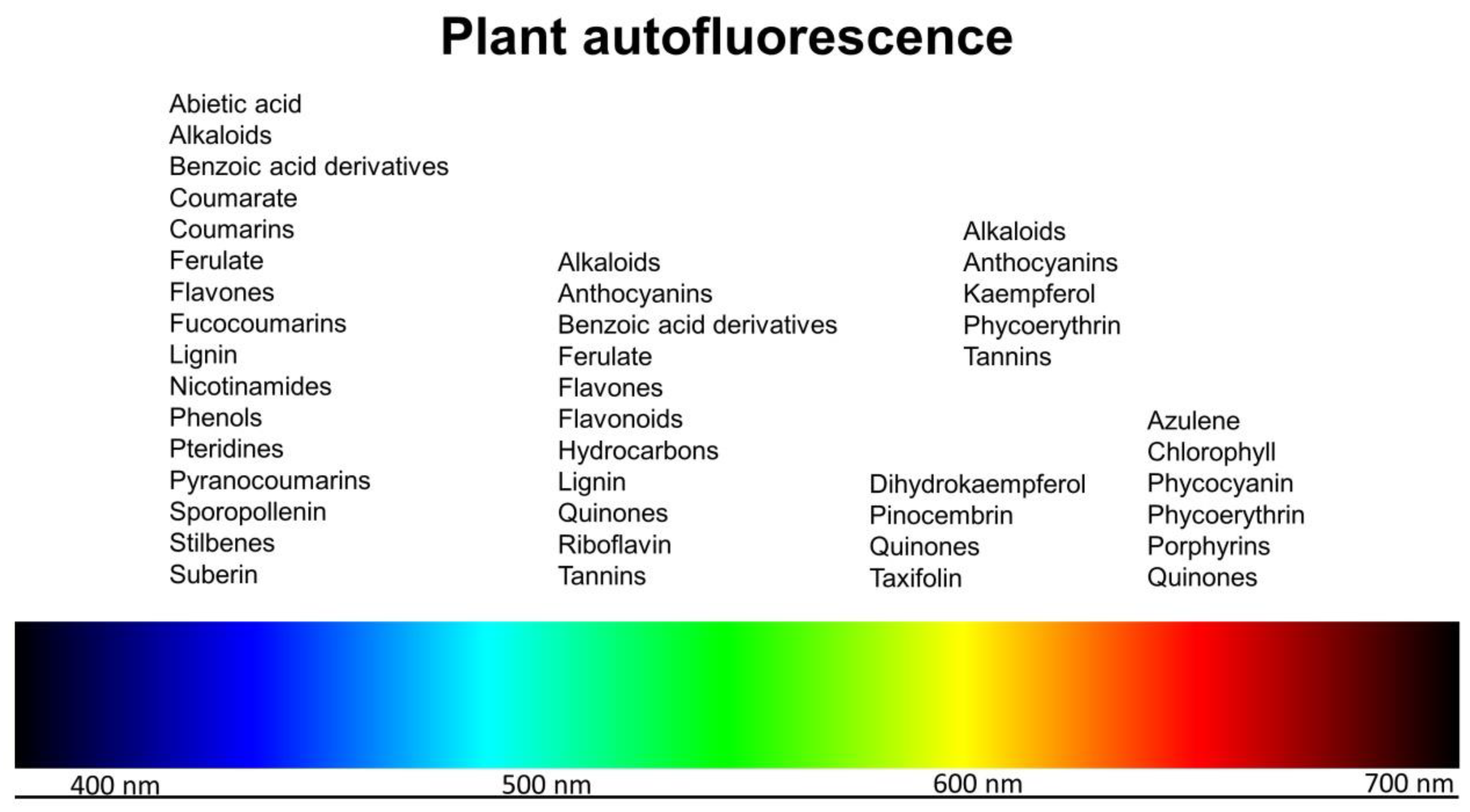
Autofluorescence in Plants
Abstract
:1. Introduction
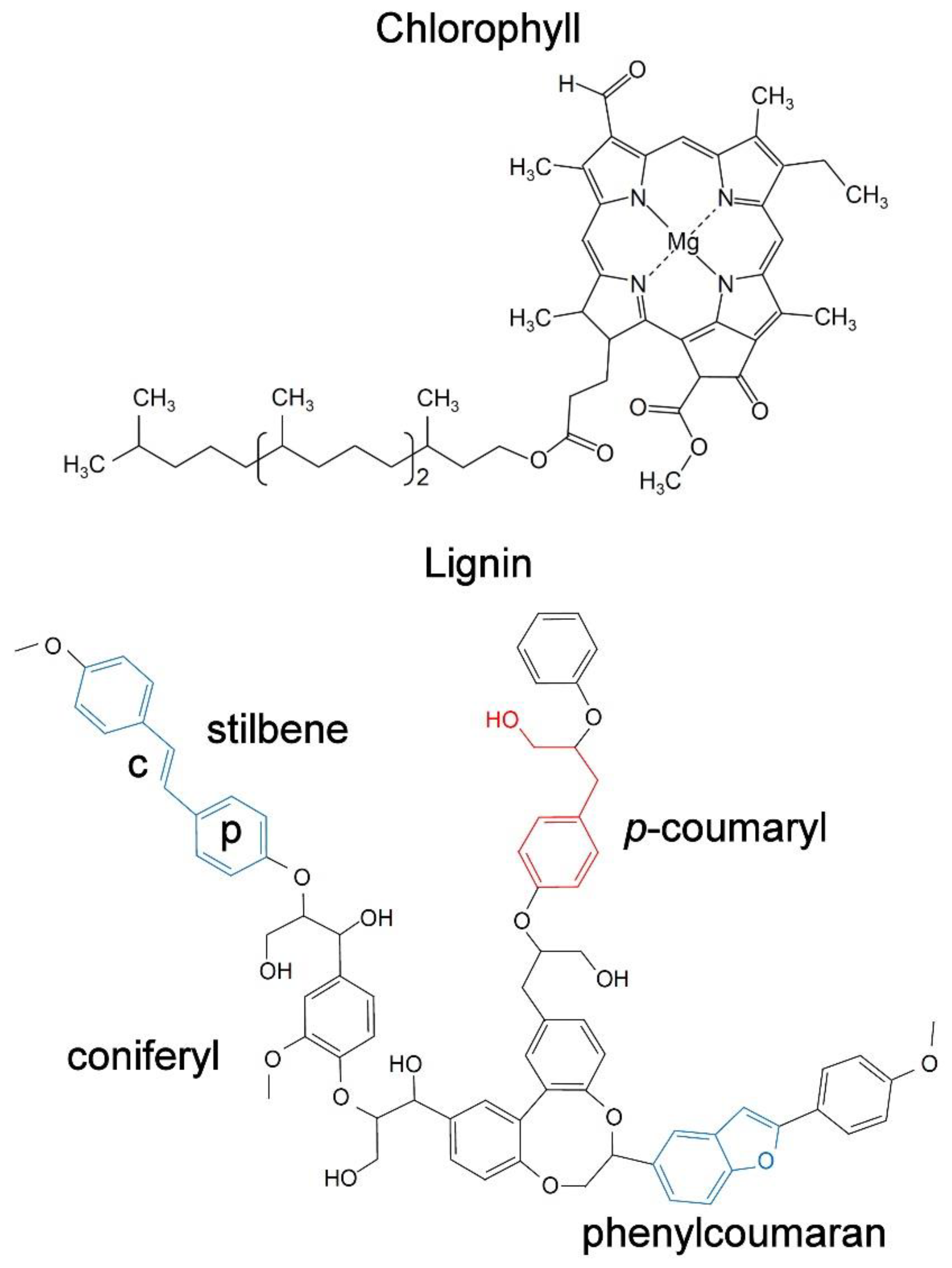
2. Chlorophyll and Other Pigments
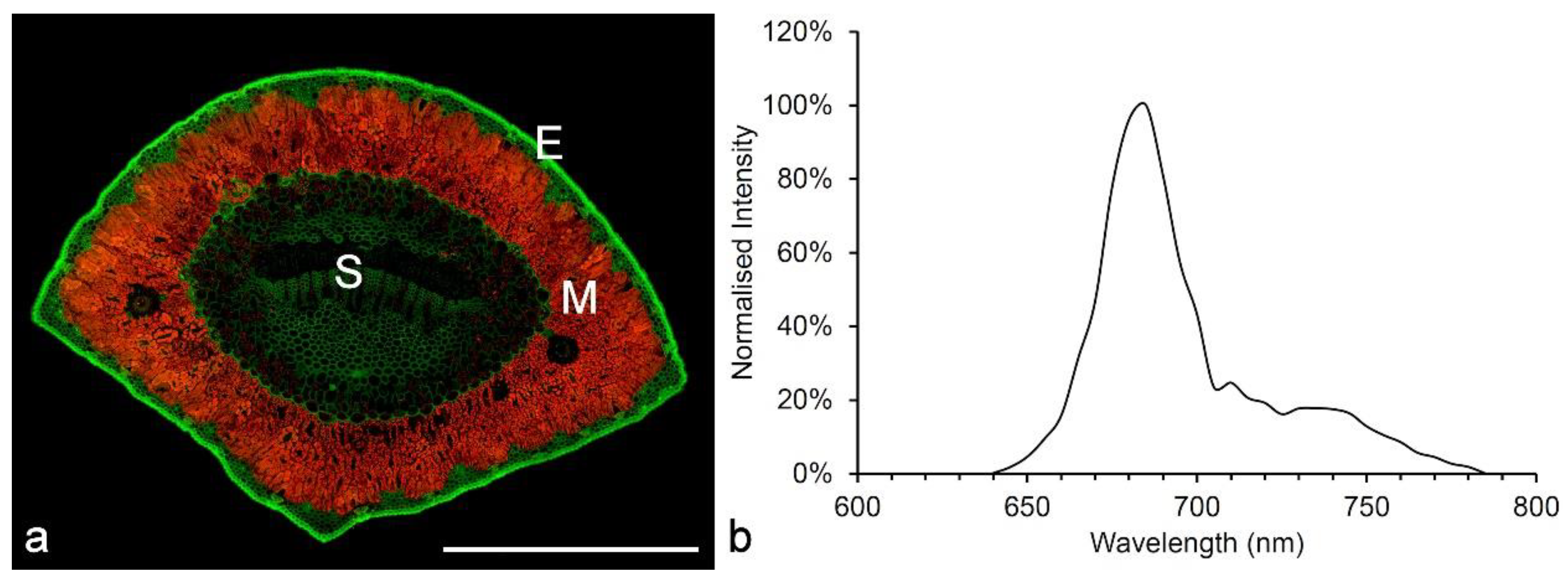
3. Lignin
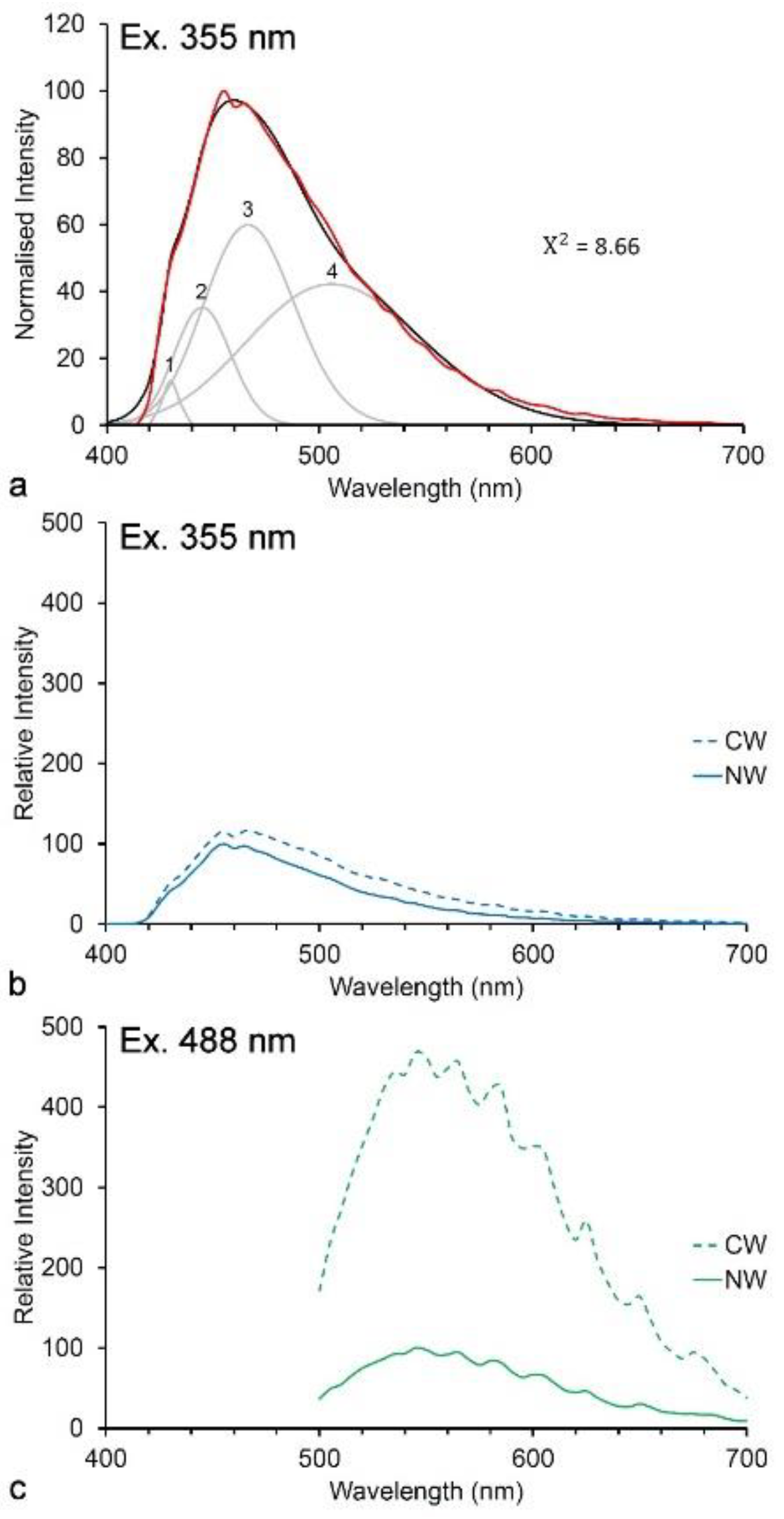
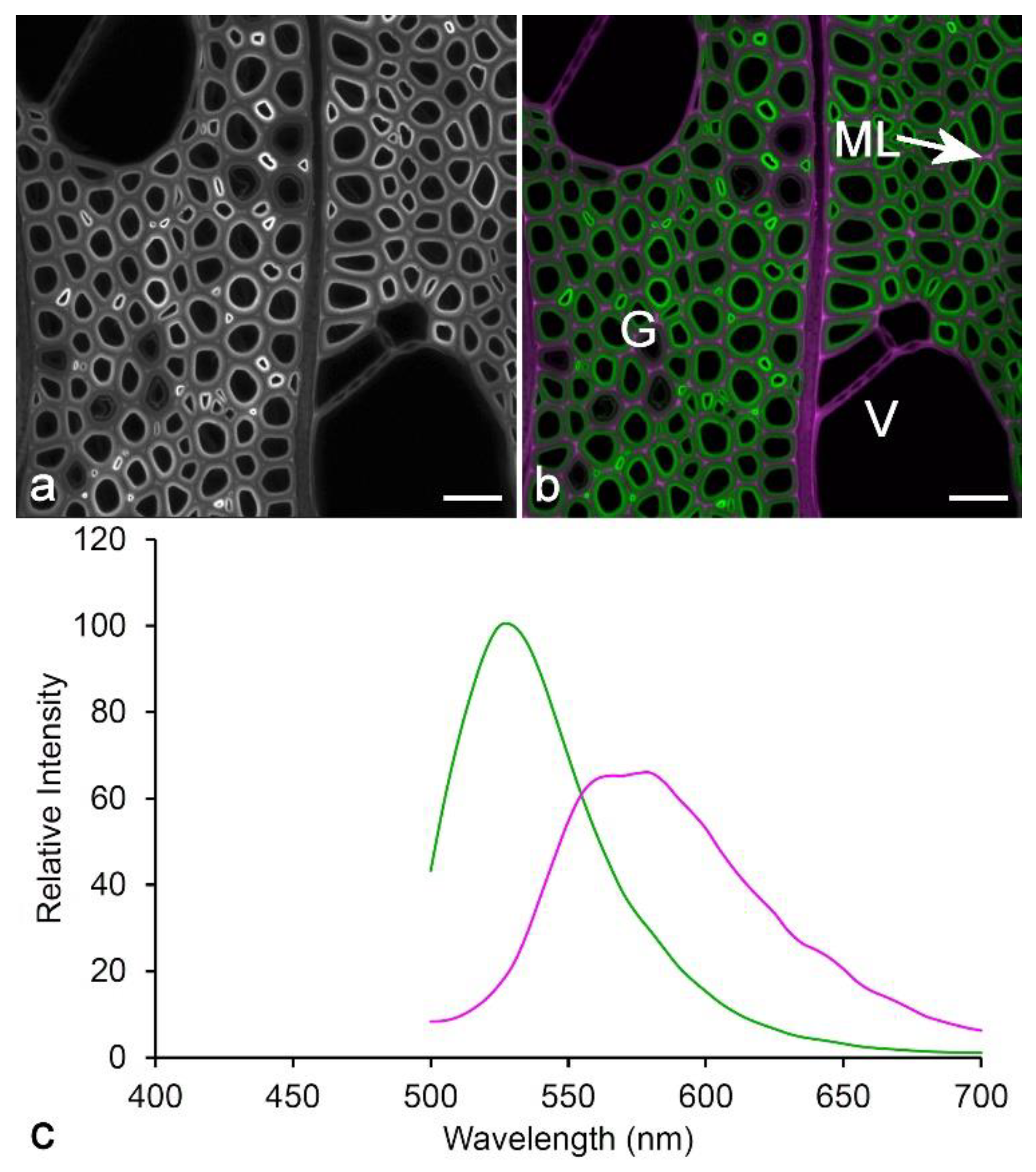
3.1. Spectroscopy
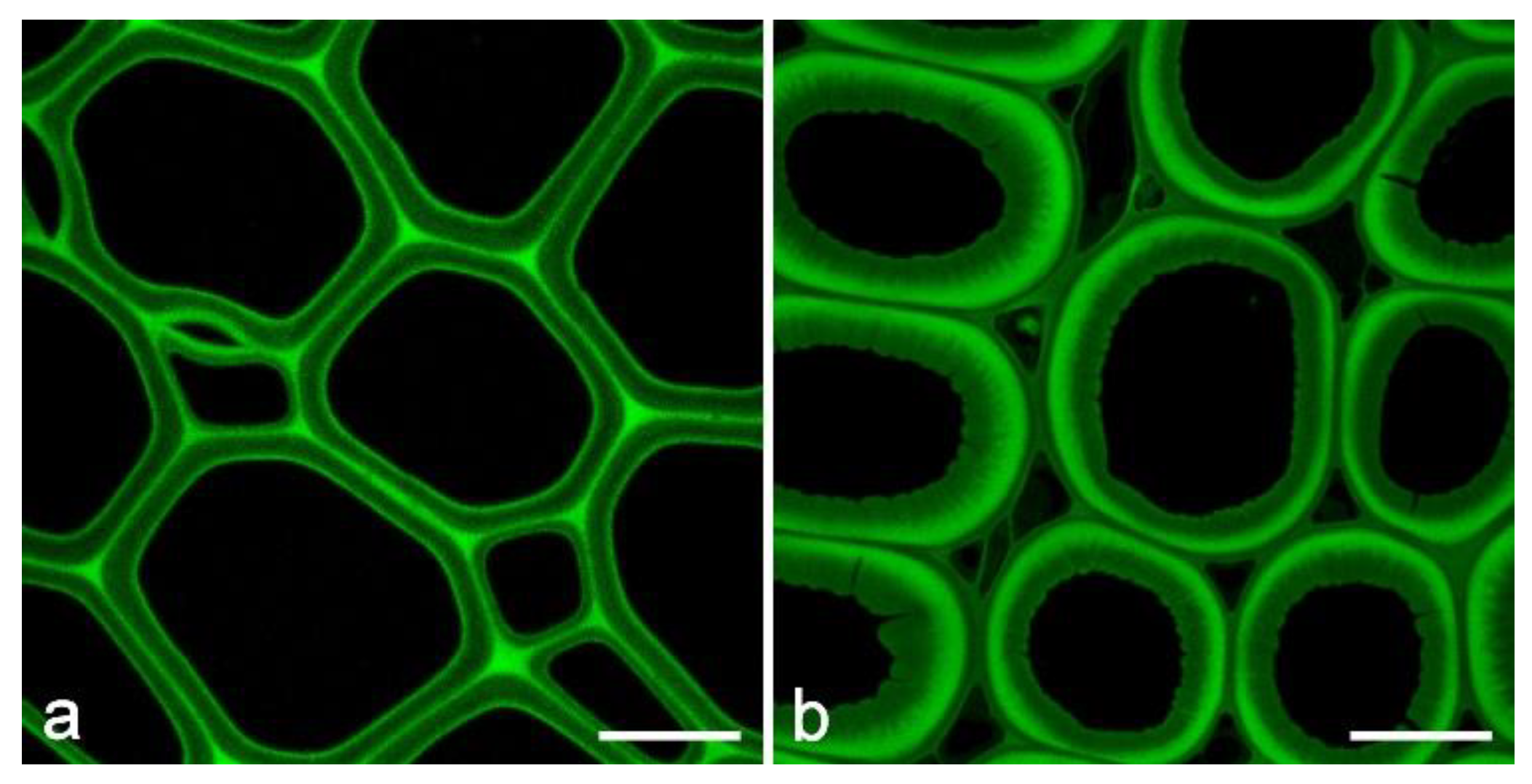
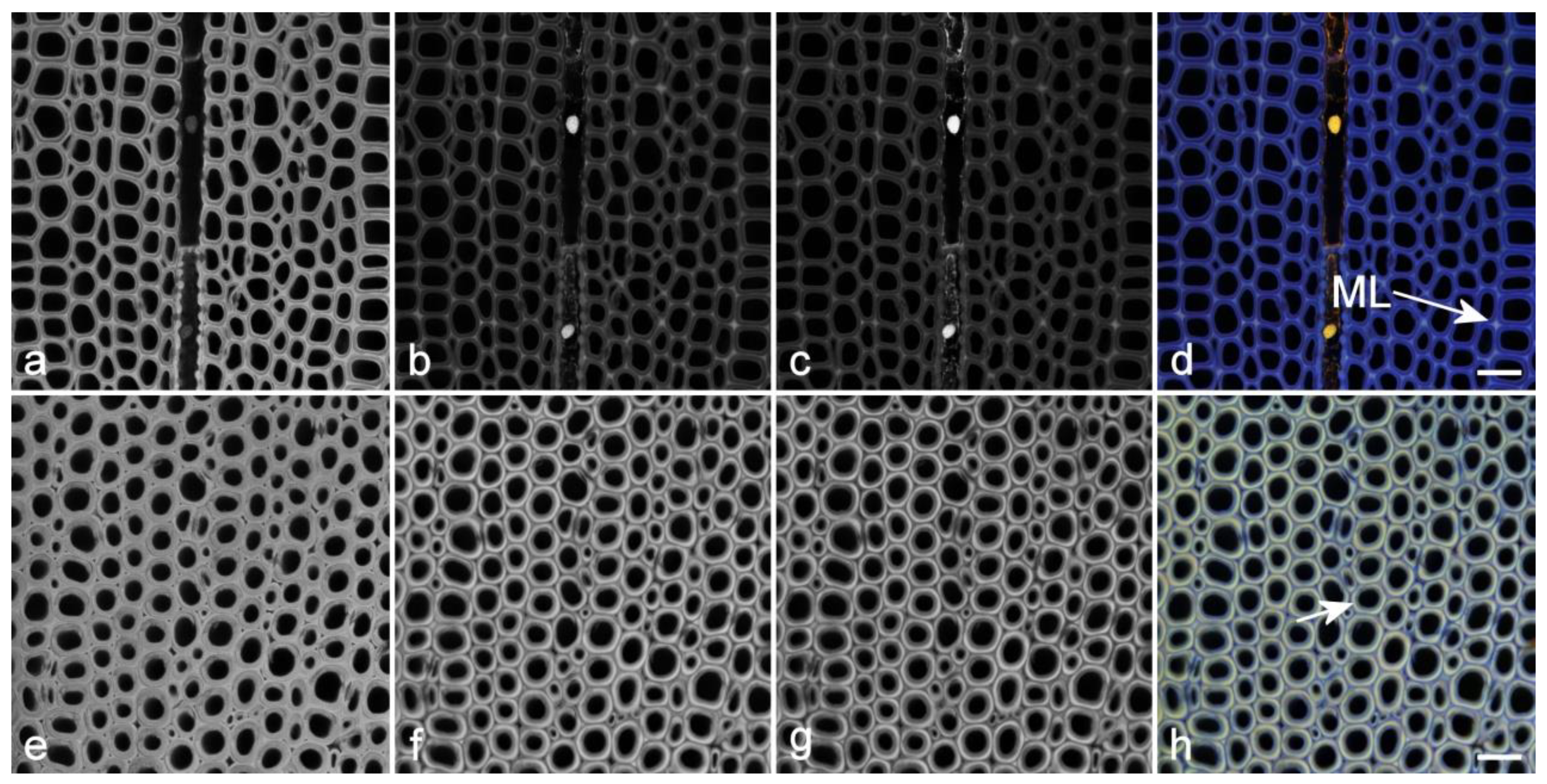
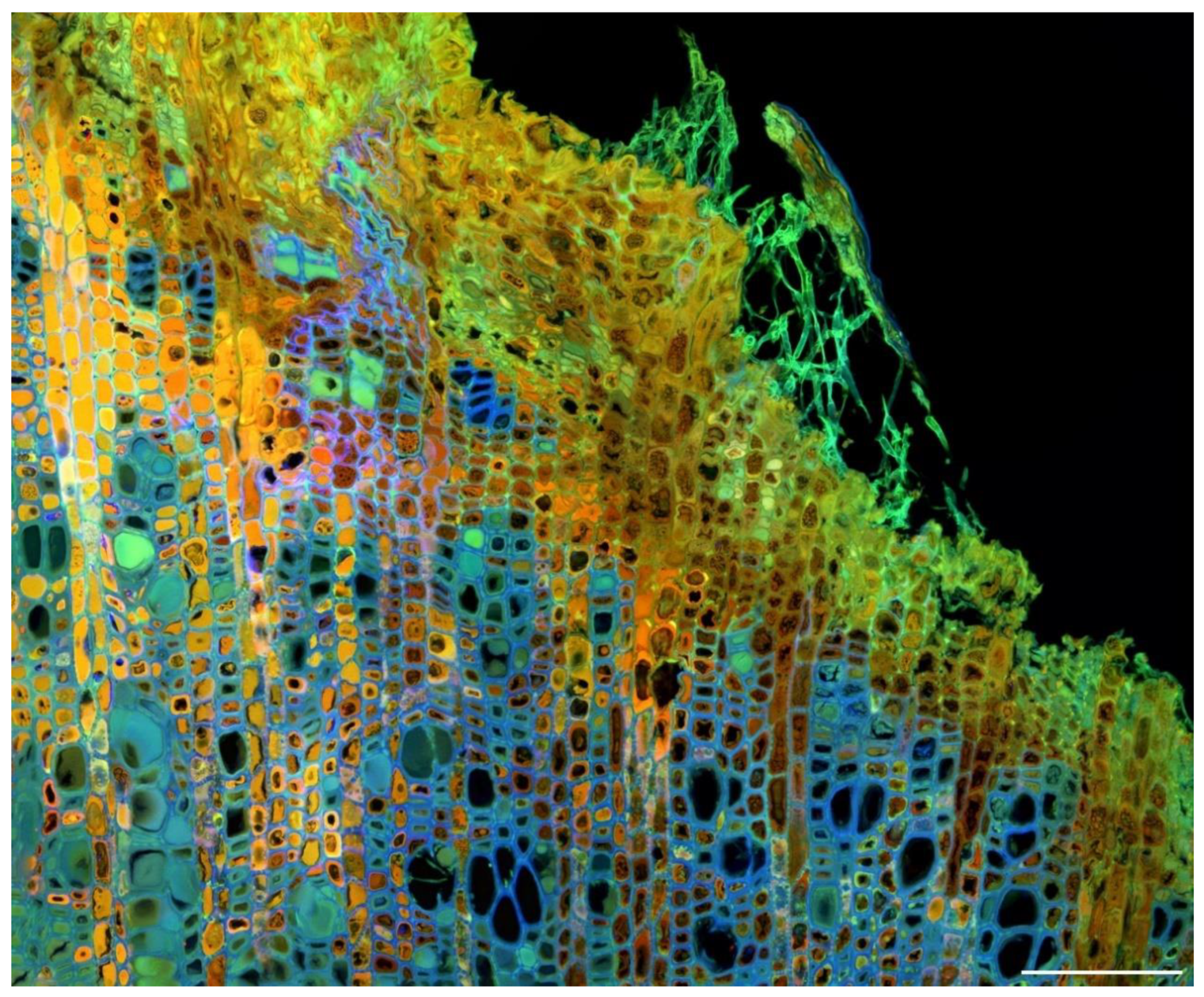
3.2. Imaging
4. Other Cell Wall-Associated Fluorophores
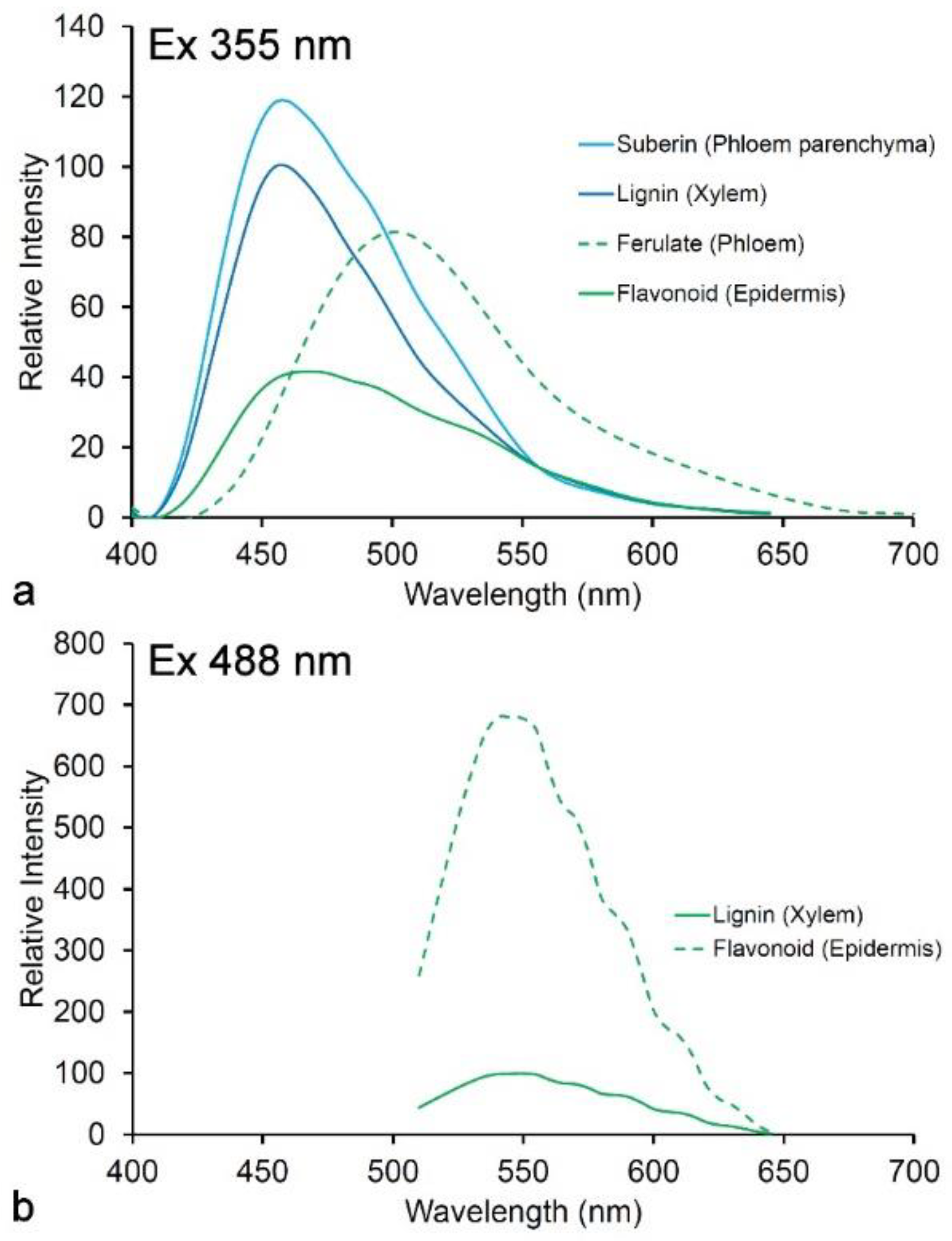
4.1. Ferulate
4.2. Cutin
4.3. Suberin
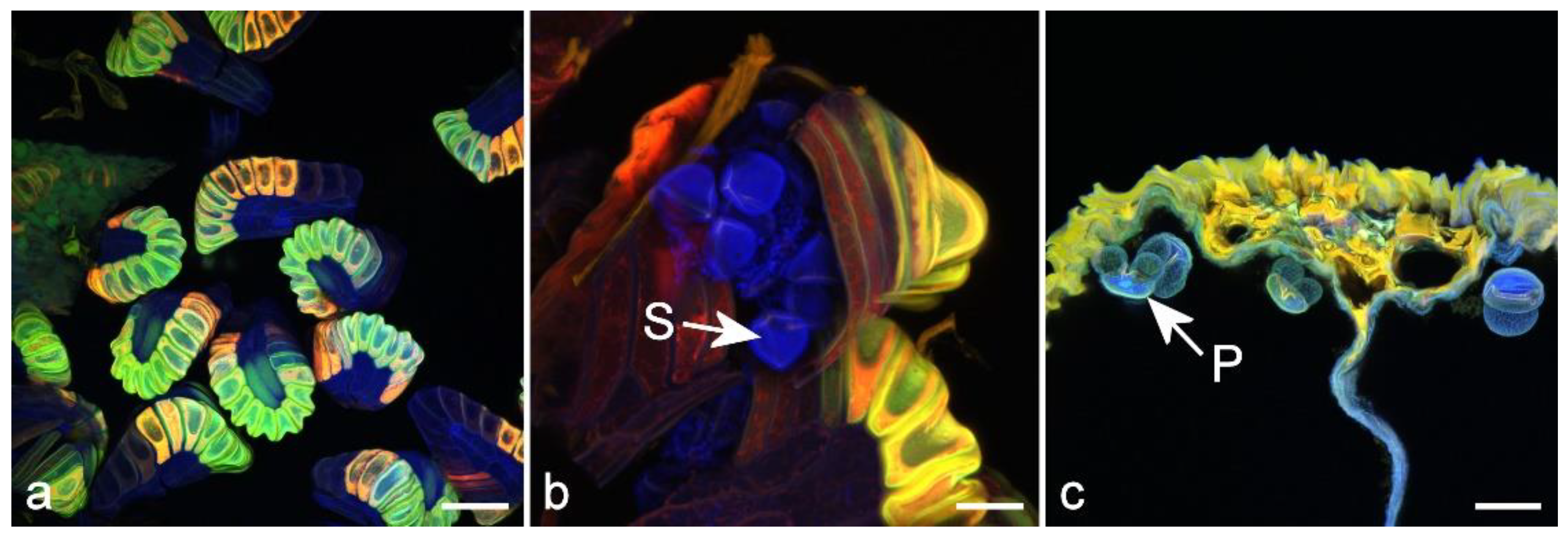
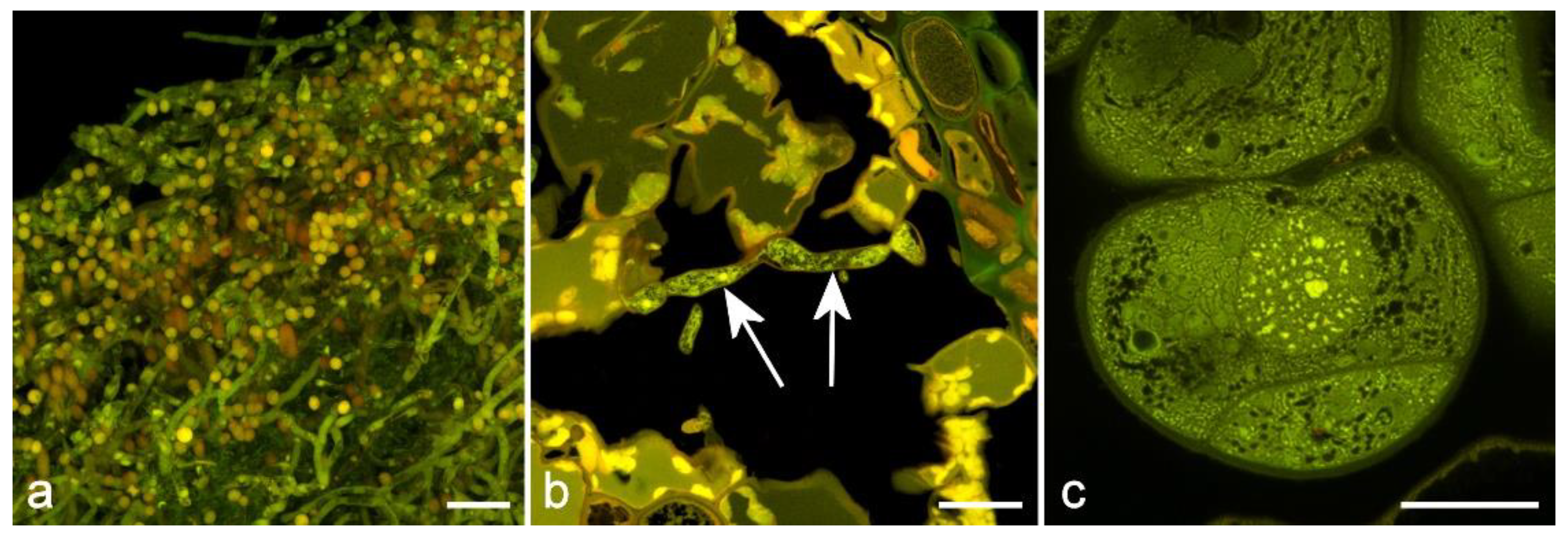
4.4. Sporopollenin
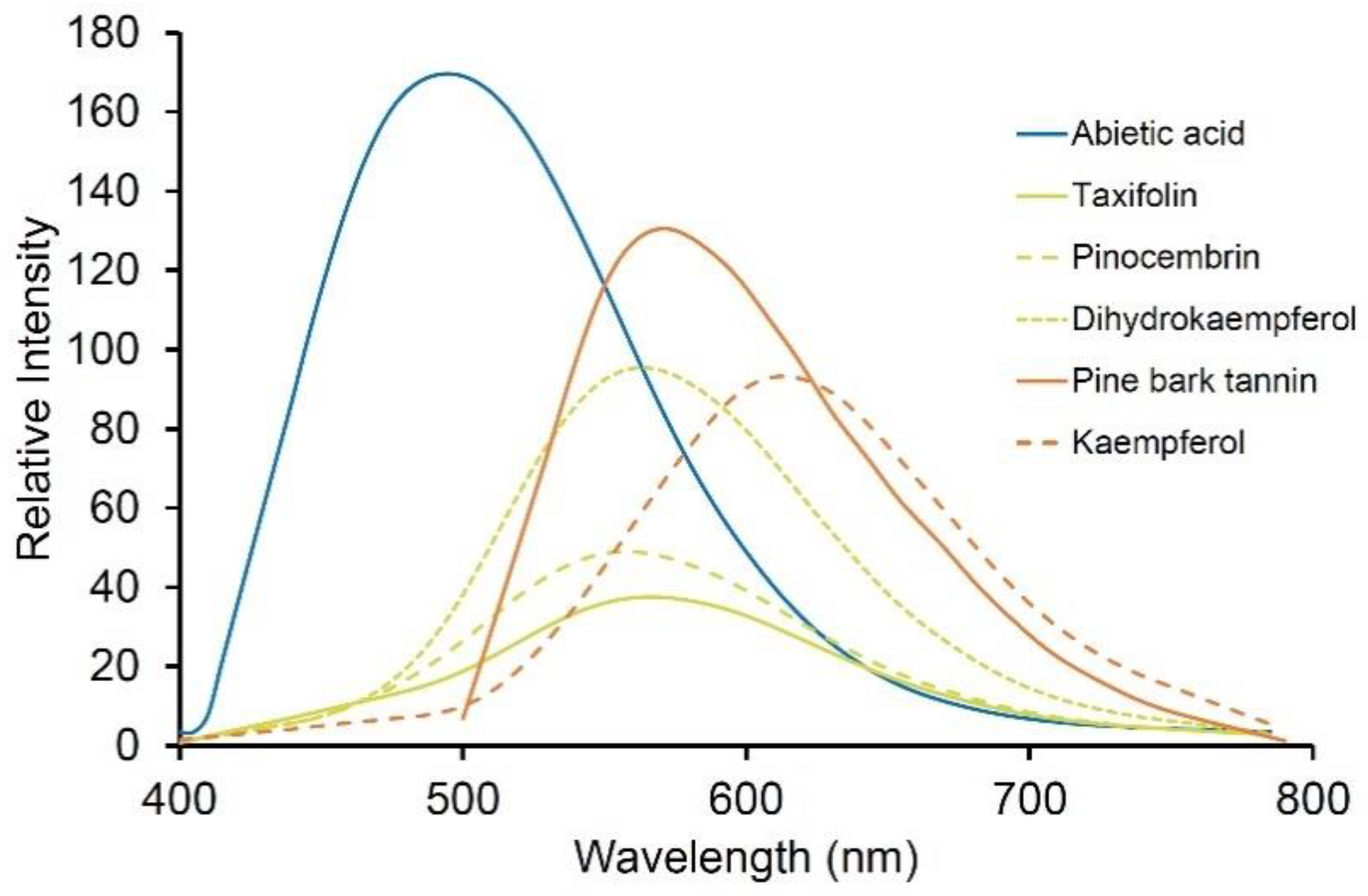
4.5. Flavonoids
5. Other Fluorophores
5.1. Stilbenes
5.2. Tannins
5.3. Terpenes
6. Induced Autofluorescence Using Glutaraldehyde
7. Deep UV Excitation
8. Spectral Imaging and Unmixing
8.1. Sequential Excitation/Emission Imaging
8.2. Unmixing
9. F-Techniques Using Autofluorescence
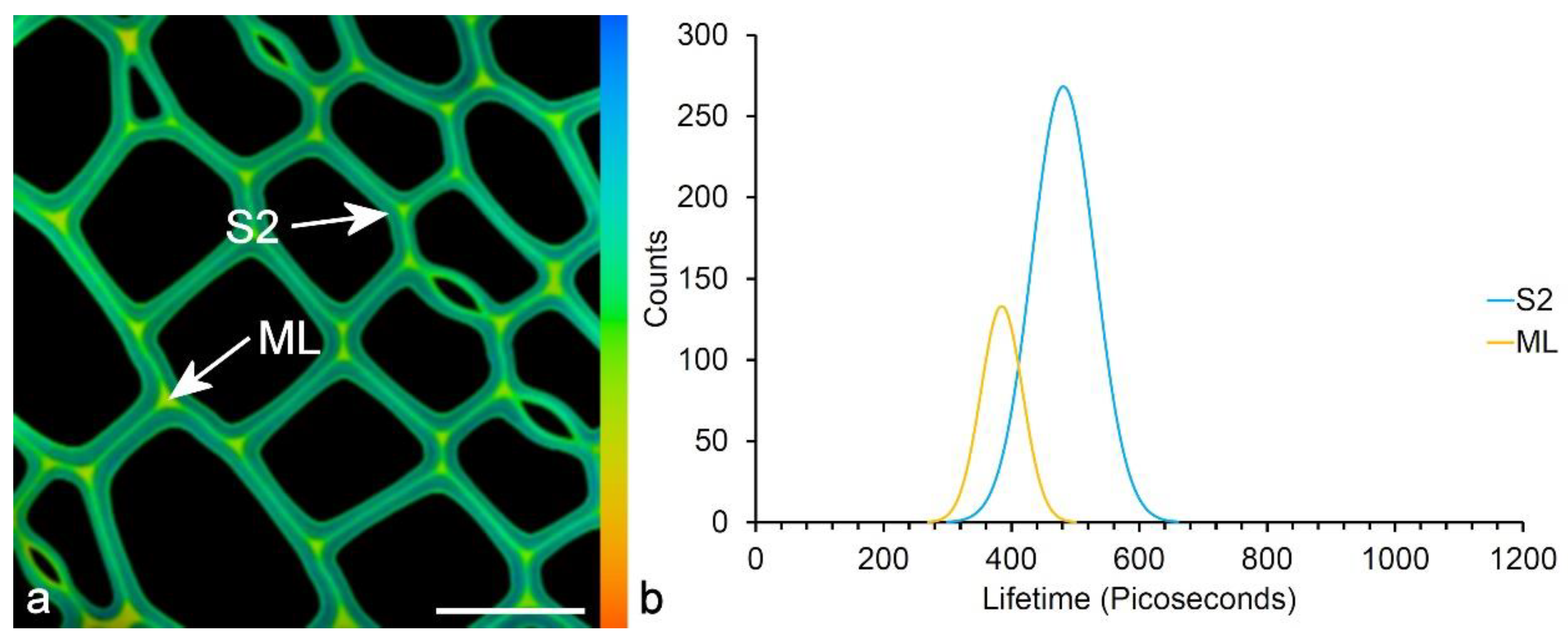
9.1. Fluorescence Lifetime Spectroscopy (FLIM)
9.2. Förster Resonance Energy Transfer (FRET)
9.3. Colocalization
10. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- García-Plazaola, J.I.; Fernández-Marína, B.; Duke, S.O.; Hernández, A.; López-Arbeloa, F.; Becerril, J.M. Autofluorescence: Biological functions and technical applications. Plant Sci. 2015, 236, 136–145. [Google Scholar] [CrossRef]
- Dumur, T.; Duncan, S.; Graumann, K.; Desset, S.; Randall, R.S.; Scheid, O.M.; Bass, H.W.; Prodanov, D.; Tatout, C.; Baroux, C. Probing the 3D architecture of the plant nucleus with microscopy approaches: Challenges and solutions. Nucleus 2019, 10, 181–212. [Google Scholar] [CrossRef] [Green Version]
- Kodama, Y. Time gating of chloroplast autofluorescence allows clearer fluorescence imaging in planta. PLoS ONE 2016, 11, e0152484. [Google Scholar] [CrossRef] [Green Version]
- Müller, S.M.; Galliardt, H.; Schneider, J.; Barisas, B.G.; Seidel, T. Quantification of Förster resonance energy transfer by monitoring sensitized emission in living plant cells. Front. Plant Sci. 2013, 4, 413. [Google Scholar] [CrossRef] [Green Version]
- Buschmann, C.; Langsdorf, G.; Lichtenthaler, H.K. Imaging of the blue, green, and red fluorescence emission of plants: An overview. Photosynthetica 2000, 38, 483–491. [Google Scholar] [CrossRef]
- Cerovic, Z.G.; Samson, G.; Morales, F.; Tremblay, N.; Moya, I. Ultraviolet-induced fluorescence for plant monitoring: Present state and prospects. Agronomie 1999, 19, 543–578. [Google Scholar] [CrossRef] [Green Version]
- Roshchina, V.V. Vital autofluorescence: Application to the study of plant living cells. Int. J. Spectrosc. 2012, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Roshchina, V.V. Autofluorescence of plant secreting cells as a biosensor and bioindicator reaction. J. Fluoresc. 2003, 13, 403–426. [Google Scholar] [CrossRef]
- Roshchina, V.V.; Yashin, V.A.; Kononov, A.V. Autofluorescence of developing plant vegetative microspores studied by confocal microscopy and microspectrofluorimetry. J. Fluoresc. 2004, 14, 745–750. [Google Scholar] [CrossRef]
