuries or for fall prevention.
Full article
►▼
Show Figures
Open AccessArticle
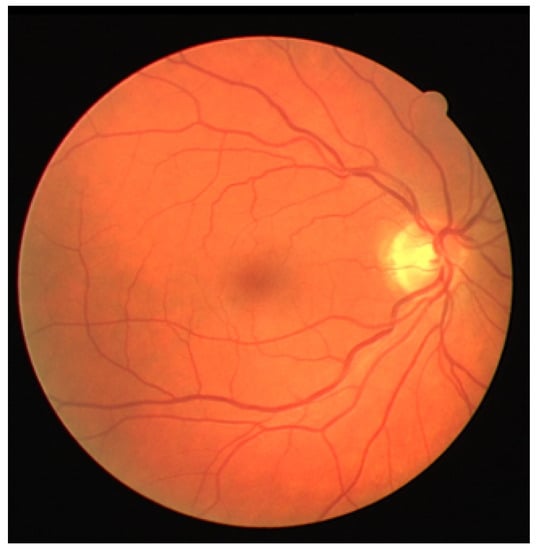
Interactive Blood Vessel Segmentation from Retinal Fundus Image Based on Canny Edge Detector
by
Alexander Ze Hwan Ooi, Zunaina Embong, Aini Ismafairus Abd Hamid, Rafidah Zainon, Shir Li Wang, Theam Foo Ng, Rostam Affendi Hamzah, Soo Siang Teoh and Haidi Ibrahim
Cited by 48 | Viewed by 7452
Abstract
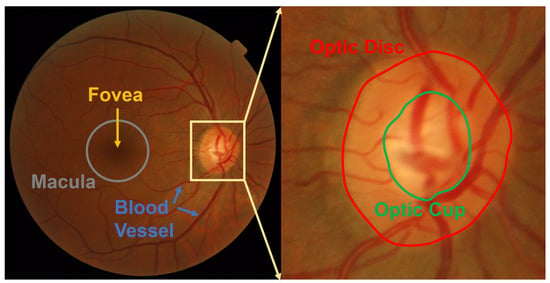
Optometrists, ophthalmologists, orthoptists, and other trained medical professionals use fundus photography to monitor the progression of certain eye conditions or diseases. Segmentation of the vessel tree is an essential process of retinal analysis. In this paper, an interactive blood vessel segmentation from retinal
[...] Read more.
Optometrists, ophthalmologists, orthoptists, and other trained medical professionals use fundus photography to monitor the progression of certain eye conditions or diseases. Segmentation of the vessel tree is an essential process of retinal analysis. In this paper, an interactive blood vessel segmentation from retinal fundus image based on Canny edge detection is proposed. Semi-automated segmentation of specific vessels can be done by simply moving the cursor across a particular vessel. The pre-processing stage includes the green color channel extraction, applying Contrast Limited Adaptive Histogram Equalization (CLAHE), and retinal outline removal. After that, the edge detection techniques, which are based on the Canny algorithm, will be applied. The vessels will be selected interactively on the developed graphical user interface (GUI). The program will draw out the vessel edges. After that, those vessel edges will be segmented to bring focus on its details or detect the abnormal vessel. This proposed approach is useful because different edge detection parameter settings can be applied to the same image to highlight particular vessels for analysis or presentation.
Full article
►▼
Show Figures
Open AccessArticle
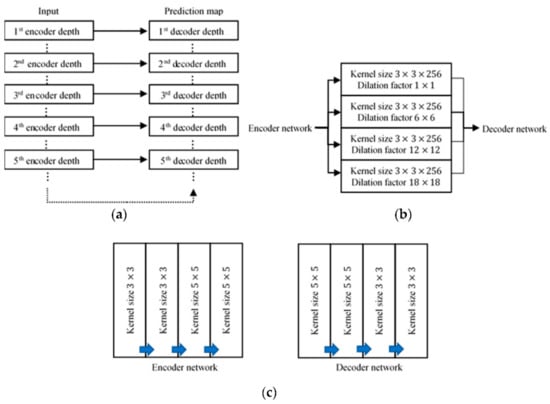
Automatic Polyp Segmentation in Colonoscopy Images Using a Modified Deep Convolutional Encoder-Decoder Architecture
by
Chin Yii Eu, Tong Boon Tang, Cheng-Hung Lin, Lok Hua Lee and Cheng-Kai Lu
Cited by 17 | Viewed by 5368
Abstract
Colorectal cancer has become the third most commonly diagnosed form of cancer, and has the second highest fatality rate of cancers worldwide. Currently, optical colonoscopy is the preferred tool of choice for the diagnosis of polyps and to avert colorectal cancer. Colon screening
[...] Read more.
