Abstract
Cultivated tomato (Solanum lycopersicum L.) is an important source of antioxidants, such as ascorbic acid, carotenoids and phenolic compounds. Epidemiological results confirm that these antioxidant molecules are associated with a reduced risk of cancer, inflammation and cardiovascular diseases. Recently, one introgression line population deriving from Solanum pennellii has been exploited to identify favorable alleles that can improve fruit quality traits in commercial varieties, including antioxidant content. The aim of this work was to evaluate growth, final yield and content of nutraceutical compounds at the ripe red fruit stage in one subline coded R182, which carries only a small region (448 Kbp) of wild genome in the cultivated genetic background (M82). Analyses carried out on R182 and on the parental line M82 demonstrated that the subline showed better performances in terms of the yield and fruit qualitative traits most considered for tomato processing. Indeed, higher yield (+28.96%), content of soluble sugars (+34.64%) and titratable acidity (+78.94%) were demonstrated for R182 compared to M82. In addition, for the nutritional traits analyzed, an increase in the content of phenols (+69.96%), ascorbic acid (+48.55%), carotenoids (+29.66%), lycopene (+31.22%) and β-carotene (+31.67%) was observed. Therefore, it is possible to assert that the subline R182 can be used as a novel germplasm resource for tomato breeding.
Keywords:
wild species; Solanum lycopersicum; fruit quality; ascorbic acid; carotenoids; phenols; total yield 1. Introduction
Tomato (Solanum lycopersicum L.) belongs to the Solanaceae family, which is one of the most economically important families and has a worldwide distribution, being present in all continents except Antarctica [1]. Tomato plays an important role in human health. It is one of the main components of the Mediterranean diet and its consumption has been associated with a reduced risk of cancer, inflammation and cardiovascular diseases [2]. These effects are attributed to the presence of antioxidant molecules, such as ascorbic acid (vitamin C), carotenoids and phenols, in the fruit [3,4].
One of the most important antioxidants is ascorbic acid because humans are unable to synthesize it on their own and have to incorporate it with the diet. Compared to wild species, cultivated tomatoes contain moderate amounts of vitamin C, as a consequence of several cycles of domestication and breeding activities [5].
In the last few years, there has been a growing interest in the introgression lines (IL) containing alleles from wild species that can be used to improve tomato productivity and nutritional quality. Previously, seventy-six introgression lines have been obtained crossing Solanum lycopersicum cv. M82 with Solanum pennellii, which are characterized by the presence in homozygosity of a portion of the genome of the wild species introduced into that of the cultivated species [6]. The S. pennellii ILs population has been used to identify more than 3000 QTLs (quantitative trait loci) affecting abiotic and biotic stress tolerance, productivity and fruit qualitative traits [7]. The S. pennellii ILs have been further broken into sublines carrying smaller introgressed regions compared to those carried by the original ILs that have been used to identify candidate genes and alleles affecting qualitative traits [8]. In particular, one subline coded R182 of the introgression line IL7-3 was demonstrated to have higher fruit qualitative traits compared to the cultivated line, M82 [6]. In the present study, analyses on the subline R182 were performed on additional traits to investigate if this line has the potential to be used as a novel germplasm resource for tomato breeding.
2. Materials and Methods
2.1. Plant Material
Plant material consisted of the cultivated genotype M82 (accession LA3475) and IL 7-3 subline (coded R182). The Tomato Genetics Resource Center (TGRC) kindly provided the seeds (http://tgrc.ucdavis.edu/; accessed on 17 May 2021) of M82 and Dr. Dani Zamir (Hebrew University, Israel) those of the subline R182. The genotypes M82 and R182 were grown in open field in the Campania Region in Italy during the year 2020 in a completely randomized design with three replicates per genotype and 10 plants per each replicate and following the standard cultural practices of the area. Tomato fruits were collected at the mature red stage. Subsequently, the seeds were removed, and fruits were ground in liquid nitrogen by a blender (FRI150, Fimar) to a fine powder, and stored at −80 °C until further analyses.
2.2. Phenotypic Analyses
At the end of the cultivation cycle, all plants of each genotype were harvested and plant height, fresh weight of biomass and yield per plant (YP) were recorded. YP was evaluated using the total weight of all collected fruits and dividing it for the number of total plants of the three replicates.