- Roshchina, V.V.; Kuchin, A.V.; Yashin, V.A. Application of autofluorescence for analysis of medicinal plants. Int. J. Spectrosc. 2017, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Rost, F.W.D. Autofluorescence in plants, fungi, and bacteria. In Fluorescence Microscopy; Rost, F.W.D., Ed.; Cambridge University Press: Cambridge, UK, 1995; Volume 2, pp. 16–36. [Google Scholar]
- Talamond, P.; Verdeil, J.-L.; Conéjéro, G. Secondary metabolite localization by autofluorescence in living plant cells. Molecules 2015, 20, 5024–5037. [Google Scholar] [CrossRef] [Green Version]
- Mottiar, Y.; Gierlinger, N.; Jeremic, D.; Master, E.R.; Mansfield, S.D. Atypical lignification in eastern leatherwood (Dirca palustris). New Phytol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Donaldson, L.A.; Singh, A.; Raymond, L.; Hill, S.; Schmitt, U. Extractive distribution in Pseudotsuga menziesii: Effects on cell wall porosity in sapwood and heartwood. IAWA J. 2019, 40, 721–740. [Google Scholar] [CrossRef]
- Donaldson, L.; Williams, N. Imaging and spectroscopy of natural fluorophores in pine needles. Plants 2018, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Lamb, J.J.; Røkke, G.; Hohmann-Marriott, M.F. Chlorophyll fluorescence emission spectroscopy of oxygenic organisms at 77 K. Photosynthetica 2018, 56, 105–124. [Google Scholar] [CrossRef] [Green Version]
- Mohammed, G.H.; Colombo, R.; Middleton, E.M.; Rascher, U.; Van der Tol, C.; Nedbal, L.; Goulas, Y.; Pérez-Priego, O.; Damm, A.; Meroni, M.; et al. Remote sensing of solar-induced chlorophyll fluorescence (SIF) in vegetation: 50 years of progress. Remote Sens. Environ. 2019, 231, 111177. [Google Scholar] [CrossRef]
- Forster, L.S.; Livingston, R. The absolute quantum yields of the fluorescence of chlorophyll solutions. J. Chem. Phys. 1952, 20, 1315–1320. [Google Scholar] [CrossRef]
- Vácha, F.; Sarafis, V.; Benediktyová, Z.; Bumba, L.; Valenta, J.; Vácha, M.; Sheue, C.-R.; Nedbal, L. Identification of photosystem I and photosystem II enriched regions of thylakoid membrane by optical microimaging of cryo-fluorescence emission spectra and of variable fluorescence. Micron 2007, 38, 170–175. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Hori, H.; Kai, S.; Ishikawa, T.; Ohnishi, A.; Tsumura, N.; Morita, N. Quality control of photosystem II: Reversible and irreversible protein aggregation decides the fate of photosystem II under excessive illumination. Front. Plant Sci. 2013, 4, 433. [Google Scholar] [CrossRef] [Green Version]
- Baker, N.R. Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 2008, 59, 89–113. [Google Scholar] [CrossRef] [Green Version]
- Galbraith, D.W.; Harkins, K.R.; Jefferson, R.A. Flow cytometric characterization of the chlorophyll contents and size distributions of plant protoplasts. Cytometry 1988, 9, 75–83. [Google Scholar] [CrossRef]
- Guanter, L.; Zhang, Y.; Jung, M.; Joiner, J.; Voigt, M.; Berry, J.A.; Frankenberg, C.; Huete, A.R.; Zarco-Tejada, P.; Lee, J.-E.; et al. Global and time-resolved monitoring of crop photosynthesis with chlorophyll fluorescence. Proc. Natl. Acad. Sci. USA 2014, E1327–E1333. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef]
- Murchie, E.H.; Lawson, T. Chlorophyll fluorescence analysis: A guide to good practice and understanding some new applications. J. Exp. Bot. 2013, 64, 3983–3998. [Google Scholar] [CrossRef] [Green Version]
