Abstract
The rapid spreading of cosmopolite breeds reduces the population sizes of Russian local sheep, possibly resulting a loss of biodiversity. Estimation of the runs of homozygosity (ROHs) in local sheep genomes is an informative tool to address their current genetic state. In this work, we aimed to address the distribution of ROHs and to estimate genome inbreeding in Russian local sheep breeds based on SNP genotyping. Medium-density SNP genotypes of twenty-three local sheep breeds (n = 332) were obtained in our previous study. We used a consecutive runs method implemented in the R package “detectRUNS” to calculate ROH which were estimated for each animal and then categorized in the ROH length classes (1–2 Mb, 2–4 Mb, 4–8 Mb, 8–16 Mb, >16 Mb). The frequency of short ROH segments (≤2 Mb) were the highest in all studied breeds (63.15–93.10%). The longest segments (>16 Mb) were the least frequent and were missing in four breeds. The genomic coefficients based on ROH estimation varied from medium (0.114) to low (0.035). Thus, we found that Russian local sheep breeds are characterized by a low level of genomic inbreeding.
1. Introduction
Genetic resources of Russian local sheep include breeds, which were specifically selected for wool and combined (wool and meat) production, and aboriginal breeds, which are adapted to extreme environments and from which all types of sheep products are used by local smallholders [1]. The rapid spreading of cosmopolite breeds decreases population sizes of Russian local sheep, resulting in a loss of biodiversity.
Runs of homozygosity (ROHs) are numbers of homozygous loci that inbred progeny inherit from parents which originated from a common ancestor. Calculation of the length of ROH segments reveals whether inbreeding in the populations was recent or ancient [2,3]. Evaluation of genomic inbreeding coefficients based on ROH calculation is not determined by allelic frequencies or sampling procedures, and is considered as a robust measure for describing levels of inbreeding in the populations with missing pedigree information [4]. In this aspect, estimation of runs of homozygosity in genomes of livestock species is an informative tool to address their current genetic state [5].
In this work, we aimed to address the distribution of the ROHs and to estimate genome inbreeding in Russian local sheep breeds based on medium-density SNP genotyping data.
2. Experiments
2.1. Materials and Methods
Materials for this study included SNP genotypes of twenty-two local sheep breeds (n = 332) which were generated with OvineSNP50 BeadChip (Illumina, San Diego, CA, USA) in our previous research [6]. The SNP profiles of the Romanov, Kuchugur, and Baikal fine-fleeced breeds were not analyzed in the present study. The list of the studied breeds and sample sizes are presented in Table 1. The details on the relevant dataset, including the sampling locations for each breed, the SNP quality control, and the phylogenetic links between the breeds, are available online at https://gsejournal.biomedcentral.com/articles/10.1186/s12711-018-0399-5 (accessed on 31 March 2021). The SNP genotypes are available from the corresponding author on reasonable request.

Table 1.
Values of inbreeding coefficient calculated based on ROHs (FROH) in Russian local breeds.
2.2. Data Processing
For ROH calculation, we used a window-free method for consecutive SNP-based detection [7] implemented in the R package “detectRUNS” [8]. One SNP with a missing genotype and up to one possible heterozygous genotype was allowed in the run. The minimum ROH length was 1000 kb.
ROH were estimated for each animal and then categorized in the corresponding ROH length classes: (1–2 Mb, 2–4 Mb, 4–8 Mb, 8–16 Mb, >16 Mb). The total number of identified ROH was calculated for each length category in each of the individuals of each breed. The mean sum of ROH was computed by adding up the length of the ROHs for each individual in the sheep populations, and then the results were averaged per breed population.
The genomic inbreeding coefficient based on ROH (FROH) was estimated as the sum of the length of all ROH per sheep as a proportion of the total autosomal SNP coverage (2.44 Gb).
3. Results
ROHs were found on all autosomes in all Russian local sheep breeds. The genome coverage by ROH was higher on OAR1 (7.88–12.10%), OAR 2 (9.03–11.94%), and OAR3 (6.28–11.42%), and lower on OAR20 (1.05–2.44%) and OAR26 (1.06–2.24%). The ROH length and ROH number varied from 93.73 ± 4.29 Mb and 68 in the Lezgin breed to 301.69 ± 19.71 Mb and 117 in the Russian long-haired breed. The individual minimum was found in the Lezgin breed (68.16 Mb and 54), while the maximum was detected in the Russian long-haired breed (469.99 Mb and 138).
