- 2.5Impact Factor
- 5.5CiteScore
- 20 daysTime to First Decision
Generative Artificial Intelligence for Clinical Decision Support System and Healthcare
This special issue belongs to the section “Applied Biosciences and Bioengineering“.
Special Issue Information
Dear Colleagues,
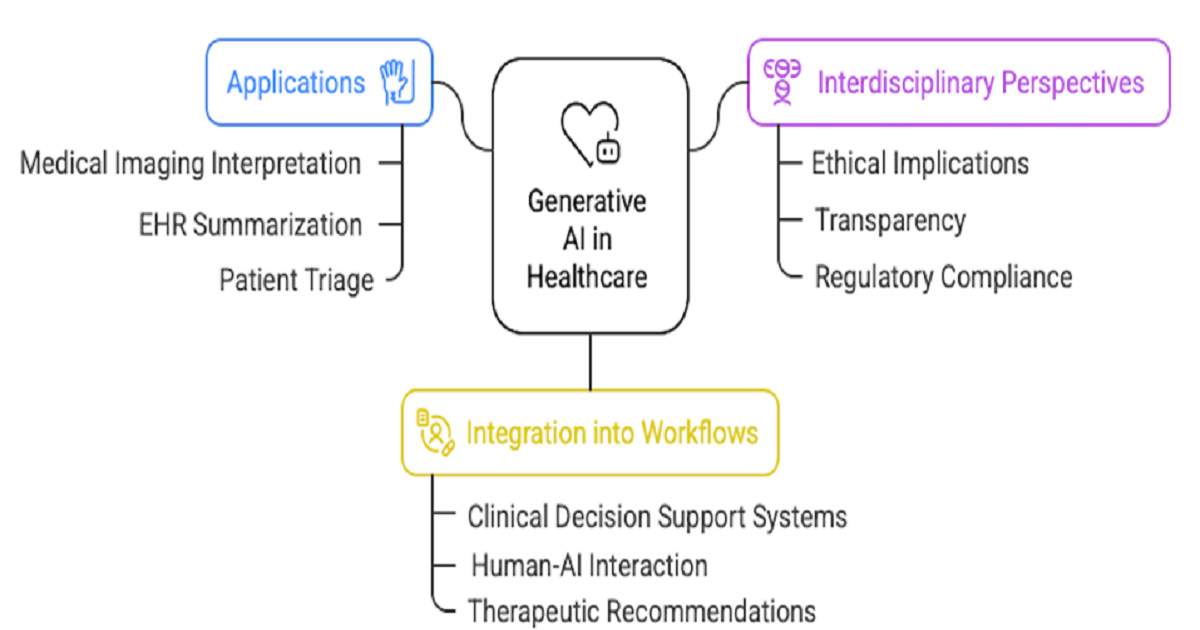
We are pleased to announce a Special Issue titled “Generative Artificial Intelligence for Clinical Decision Support System and Healthcare”. As healthcare continues to evolve with the integration of cutting-edge digital technologies, Generative Artificial Intelligence (GenAI) is emerging as a key enabler in transforming clinical workflows, enhancing decision-making, and personalizing patient care.
This Special Issue aims to highlight innovative methodologies, formative use cases, and impactful applications of GenAI in clinical decision support systems (CDSS) and across the healthcare continuum. We welcome original research and review contributions that explore how GenAI—through models such as large language models, generative adversarial networks, and diffusion models—is being integrated into healthcare environments to improve efficiency, accuracy, and outcomes.
By contributing to this Special Issue, you will have the opportunity to disseminate your work to a multidisciplinary audience of researchers, clinicians, and healthcare technology experts. This issue aims to foster meaningful dialog across disciplines, encourage the translation of generative AI innovations into clinical practice, and support the development of robust, ethical, and scalable solutions that address real-world healthcare challenges.
We invite contributions that explore the transformative role of GenAI in healthcare. All submissions will undergo a rigorous peer-review process to ensure scientific excellence.
Topics of interest include, but are not limited to:
- GenAI-powered systems for clinical note generation and EHR summarization;
- AI copilots in diagnostic workflows and decision support;
- Virtual assistants for patient communication, triage, or education;
- Integration of GenAI into multidisciplinary clinical settings;
- Use of GenAI for precision medicine;
- Educational and training tools enhanced by GenAI technologies;
- Ethical, regulatory, and transparency considerations in GenAI applications;
- Explainable generative models for clinical interpretability and trust.
Dr. Giacomo Cappon
Guest Editor
Dr. Luca Cossu
Guest Editor Assistant
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the special issue website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Applied Sciences is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2400 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- generative AI
- GPT in healthcare
- decision support system
- clinical decision-making
- large language models
- explainable AI in medicine
- AI in clinical workflows
- human-AI collaboration in medicine
Benefits of Publishing in a Special Issue
- Ease of navigation: Grouping papers by topic helps scholars navigate broad scope journals more efficiently.
- Greater discoverability: Special Issues support the reach and impact of scientific research. Articles in Special Issues are more discoverable and cited more frequently.
- Expansion of research network: Special Issues facilitate connections among authors, fostering scientific collaborations.
- External promotion: Articles in Special Issues are often promoted through the journal's social media, increasing their visibility.
- e-Book format: Special Issues with more than 10 articles can be published as dedicated e-books, ensuring wide and rapid dissemination.