- Drabent, R.; Pliszka, B.; Olzewska, T. Fluorescence properties of plant anthocyanin pigments. I Fluorescence of anthocyanins in Brassica oleracea L. extracts. J. Photochem. Photobiol. 1999, 50, 53–58. [Google Scholar] [CrossRef]
- Chanoca, A.; Burkel, B.; Kovinich, N.; Grotewold, E.; Eliceiri, K.W.; Otegui, M.S. Using fluorescence lifetime microscopy to study the subcellular localization of anthocyanins. Plant J. 2016, 88, 895–903. [Google Scholar] [CrossRef]
- Zaghdoudi, K.; Ngomo, O.; Vanderesse, R.; Arnoux, P.; Myrzakhmetov, B.; Frochot, C.; Guiavarc’h, Y. Extraction, identification and photo-physical characterization of persimmon (Diospyros kaki L.) carotenoids. Foods 2017, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Merzlyak, M.N.; Melø, T.B.; Naqvi, K.R. Effect of anthocyanins, carotenoids, and flavonols on chlorophyll fluorescence excitation spectra in apple fruit: Signature analysis, assessment, modeling, and relevance to photoprotection. J. Exp. Bot. 2008, 59, 349–359. [Google Scholar] [CrossRef]
- Kleinegris, D.M.M.; Van Es, M.A.; Janssen, M.; Brandenburg, W.A.; Wijffels, R.H. Carotenoid fluorescence in Dunaliella salina. J. Appl. Phycol. 2010, 22, 645–649. [Google Scholar] [CrossRef] [Green Version]
- Castellan, A.; Ruggiero, R.; Frollini, E.; Ramos, L.A.; Chirat, C. Studies on fluorescence of cellulosics. Holzforschung 2007, 61, 504–508. [Google Scholar] [CrossRef]
- Donaldson, L.A. Softwood and hardwood lignin fluorescence spectra of wood cell walls in different mounting media. IAWA J. 2013, 34, 3–19. [Google Scholar] [CrossRef]
- Hafren, J. Excitation wavelength-specific changes in lignocellulosic autofluorescence. J. Wood Sci. 2007, 53, 358–360. [Google Scholar] [CrossRef]
- Donaldson, L.A.; Knox, J.P. Localisation of cell wall polysaccharides in normal and compression wood of radiata pine—Relationships with lignification and microfibril orientation. Plant Physiol. 2012, 158, 642–653. [Google Scholar] [CrossRef] [Green Version]
- Donaldson, L.A.; Radotić, K.; Kalauzi, A.; Djikanović, D.; Jeremić, M. Quantification of compression wood severity in tracheids of Pinus radiata D. Don using confocal fluorescence imaging and spectral deconvolution. J. Struct. Biol. 2010, 169, 106–115. [Google Scholar] [CrossRef]
- Radotić, K.; Kalauzi, A.; Djikanović, D.; Jeremić, M.; Leblanc, R.M.; Cerović, Z.G. Component analysis of the fluorescence spectra of a lignin model compound. J. Photochem. Photobiol. B Biol. 2006, 83, 1–10. [Google Scholar] [CrossRef]
- Albinsson, B.; Li, S.; Lundquist, K.; Stomberg, R. The origin of lignin fluorescence. J. Mol. Struct. 1999, 508, 19–27. [Google Scholar] [CrossRef]
- Castellan, A.; Davidson, R.S. Steady-state and dynamic fluorescence emission from Abies wood. J. Photochem. Photobiol. A Chem. 1994, 78, 275–279. [Google Scholar] [CrossRef]
- Lundquist, K.; Josefsson, B.; Nyquist, G. Analysis of lignin products by fluorescence spectroscopy. Holzforshung 1978, 32, 27–32. [Google Scholar] [CrossRef]
- Xue, Y.; Qiu, X.; Wu, Y.; Qian, Y.; Zhou, M.; Deng, Y.; Li, Y. Aggregation-induced emission: The origin of lignin fluorescence. Polym. Chem. 2016, 7, 3502–3508. [Google Scholar] [CrossRef]