2.3. Qualitative Analyses
For each genotype and biological replicate, fruits were collected at the mature red stage to evaluate soluble solids content, titratable acidity and firmness. The soluble solids content was measured as °Brix in the homogenized juice from ripe fruit by a digital refractometer (Hanna). Titratable acidity or total acidity was measured by titration to a pH end point with a standard base (NaOH 0.1 N). Firmness was measured by using a penetrometer PCE_PTR200 with a surface needle of 8 mm.
The evaluation of lipophilic fraction was carried out according to the method reported by Wellburn [9] and Zouari et al. [10] as modified by Rigano et al. [11]. To extract the carotenoids, 500 mg of frozen sample powder was extracted with 8 mL of acetone/hexane (40/60, v/v). The mixture was stirred at 300 rpm for 20 min at room temperature and then centrifuged at 5000 rpm for 5 min at 4 °C. This procedure was repeated three consecutive times and each time the supernatant was collected in a falcon tube and ultimately stored at −20 °C until further analysis. All biological replicates of the samples were analyzed in triplicate. The absorbance was measured at 470 nm for carotenoids and at 453, 505, 645 and 663 nm for lycopene and β-carotene, with a NanoPhotometerTM (Implen, Munich, Germany), using acetone/hexane as reference. The total carotenoids, lycopene and β-carotene were determined by the equation reported by Wellburn [9]. The concentration was expressed as mg/100 g of fresh weight (FW).
Reduced ascorbic acid (reduced AsA), and total ascorbic acid (AsA + DHA, total AsA) determination was carried out by a colorimetric method [12] with modifications reported by Rigano et al. [13]. Briefly, 500 mg of frozen fruit powder were extracted with 300 µL of ice cold 6% TCA (trichloacetic acid). The mixture was vortexed for 10 s, incubated for 15 min on ice and centrifuged at 14,000 rpm for 20 min at 4 °C.
For the reduced ascorbic acid determination, twenty microliters of supernatant for each assay were transferred in a 1.5 mL Eppendorf tube with 20 µL of 0.4 M phosphate buffer (pH 7.4) and 10 µL of double distilled (dd) H2O. Then, 80 µL of color reagent solution were added. This solution was prepared by mixing solution A [31% H3PO4, 4.6% (w/v) TCA and 0.6% (w/v) FeCl3] with solution B [4% 2,2′- dipyridil (w/v; made up in 70% ethanol)].
For the total ascorbic acid determination, twenty microliters of sample, 20 µL of 5 mM dithiotreitol in 0.4 M phosphate buffer (pH 7.4), were added to reduce the oxidized ascorbate, and the mixture was incubated for 20 min at 37 °C. Ten microliters of N-ethyl maleimide (NEM; 0.5% (w/v) in water) were added, mixed, and left for 1 min at room temperature. Eighty microliters of color reagent were added as previously described for reduced AsA.
Both the final mixtures for the tests of reduced and total AsA were incubated at 37 °C for 40 min and measured at 525 nm by a NanoPhotometerTM (Implen, Munich, Germany) using 6% TCA as reference. Three separated biological replicates for each sample and three technical assays for each biological repetition were measured. The concentration was expressed as mg/100 g of fresh weight (FW).
The extraction of hydrophilic fraction was carried out according to the Folin–Ciocalteu’s test with minor changes [13]. Briefly, frozen ground tissue (250 mg) was weighed, and the phenols were extracted with 1 mL of 60% methanol. The volume of the extracts was increased to 5 mL adding 60% methanol. The samples were centrifuged at 14,000 rpm for 15 min at 4 °C, and the supernatant was stored at −20 °C until further analysis. Three separate biological replicates were measured for each sample and three technical analyses were performed for each biological replicate. Later, 62.5 μL of supernatant, 62.5 μL of Folin–Ciocalteu reagent (Sigma, St. Louis, MO, USA) and 250 μL of deionized water were mixed and incubated for 6 min in the dark; 625 μL of 7.5% sodium carbonate and 500 μL of deionized water were subsequently added and incubated for 90 min at room temperature in the dark. The absorbance was measured at 760 nm by a NanoPhotometerTM (Implen, Munich, Germany). Total phenol concentration was expressed as mg GAE/100 g of fresh weight (FW). Results were expressed as the mean value ± SD (n = 9).