Colorectal cancer has become the third most commonly diagnosed form of cancer, and has the second highest fatality rate of cancers worldwide. Currently, optical colonoscopy is the preferred tool of choice for the diagnosis of polyps and to avert colorectal cancer. Colon screening is time-consuming and highly operator dependent. In view of this, a computer-aided diagnosis (CAD) method needs to be developed for the automatic segmentation of polyps in colonoscopy images. This paper proposes a modified SegNet Visual Geometry Group-19 (VGG-19), a form of convolutional neural network, as a CAD method for polyp segmentation. The modifications include skip connections, 5 × 5 convolutional filters, and the concatenation of four dilated convolutions applied in parallel form. The CVC-ClinicDB, CVC-ColonDB, and ETIS-LaribPolypDB databases were used to evaluate the model, and it was found that our proposed polyp segmentation model achieved an accuracy, sensitivity, specificity, precision, mean intersection over union, and dice coefficient of 96.06%, 94.55%, 97.56%, 97.48%, 92.3%, and 95.99%, respectively. These results indicate that our model performs as well as or better than previous schemes in the literature. We believe that this study will offer benefits in terms of the future development of CAD tools for polyp segmentation for colorectal cancer diagnosis and management. In the future, we intend to embed our proposed network into a medical capsule robot for practical usage and try it in a hospital setting with clinicians.
Full article
►▼
Show Figures
Open AccessArticle
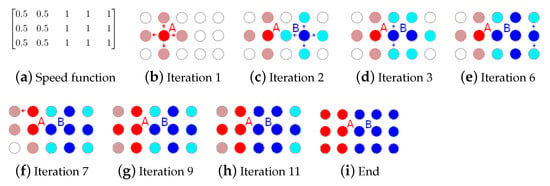
Lung Nodule Segmentation with a Region-Based Fast Marching Method
by
Marko Savic, Yanhe Ma, Giovanni Ramponi, Weiwei Du and Yahui Peng
Cited by 44 | Viewed by 7463
Abstract
When dealing with computed tomography volume data, the accurate segmentation of lung nodules is of great importance to lung cancer analysis and diagnosis, being a vital part of computer-aided diagnosis systems. However, due to the variety of lung nodules and the similarity of
[...] Read more.
When dealing with computed tomography volume data, the accurate segmentation of lung nodules is of great importance to lung cancer analysis and diagnosis, being a vital part of computer-aided diagnosis systems. However, due to the variety of lung nodules and the similarity of visual characteristics for nodules and their surroundings, robust segmentation of nodules becomes a challenging problem. A segmentation algorithm based on the fast marching method is proposed that separates the image into regions with similar features, which are then merged by combining regions growing with k-means. An evaluation was performed with two distinct methods (objective and subjective) that were applied on two different datasets, containing simulation data generated for this study and real patient data, respectively. The objective experimental results show that the proposed technique can accurately segment nodules, especially in solid cases, given the mean Dice scores of 0.933 and 0.901 for round and irregular nodules. For non-solid and cavitary nodules the performance dropped—0.799 and 0.614 mean Dice scores, respectively. The proposed method was compared to active contour models and to two modern deep learning networks. It reached better overall accuracy than active contour models, having comparable results to DBResNet but lesser accuracy than 3D-UNet. The results show promise for the proposed method in computer-aided diagnosis applications.
Full article
►▼
Show Figures