- Donaldson, L.A.; Kroese, H.W.; Hill, S.J.; Franich, R.A. Detection of wood cell wall porosity using small carbohydrate molecules and confocal fluorescence microscopy. J. Microsc. 2015, 259, 228–236. [Google Scholar] [CrossRef]
- Kim, J.S.; Gao, J.; Terziev, N.; Cuccui, I.; Daniel, G. Chemical and ultrastructural changes of ash wood thermally modified using the thermo-vacuum process: I. Histo/cytochemical studies on changes in the structure and lignin chemistry. Holzforschung 2015, 69, 603–613. [Google Scholar] [CrossRef]
- Thygesen, L.G.; Barsberg, S.; Venås, T.M. The fluorescence characteristics of furfurylated wood studied by fluorescence spectroscopy and confocal laser scanning microscopy. Wood Sci. Technol. 2010, 44, 51–65. [Google Scholar] [CrossRef]
- Irbe, I.; Noldt, G.; Koch, G.; Andersone, I.; Andersons, B. Application of scanning UV microspectrophotometry for the topochemical detection of lignin within individual cell walls of brown-rotted Scots pine (Pinus sylvestris L.) sapwood. Holzforschung 2006, 60, 601–607. [Google Scholar] [CrossRef]
- Cox, G.; Moreno, N. Second-harmonic imaging of plant polysaccharides. J. Biomed. Opt. 2005, 10, 024013. [Google Scholar] [CrossRef]
- Kushida, S.; Braam, D.; Pan, C.; Dao, T.D.; Tabata, K.; Sugiyasu, K.; Takeuchi, M.; Ishii, S.; Nagao, T.; Lorke, A.; et al. Whispering gallery resonance from self-assembled microspheres of highly fluorescent isolated conjugated polymers. Macromolecules 2015, 48, 3928–3933. [Google Scholar] [CrossRef]
- Wagner, A.; Donaldson, L.A.; Kim, H.; Phillips, L.; Flint, H.; Steward, D.; Torr, K.; Koch, G.; Schmitt, U.; Ralph, J. Silencing of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol. 2008, 149, 370–383. [Google Scholar] [CrossRef] [Green Version]
- Donaldson, L.A.; Grace, J.C.; Downes, G. Within tree variation in anatomical properties of compression wood in radiata pine. IAWA J. 2004, 25, 253–271. [Google Scholar] [CrossRef] [Green Version]
- Donaldson, L.A.; Nanayakkara, B.; Radotić, K.; Djikanovic-Golubovic, D.; Mitrovic, A.; Bogdanovic, J.; Simonovic, J.; Kalauzi, A. Xylem parenchyma cell walls lack a gravitropic response in conifer compression wood. Planta 2015, 242, 1413–1424. [Google Scholar] [CrossRef]
- Donaldson, L.A.; Cairns, M.; Hill, S.J. Comparison of micropore distribution in cell walls of softwood and hardwood xylem. Plant Physiol. 2018, 178, 1142–1153. [Google Scholar] [CrossRef] [Green Version]
- Auxenfans, T.; Terryn, C.; Paës, G. Seeing biomass recalcitrance through fluorescence. Sci. Rep. 2017, 7, 8838. [Google Scholar] [CrossRef] [Green Version]
- Chabbert, B.; Terryn, C.; Herbaut, M.; Vaidya, A.; Habrant, A.; Paës, G.; Donaldson, L. Fluorescence techniques can reveal cell wall organization and predict saccharification in pretreated wood biomass. Ind. Crops. Prod. 2018, 123, 84–92. [Google Scholar] [CrossRef]
- Chimenez, T.A.; Gehlen, M.H.; Marabezi, K.; Curvelo, A.A.S. Characterization of sugarcane bagasse by autofluorescence microscopy. Cellulose 2014, 21, 653–664. [Google Scholar] [CrossRef]
- Coletta, V.C.; Rezende, C.A.; Da Conceição, F.A.; Polikarpov, I.; Guimarães, F.E.G. Mapping the lignin distribution in pretreated sugarcane bagasse by confocal and fluorescence lifetime imaging microscopy. Biotechnol. Biofuels 2013, 6, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donaldson, L.A.; Newman, R.H.; Vaidya, A. Nanoscale interactions of polyethylene glycol with thermo-mechanically pre-treated Pinus radiata biofuel substrate. Biotechnol. Bioeng. 2014, 111, 719–725. [Google Scholar] [CrossRef]