2.4. Statistical Analysis
In the manuscript, the comparison between the genotypes was carried out by a Student’s t-test. Differences at p < 0.05 were considered to be significant. The data were also subjected to analysis of variance using a one-way ANOVA. Differences at p < 0.05 were considered to be significant. ANOVA was performed using a Statistical Package for Social Sciences (SPSS) Package 6, version 23.0. The percentage of variations of each parameter in subline R182 compared to M82 was calculated by using the following formula:
Increase or Decrease (%) = [(value of R182 − value of M82) × value of M82−1] × 100
3. Results and Discussion
Calafiore et al. [8] studied one Solanum pennellii introgression line (IL7-3) and demonstrated that this line had an increased content of ascorbic acid and carotenoids in ripe fruit compared to the cultivated line M82. Additionally, a subline (coded R182) of IL7-3 was identified, which showed a higher antioxidants content compared to the cultivated line M82 [14]. In the present work, in order to further analyze the potential of the R182 subline, we evaluated titratable acidity, the content of soluble solids and of the main nutraceutical compounds, such as ascorbic acid, carotenoids and phenols, in the subline and in the cultivated line M82. Moreover, a phenotypic analysis was performed to evaluate the productivity of each genotype.
The cultivated line M82 and subline R182 were analyzed by measuring height, fresh weight of biomass and yield per plant (Table 1). Moreover, fruit quality was evaluated by measuring nine different traits (Table 2 and Figure 1).

Table 1.
Biometric parameters of M82 and R182 grown in open field. The values are means ± SD (n = 9). The differences between the two genotypes (M82 vs. R182) were evaluated by the Student’s t-test (* p < 0.05).

Table 2.
Fruit quality traits of M82 and R182 grown in open field. The values are means ± SD (n = 9). The differences between the two genotypes (M82 vs. R182) were evaluated by the Student’s t-test (*** p < 0.001).
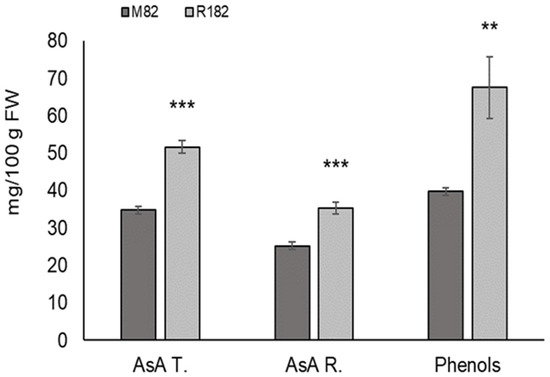
Figure 1.
Content of total ascorbic acid (AsA T.), reduced ascorbic acid (AsA R.) and total phenols in two tomato genotypes analyzed. All are expressed as mg/100 g FW. Values are means ± SD (n = 9). Asterisks indicate statistically differences of R182 compared to M82 by Student’s t-test (** p < 0.01; *** p < 0.001).
Yield per plant and height were higher in R182 compared to M82, increasing respectively by 28.96% and 11.84%. On the other hand, no differences were evidenced in the fresh weight of the biomass between the two genotypes (Table 1). These results are interesting considering that previous analyses [8] did not evidence a higher productivity in R182 compared to M82. This could be due to the different environmental conditions of the experimental fields used in these different studies.
No differences in terms of fruit firmness were evidenced herein between R182 and M82 (Table 3), in contrast with results obtained by Calafiore et al. [8].

Table 3.
Analyses of variance for all measurements of two tomato genotypes ((M82 vs. R182).