Figure 1 shows the distribution of the ROH segments in length classes in Russian local sheep breeds. The frequency of short ROH segments (≤2 Mb) was the highest in all studied breeds and varied from 63.15% in the Russian long-haired breed to 93.10% in the Lezgin breed. The Tushin breed had the minimum of the ROH segments of 2–4 Mb length class (6.09%), while the Russian long-haired breed had the maximum (24.62%). The frequencies of the ROH segments within length class (4–8 Mb) varied from 0.57% in the Lezgin breed to 10.01% in the Russian long-haired breed. Long ROH segments (8–16 Mb) ranged from 0.19% in the Tuva breed to 3.46% in the Andean breed. The longest ROH segments (>16 Mb) were the least frequent (0.07–1.22%) and were missing in four breeds.
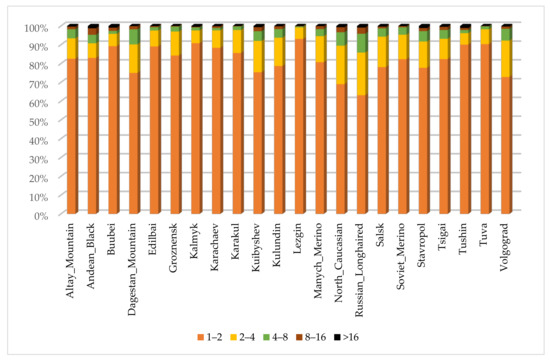
Figure 1.
Distribution of the runs of homozygosity in length classes (1–2 Mb, 2–4 Mb, 4–8 Mb, 8–16 Mb, >16 Mb) in Russian local sheep breeds.
Estimates of inbreeding coefficient calculated based on ROH in Russian local breeds are given in Table 1. The values of genomic inbreeding coefficient varied from medium to low. The minimum FROH was calculated in the Lezgin breed (FROH = 0.035) and the maximum was detected in the Russian long-haired breed (FROH = 0.114). The range of individual FROH variability was more noticeable in several breeds. including Andean Black (FROH from 0.03 to 0.15), Buubei (FROH from 0.03 to 0.15), Kalmyk (FROH from 0.03 to 0.11), North Caucasian (FROH from 0.06 to 0.11), Russian long-haired (FROH from 0.08 to 0.18), and Stavropol (FROH from 0.04 to 0.16).
4. Discussion
There are several approaches to address biodiversity and its dynamics in the populations of livestock species: effective population size, heterozygosity, and runs of homozygosity [9]. In our previous study, we calculated and analyzed effective population sizes and heterozygosity to unlock current state of genetic diversity in Russian local sheep breeds [6]. Nonetheless, estimating runs of homozygosity is a useful tool to reveal the presence of long-term inbreeding in livestock populations [3].
The prevalence of short ROH segments = in Russian local sheep breeds is compatible with the relevant patterns detected in Swiss [4] and Italian local sheep breeds [10], as well as in commercial sheep breeds including Charollais, Suffolk, Texel [11], Border Leicester, Merino, and Poll Dorset [9].
The average values of ROH length calculated in our study were higher than those obtained in commercial sheep breeds (93.73–301.69 Mb versus 92.61–128.31 Mb [11]/94.88–126.06 Mb [9], respectively). The maximum individual ROH length values estimated in our study (400 and 469.99 Mb) were close to those obtained in Australian populations of Border Leicester, Merino, and Poll Dorset breeds (427.2, 410.5, and 396.45 Mb, respectively) [9].
The mean FROH varied from 0.016 to 0.099 in Italian local breeds, including Valle del Belice and Comisana [10], and from 0.022 to 0.153 in Swiss local breeds [4]. These estimates are compatible with the FROH variation detected in Russia local sheep breeds.
The highest values of genome coverage by ROH segments and the maximum inbreeding coefficient calculated based on ROH, which were found in the Russian long-haired breed, might correspond to the small population size (1400 heads at the end of 2019) [12] and the use of a limited number of sires.
5. Conclusions
Here, we presented a detailed analysis of the patterns of distribution of the runs of homozygosity in major Russian local sheep breeds based on 50K SNP profiles. Our findings provide an evidence of a low genomic inbreeding in local sheep populations (except for the Russian long-haired breed). The study results provide useful information to design conservation programs for local genetic resources of sheep.