- Donaldson, L.A.; Vaidya, A. Visualising recalcitrance by colocalization of cellulase, lignin and cellulose in pretreated pine biomass using fluorescence microscopy. Sci. Rep. 2017, 7, 44386. [Google Scholar] [CrossRef] [Green Version]
- Terryn, C.; Paës, G.; Spriet, C. FRET-SLiM on native autofluorescence: A fast and reliable method to study interactions between fluorescent probes and lignin in plant cell wall. Plant Methods 2018, 14, 74. [Google Scholar] [CrossRef]
- Wightman, R.; Busse-Wicher, M.; Dupree, P. Correlative FLIM-confocal-Raman mapping applied to plant lignin composition and autofluorescence. Micron 2019, 126, 102733. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhao, S.; Wei, H.; Tucker, M.P.; Himmel, M.E.; Mosier, N.S.; Meilan, R.; Ding, S.-Y. In situ micro-spectroscopic investigation of lignin in poplar cell walls pretreated by maleic acid. Biotechnol. Biofuels 2015, 8, 126. [Google Scholar] [CrossRef] [Green Version]
- Harris, P.J.; Hartley, R.D. Detection of bound ferulic acid in cell walls of the Gramineae by ultraviolet fluorescence microscopy. Nature 1976, 259, 508–510. [Google Scholar] [CrossRef]
- Cappelli, A.; Paolino, M.; Reale, A.; Razzano, V.; Grisci, G.; Giuliani, G.; Donati, A.; Bonechi, C.; Lamponi, S.; Mendichi, R.; et al. Hyaluronan-based graft copolymers bearing aggregation-induced emission fluorogens. RSC Adv. 2018, 8, 5864–5881. [Google Scholar] [CrossRef] [Green Version]
- Carnachan, S.M.; Harris, P.J. Ferulic acid is bound to the primary cell walls of all gymnosperm families. Biochem. Syst. Ecol. 2000, 28, 865–879. [Google Scholar] [CrossRef]
- Berg, R.H. Evaluation of spectral imaging for plant cell analysis. J. Microsc. 2004, 214, 174–181. [Google Scholar] [CrossRef]
- Buda, G.J.; Isaacson, T.; Matas, A.J.; Paolillo, D.J.; Rose, J.K.C. Three-dimensional imaging of plant cuticle architecture using confocal scanning laser microscopy. Plant J. 2009, 60, 378–385. [Google Scholar] [CrossRef]
- Fernández, S.; Osorio, S.; Heredia, A. Monitoring and visualising plant cuticles by confocal laser scanning microscopy. Plant Physiol. Biochem. 1999, 37, 789–794. [Google Scholar] [CrossRef]
- Philippe, G.; Sørensen, I.; Jiao, C.; Sun, X.; Fei, Z.; Domozych, D.S.; Rose, J.K.C. Cutin and suberin: Assembly and origins of specialized lipidic cell wall scaffolds. Curr. Opin. Plant Biol. 2020, 55, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Correia, V.G.; Bento, A.; Pais, J.; Rodrigues, R.; Haliński, Ł.P.; Frydrych, M.; Greenhalgh, A.; Stepnowski, P.; Vollrath, F.; King, A.W.T.; et al. The molecular structure and multifunctionality of the cryptic plant polymer suberin. Mater. Today Bio 2020, 5, 100039. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-S.; Phyo, P.; Jacobowitz, J.; Hong, M.; Weng, J.-K. The molecular structure of plant sporopollenin. Nat. Plants 2019, 5, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Willemse, M.T.M. Changes in the autofluorescence of the pollen wall during microsporogenesis and chemical treatments. Acta Bot. Neerl. 1972, 21, 1–16. [Google Scholar] [CrossRef]