Soluble solids (°Brix) and titratable acidity were significantly higher in R182 compared to M82 (Table 3). In particular, titratable acidity increased by 78.94% in R182 compared to the cultivated genotype and an increase in °Brix of 34.64% was evidenced, as previously demonstrated [8] (Table 2). Degrees Brix are important for tomato fruit quality and increasing them is one of the main objectives in tomato breeding. This parameter is determined mostly by the accumulation of soluble sugars involved in primary metabolism, that is controlled by QTLs. Previously, Ikeda et al. [15] revealed that different S. pennellii introgression lines (IL8-3, IL 4-4, IL7-3) had increased °Brix compared to the parental line M82. This is interesting considering that IL7-3 is one of the parental line of R182.
Significantly higher accumulation of nutraceutical compounds, such as lipophilic and hydrophilic antioxidants, were demonstrated at the stage of ripe red fruits in the subline R182 compared to M82, according to ANOVA analyses (Table 3).
Assessing the content of lipophilic antioxidant, the R182 genotype demonstrated a 30% increase in the content of carotenoids, lycopene and β-carotene compared to M82. This result is in contrast with previous results that showed no difference in terms of carotenoid content between the two lines and should be further verified [8].
The content of hydrophilic antioxidants was higher in the subline R182 compared to the cultivated line M82. Results obtained are reported in Figure 1. Genotype R182 showed an increase of 48.55% in total ascorbic acid content and 40.09% in reduced ascorbic acid content compared to M82, confirming data previously obtained in our laboratories [14]. Ascorbic acid values determined in R182 exceeded those usually reported in commercial varieties that normally range between 10 and 40 mg/100 g FW, confirming the potential of this line for the improvement of current varieties [16].
The average of total phenolic compounds of M82 was 39.71 mg GAE/100 g FW. The subline R182 showed an increase of 70% compared to the cultivated line. It is the first time that an increase in the amount of this compound was evidenced in this subline and this finding should be further confirmed in additional field trials in subsequent years.
The increased performances demonstrated for R182 in this study are probably due to the activity of genes, regulatory factors and other elements present in the wild introgressed region. For this reason, in a previous study by using a dense single nucleotide polymorphism (SNP) map obtained by RNA sequencing (RNA-Seq), the introgressed region of the subline R182 was finely identified. This previous study allowed us to define the wild genes that replaced the genes of the cultivated background (cv. M82) [14]. Few wild genes were also identified in the introgressed region of R182 that could putatively control ascorbic acid content in the fruit of the subline and are indirectly or directly involved in sugar and/or hormones pathways [14]. As an example, one gene coding for a pyrophosphate-fructose 6-phosphate 1-phosphotransferase (PFP) and two gene codings for major facilitator superfamily (MFS) membrane proteins were identified.
These wild genes will be further analyzed trough functional genomic approaches by using the Crispr/Cas9 technology.
4. Conclusions
Tomato fruit is one major source of antioxidant for the benefit of nutrition and human health, thus it is very useful to select genotypes with high content of antioxidant molecules. From an agronomic point of view, it is important that these genotypes also have good productivity. The results of this study demonstrated a better performance of one S. pennellii subline named R182 compared to that of the control line M82. Our results suggested that it could be possible to use this subline as a novel germplasm resource for tomato breeding. Now, additional studies are necessary to identify the genes controlling the quality traits evidenced in this line.
Supplementary Materials
The following are available online at: https://www.mdpi.com/article/10.3390/IECAG2021-10013/s1.
Author Contributions
Conceptualization, M.M.R., A.B. and L.P.; formal analysis, L.P. and S.F.; investigation, L.P. and S.F.; resources, M.M.R., A.B. and S.F.; data curation, L.P. and S.F.; writing—original draft preparation, L.P.; writing—review and editing, M.M.R., A.B. and S.F.; visualization, L.P.; project administration, M.M.R., A.B.; funding acquisition, M.M.R. and A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no founding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in figures and tables within this article (Supplementary Materials).