Author Contributions
N.Z., G.B. and T.D. conceived and designed the experiments; T.D. and H.R. performed the experiments; A.D. and H.R. analyzed the data; M.S. and K.W. contributed reagents/materials/analysis tools; T.D. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by Ministry of Science and Higher Education of the Russian Federation.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of L.K. Ernst Federal Research Center for Animal Husbandry (protocol No. 4 from the 19 January 2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
The SNP genotypes presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. Ministry of Science and Higher Education of the Russian Federation had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| SNP | Single Nucleotide Polymorphisms |
| ROH | Runs of homozigosity |
| FROH_Mean | inbreeding coefficient calculated based on ROH with a minimum length of 1 Mb |
References
- Dunin, I.M.; Dankvert, A.G. Spravochnik Porod i tipov Sel`skokhozyastvennykh Zhivotnykh, Razvodimykh v Rossiiskoi Federatsii; VNIIPLEM: Moskva, Russia, 2013. (In Russian) [Google Scholar]
- McQuillan, R.; Leutenegger, A.L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008, 83, 359–372. Available online: https://www.cell.com/ajhg/fulltext/S0002-929700445-X (accessed on 10 February 2021). [CrossRef] [PubMed] [Green Version]
- Curik, I.; Ferenčaković, M.; Sölkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Signer-Hasler, H.; Burren, A.; Ammann, P.; Drögemüller, C.; Flury, C. Runs of homozygosity and signatures of selection: A comparison among eight local Swiss sheep breeds. Anim. Genet. 2019, 50, 512–525. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1111/age.12828 (accessed on 12 February 2021). [CrossRef]
- Sölkner, J.; Ferenčaković, M.; Gredler, B.; Curik, I. Genomic metrics of individual autozygosity, applied to a cattle population. In Proceedings of the 61st Annual Meeting of the European Association of Animal Production, Heraklion, Crete Island, Greece, 23–27 August 2010; Wageningen Academic Publishers: Wageningen, The Netherlands, 2010. [Google Scholar]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Barbato, M.; Traspov, A.A.; Brem, G.; et al. Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. GSE 2018, 50, 29. Available online: https://gsejournal.biomedcentral.com/articles/10.1186/s12711-018-0399-5 (accessed on 10 February 2021). [CrossRef] [PubMed] [Green Version]
- Marras, G.; Gaspa, G.; Sorbolini, S.; Dimauro, C.; Ajmone-Marsam, P.; Valentini, A.; Williams, J.L.; Macciotta, N.P. Analysis of runs of homozygosity and their relationship with inbreeding in five cattle breeds farmed in Italy. Anim. Genet. 2014, 46, 110–121. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1111/age.12259 (accessed on 10 February 2021). [CrossRef] [PubMed]
- Cran.R-project.org: detectRUNS: Detect Runs of Homozygosity and Runs of Heterozygosity in Diploid Genomes. Available online: https://cran.r-project.org/web/packages/detectRUNS/index.html (accessed on 25 January 2021).
- Al-Mamun, H.A.; Clark, S.A.; Kwan, P.; Gondro, C. Genome-wide linkage disequilibrium and genetic diversity in five populations of Australian domestic sheep. Genet. Sel. Evol. GSE 2015, 47, 90. Available online: https://gsejournal.biomedcentral.com/articles/10.1186/s12711-015-0169-6 (accessed on 12 February 2021). [CrossRef] [PubMed] [Green Version]
- Mastrangelo, S.; Ciani, E.; Sardina, M.T.; Sottile, G.; Pilla, F.; Portolano, B.; Bi.Ov. Ita Consortium. Runs of homozygosity reveal genome-wide autozygosity in Italian sheep breeds. Anim. Genet. 2018, 49, 71–81. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1111/age.12634 (accessed on 12 February 2021). [CrossRef] [PubMed] [Green Version]
- Purfield, D.C.; McParland, S.; Wall, E.; Berry, D.P. The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds. PLoS ONE 2017, 12, e0176780. Available online: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0176780 (accessed on 12 February 2021). [CrossRef] [PubMed] [Green Version]
- Dunin, I.M.; Amerhanov, H.A.; Safina, G.F.; Grigoryan, L.N.; Hatataev, S.A.; Hmelevskaya, G.N.; Pavlov, M.B.; Stepanova, N.G. Ezhegodnik po Plemennoj Rabote v Ovcevodstve i Kozovodstve v Hozyajstvah Rossijskoj Federacii (2019 God); FGBNU Vserossiiskii Nauchno-Issledovatelskii Institut Plemennogo Dela Lesnye Poliany: Moskva, Russia, 2019. (In Russian) [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).