- Roshchina, V.V.; Melnikova, E.V.; Iashin, V.A.; Kamaukhov, V.N. Autofluorescence of intact Equisetum arvense L. spores during their development. Biofizika 2002, 47, 318–324. (In Russian) [Google Scholar]
- Castro, A.J.; Rejón, J.D.; Fendri, M.; Jiménez-Quesada, M.J.; Zafra, A.; Jiménez-López, J.C.; Rodríguez-García, M.I.; Alché, J.D. Taxonomical discrimination of pollen grains by using confocal laser scanning microscopy (CLSM) imaging of autofluorescence. In Microscopy: Science, Technology, Applications and Education; Méndez-Vilas, A., Díaz, J., Eds.; Formatex: Badajoz, Spain, 2010; pp. 607–613. [Google Scholar]
- Urbanczyk, J.; Casado, M.A.F.; Díaz, T.E.; Heras, P.; Infante, M.; Borrego, A.G. Reprint of “Spectral fluorescence variation of pollen and spores from recent peat-forming plants”. Int. J. Coal Geol. 2015, 139, 206–216. [Google Scholar] [CrossRef]
- Acuña, A.U.; Amat-Guerri, F.; Morcillo, P.; Liras, M.; Rodriguez, B. Structure and formation of the fluorescent compound of Lignum nephriticum. Org. Lett. 2009, 11, 3020–3023. [Google Scholar] [CrossRef]
- Schnitzler, J.-P.; Jungblut, T.P.; Heller, W.; Köfferlein, M.; Hutzler, P.; Heinzmann, U.; Schmelzer, E.; Ernst, D.; Langebartels, C.; Sandermann, H. Tissue localization of UV-B screening pigments and of chalcone synthase mRNA in needles of Scots pine seedlings. New Phytol. 1996, 132, 247–258. [Google Scholar] [CrossRef]
- Weston, L.A.; Mathesius, U. Flavonoids: Their structure, biosynthesis and role in the rhizosphere, including allelopathy. J. Chem. Ecol. 2013, 39, 283–297. [Google Scholar] [CrossRef]
- Hutzler, P.; Fischbach, R.; Heller, W.; Jungblut, T.P.; Reuber, S.; Schmitz, R.; Veit, M.; Weissenböck, G.; Schnitzler, J.-P. Tissue localization of phenolic compounds in plants by confocal laser scanning microscopy. J. Exp. Bot. 1998, 49, 953–965. [Google Scholar] [CrossRef]
- Sudo, E.; Teranishi, M.; Hidema, J.; Taniuchi, T. Visualization of flavonol distribution in the abaxial epidermis of onion scales via detection of its autofluorescence in the absence of chemical processes. Biosci. Biotechnol. Biochem. 2009, 73, 2107–2109. [Google Scholar] [CrossRef]
- Monago-Maraña, O.; Durán-Merás, I.; Galeano-Díaz, T.; De la Peña, A.M. Fluorescence properties of flavonoid compounds. Quantification in paprika samples using spectrofluorimetry coupled to second-order chemometric tools. Food Chem. 2016, 196, 1058–1065. [Google Scholar] [CrossRef]
- Buer, C.S.; Muday, G.K. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 2004, 16, 1191–1205. [Google Scholar] [CrossRef] [Green Version]
- Peer, W.A.; Brown, D.E.; Tague, B.W.; Muday, G.K.; Taiz, L.; Murphy, A.S. Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol. 2001, 126, 536–548. [Google Scholar] [CrossRef] [Green Version]
- Bellow, S.; Latouche, G.; Brown, S.C.; Poutaraud, A.; Cerovic, Z.G. In vivo localization at the cellular level of stilbene fluorescence induced by Plasmopara viticola in grapevine leaves. J. Exp. Bot. 2012, 63, 3697–3708. [Google Scholar] [CrossRef] [Green Version]