Acknowledgments
The authors would like to thank D Fabrizio Olivieri and Salvatore Graci for their contribution in the conduct of some experiments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yadav, R.; Rathi, M.; Pednekar, A.; Rewachandani, Y. A Detailed Review on Solanaceae Family. Eur. J. Pharm. Med. Res. 2016, 3, 369–378. [Google Scholar]
- Francesca, S.; Arena, C.; Hay Mele, B.; Schettini, C.; Ambrosino, P.; Barone, A.; Rigano, M.M. The use of a plant-based biostimulant improves plant performances and fruit quality in tomato plants grown at elevated temperatures. Agronomy 2020, 10, 363. [Google Scholar] [CrossRef] [Green Version]
- Calafiore, R.; Ruggieri, V.; Raiola, A.; Rigano, M.M.; Sacco, A.; Hassan, M.; Frusciante, L.; Barone, A. Exploiting Genomics Resources to Identify Candidate Genes Underlying Antioxidants Content in Tomato Fruit. Front. Plant Sci. 2016, 7, 397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruggieri, V.; Calafiore, R.; Schettini, C.; Rigano, M.M.; Olivieri, F.; Frusciante, L.; Barone, A. Exploiting Genetic and Genomic Resources to Enhance Heat-Tolerance in Tomatoes. Agronomy 2019, 9, 22. [Google Scholar] [CrossRef] [Green Version]
- Raiola, A.; Rigano, M.M.; Calafiore, R.; Frusciante, L.; Barone, A. Enhancing the health-promoting effects of tomato fruit for biofortified food. Mediat. Inflamm. 2014, 2014, 16. [Google Scholar] [CrossRef] [PubMed]
- Eshed, Y.; Zamir, D. A genomic library of Lycopersicon pennellii in L. esculentum: A tool for fine mapping of genes. Euphytica 1994, 79, 175–179. [Google Scholar] [CrossRef]
- Uluisik, S. Chemical and structural quality traits during postharvest ripening regulated by chromosome segments from a wild relative of tomato Solanum pennellii IL4-2 and IL5-1. J. Food Biochem. 2021, 45, e13858. [Google Scholar] [CrossRef] [PubMed]
- Calafiore, R.; Aliberti, A.; Ruggieri, V.; Olivieri, F.; Rigano, M.M.; Barone, A. Phenotypic and Molecular Selection of a Superior Solanum pennellii Introgression Sub-Line Suitable for Improving Quality Traits of Cultivated Tomatoes. Front. Plant Sci. 2019, 10, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wellburn, A.R. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Zouari, I.; Salvioli, A.; Chialva, M.; Novero, M.; Miozzi, L.; Tenore, G.C.; Bagnaresi, P.; Bonfante, P. From root to fruit: RNA-Seq analysis shows that arbuscular mycorrhizal symbiosis may affect tomato fruit metabolism. BMC Genom. 2014, 15, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigano, M.M.; Arena, C.; Di Matteo, A.; Sellitto, S.; Frusciante, L.; Barone, A. Eco-physiological response to water stress of drought-tolerant and drought-sensitive tomato genotypes. Plant Biosyst. 2016, 150, 682–691. [Google Scholar] [CrossRef] [Green Version]
- Stevens, R.; Buret, M.; Garchery, C.; Carretero, Y.; Causse, M. Technique for rapid small-scale analysis of vitamin C levels in fruit and application to a tomato mutant collection. J. Agric. Food Chem. 2006, 54, 6159–6165. [Google Scholar] [CrossRef] [PubMed]
- Rigano, M.M.; Raiola, A.; Tenore, G.C.; Monti, D.M.; Del Giudice, R.; Frusciante, L.; Barone, A. Quantitative trait loci pyramiding can improve the nutritional potential of tomato (Solanum lycopersicum) fruits. J. Agric. Food Chem. 2014, 62, 11519–11527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aliberti, A.; Olivieri, F.; Graci, S.; Rigano, M.M.; Barone, A.; Ruggieri, V. Genomic Dissection of a Wild Region in a Superior Solanum pennellii Introgression Sub-Line with High Ascorbic Acid Accumulation in Tomato Fruit. Genes 2020, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Hiraga, M.; Shirasawa, K.; Nishiyama, M.; Kanahama, K.; Kanayama, Y. Analysis of a tomato introgression line, IL8-3, with increased Brix content. Sci. Hortic. 2013, 153, 103–108. [Google Scholar] [CrossRef]
- Ruggieri, V.; Bostan, H.; Barone, A.; Frusciante, L.; Chiusano, M.L. Integrated bioinformatics to decipher the ascorbic acid metabolic network in tomato. Plant Mol. Biol. 2016, 91, 397–412. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).