- Dubouzet, J.G.; Donaldson, L.; Black, M.A.; McNoe, L.; Liu, V.; Lloyd-Jones, G. Heterologous hybridization to a Pinus microarray: Profiling of gene expression in Pinus radiata saplings exposed to ethephon. NZ J. For. Sci. 2014, 44, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Mishra, G.; Collings, D.A.; Altaner, C.M. Cell organelles and fluorescence of parenchyma cells in Eucalyptus bosistoana sapwood and heartwood investigated by microscopy. NZ J. For. Sci. 2018, 48, 13. [Google Scholar] [CrossRef]
- Brillouet, J.M.; Romieu, C.; Schoefs, B.; Solymosi, K.; Cheynier, V.; Fulcrand, H.; Verdeil, J.-L.; Conéjéro, G. The tannosome is an organelle forming condensed tannins in the chlorophyllous organs of tracheophyta. Ann. Bot. 2013, 112, 1003–1014. [Google Scholar] [CrossRef] [Green Version]
- Engelbrecht, L.; Frazenburg, M.; Laurie, A. The South African persimmon under the microscope. S. Afr. Fruit J. 2019, 18, 53–55. [Google Scholar]
- Binsch, G.; Heilbronner, E.; Jankow, R.; Schmidt, D. On the fluorescence anomaly of azulene. Chem. Phys. Lett. 1967, 1, 135–138. [Google Scholar] [CrossRef]
- Collins, J.S.; Goldsmith, T.H. Spectral properties of fluorescence induced by glutaraldehyde fixation. J. Histochem. Cytochem. 1981, 29, 411–414. [Google Scholar] [CrossRef]
- Prior, D.A.M.; Oparka, K.J.; Roberts, I.M. En-bloc optical sectioning of resin-embedded specimens using a confocal laser scanning microscope. J. Microsc. 1999, 193, 20–27. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.H.; Warren, H.L. Natural autofluorescence in fungi and its correlation with viability. Mycologia 1984, 76, 1049–1058. [Google Scholar] [CrossRef]
- Culley, S.; Kalina, L.; Tosheva, K.L.; Pereira, P.M.; Henriques, R. SRRF: Universal live-cell super-resolution microscopy. Int. J. Biochem. Cell Biol. 2018, 101, 74–79. [Google Scholar] [CrossRef]
- Jamme, F.; Kascakova, S.; Villette, S.; Allouche, F.; Pallu, S.; Rouam, V.; Réfrégiers, M. DUV Autofluorescence microscopy for cell biology and tissue histology. Biol. Cell. 2013, 105, 277–288. [Google Scholar] [CrossRef]
- Devaux, M.-F.; Jamme, F.; André, W.; Bouchet, B.; Alvarado, C.; Durand, S.; Robert, P.; Saulnier, L.; Bonnin, E.; Guillon, F. Synchrotron time-lapse imaging of lignocellulosic biomass hydrolysis: Tracking enzyme localization by protein autofluorescence and biochemical modification of cell walls by microfluidic infrared microspectroscopy. Front. Plant Sci. 2018, 9, 200. [Google Scholar] [CrossRef] [Green Version]
- Neher, R.A.; Mitkovski, M.; Kirchhoff, F.; Neher, E.; Theis, F.J.; Zeug, A. Blind source separation techniques for the decomposition of multiply labeled fluorescence images. Biophys. J. 2009, 96, 3791–3800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, R.H.; Beachy, R.N. Fluorescent protein applications in plants. Method. Cell Biol. 2008, 85, 153–177. [Google Scholar] [CrossRef]
- Wahl, M. Time-Correlated Single-Photon Counting; Picoquant Technical Note: Berlin, Germany, 2014; pp. 1–14. [Google Scholar]
- Heskes, A.M.; Lincoln, C.N.; Goodger, J.Q.D.; Woodrow, I.E.; Smith, T.A. Multiphoton fluorescence lifetime imaging shows spatial segregation of secondary metabolites in Eucalyptus secretory cavities. J. Microsc. 2012, 247, 33–42. [Google Scholar] [CrossRef]
- Brody, S.S. Fluorescence lifetime, yield, energy transfer and spectrum in photosynthesis, 1950-1960. In Discoveries in Photosynthesis; Govindjee, Beatty, J.T., Gest, H., Allen, J.F., Eds.; Advances in Photosynthesis and Respiration; Springer: Dordrecht, The Netherlands, 2005; Volume 20, pp. 165–170. [Google Scholar] [CrossRef]
- Holub, O.; Seufferheld, M.J.; Gohlke, C.; Govindjee; Clegg, R.M. Fluorescence lifetime imaging (FLI) in real-time—A new technique in photosynthesis research. Photosynthetica 2000, 38, 581–599. [Google Scholar] [CrossRef]
- Amarnath, K.; Zaks, J.; Park, S.D.; Niyogi, K.K.; Fleming, G.R. Fluorescence lifetime snapshots reveal two rapidly reversible mechanisms of photoprotection in live cells of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2012, 109, 8405–8410. [Google Scholar] [CrossRef] [Green Version]
- Holub, O.; Seufferheld, M.J.; Gohlke, C.; Govindjee; Heiss, G.J.; Clegg, R.M. Fluorescence lifetime imaging microscopy of Chlamydomonas reinhardtii: Non-photochemical quenching mutants and the effect of photosynthetic inhibitors on the slow chlorophyll fluorescence transient. J. Microsc. 2007, 226, 90–120. [Google Scholar] [CrossRef]
- Moise, N.; Moya, I. Correlation between lifetime heterogeneity and kinetics heterogeneity during chlorophyll fluorescence induction in leaves: 2. Multi-frequency phase and modulation analysis evidences a loosely connected PSII pigment-protein complex. Biochim. Biophys. Acta 2004, 1657, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Lei, R.; Jiang, H.; Hu, F.; Yan, J.; Zhu, S. Chlorophyll fluorescence lifetime imaging provides new insight into the chlorosis induced by plant virus infection. Plant Cell Rep. 2017, 36, 327–341. [Google Scholar] [CrossRef]
- Bilger, W. Desiccation-induced quenching of chlorophyll fluorescence in cryptogams. In Non-Photochemical Quenching and Energy Dissipation in Plants, Algae and Cyanobacteria; Demmig-Adams, B., Garab, G., Adams, W., III, Govindjee, Eds.; Advances in Photosynthesis and Respiration (Including Bioenergy and Related Processes); Springer: Dordrecht, The Netherlands, 2014; Volume 40, pp. 409–420. [Google Scholar] [CrossRef]
- Zeng, Y.; Wu, Y.; Li, D.; Zheng, W.; Wang, W.-X.; Qu, J.Y. Two-photon excitation chlorophyll fluorescence lifetime imaging: A rapid and non-invasive method for in vivo assessment of cadmium toxicity in a marine diatom Thalassiosira weissflogii. Planta 2012, 236, 1653–1663. [Google Scholar] [CrossRef]
- Donaldson, L.A.; Radotic, K. Fluorescence lifetime imaging of lignin autofluorescence in normal and compression wood. J. Microsc. 2013, 251, 178–187. [Google Scholar] [CrossRef]
- Periasamy, A.; Day, R.N. Molecular Imaging, FRET Microscopy and Spectroscopy; Oxford University Press: New York, NY, USA, 2005; p. 312. [Google Scholar] [CrossRef]
- Weidtkamp-Peters, S.; Stahl, Y. The use of FRET/FLIM to study proteins interacting with plant receptor kinases. In Plant Receptor Kinases; Aalen, R., Ed.; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1621, pp. 163–175. [Google Scholar] [CrossRef]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donaldson, L. Autofluorescence in Plants. Molecules 2020, 25, 2393. https://doi.org/10.3390/molecules25102393
Donaldson L. Autofluorescence in Plants. Molecules. 2020; 25(10):2393. https://doi.org/10.3390/molecules25102393
Chicago/Turabian StyleDonaldson, Lloyd. 2020. "Autofluorescence in Plants" Molecules 25, no. 10: 2393. https://doi.org/10.3390/molecules25102393





