Abstract
The deamidation of glutamine (Gln) residues, which occurs non-enzymatically under physiological conditions, triggers protein denaturation and aggregation. Gln residues are deamidated via the cyclic glutarimide intermediates to l-α-, d-α-, l-β-, and d-β-glutamate residues. The production of these biologically uncommon amino acid residues is implicated in the pathogenesis of autoimmune diseases. The reaction rate of Gln deamidation is influenced by the C-terminal adjacent (N +1) residue and is highest in the Gln-glycine (Gly) sequence. Here, we investigated the effect of the (N + 1) Gly on the mechanism of Gln deamidation and the activation barrier using quantum chemical calculations. Energy-minima and transition-state geometries were optimized by the B3LYP density functional theory, and MP2 calculations were used to obtain the single-point energy. The calculated activation barrier (85.4 kJ mol−1) was sufficiently low for the reactions occurring under physiological conditions. Furthermore, the hydrogen bond formation between the catalytic ion and the main chain of Gly on the C-terminal side was suggested to accelerate Gln deamidation by stabilizing the transition state.
1. Introduction
The nonenzymatic deamidation of glutamine (Gln) and asparagine (Asn) residues, which is a post-translational modification observed in the human body, causes structural changes in proteins; thus, it is related to protein aggregation and hypofunction [1,2,3,4,5]. For example, deamidation accumulates in the ocular crystallin proteins because crystallin is not degraded by protein turnover; this accumulation leads to the aggregation of crystallin, resulting in cataracts [6,7,8]. Deamidation is also related to the formation of biologically uncommon amino acids, such as β-Asp and γ-Gln [9,10,11]. The production of such uncommon residues triggers the onset and exacerbation of autoimmune diseases [12]. Nonenzymatic post-translational modifications in the histone H2B render it immunogenic and induce the production of autoantibodies against normal H2B; this process is associated with systemic lupus erythematosus (SLE) [13,14,15]. Deamidated gliadin peptides were shown to have the potential to produce autoantibodies in celiac disease [16,17]. The deamidation of Gln in preproinsulin, InsB30–C13, can bind to HLA-DQ2cis, HLA-DQ2trans, HLA-DQ8trans, and HLA-DQ8cis and enhance T-cell autoreactivity [18]. Therefore, post-translational deamidation can cause a lack of central tolerance in type 1 diabetic patients. Deamidation of complementarity-determining regions in monoclonal antibodies changes binding affinity and specificity for antigens [19,20,21,22]. The deamidation in the Fc region is related to the clearance of endogenous antibodies [23]. Therefore, deamidation is a relevant reaction in age-related diseases, autoimmune diseases, and peptide/protein-based drug design.
Asn deamidation is suggested to be initiated by the nucleophilic attack of main-chain nitrogen on the C-terminal side to the amide carbon of Asn side chain and proceeds via an intermediate, including five-membered succinimide ring. Next, this intermediate is partially hydrolyzed to l-α- or l-β-aspartate (l-α- or l-β-Asp) residue [24]. As the succinimide intermediate is racemization prone, the intermediate is also converted to d-α-/d-β-Asp by deamidation. The Gln deamidation is suggested to occur by the similar mechanism to the Asn deamidation and proceeds via the six-membered glutarimide ring intermediate (Scheme 1) [24]. Therefore, l-α-Gln is proposed to be transformed to l-α-, d-α-, l-β-, and d-β-Glu. However, the deamidation rate of Gln is much slower than that of Asn [25], and the detailed mechanism is not well understood. Various external conditions, such as buffer anions, ionic strength, pH, and temperature affect the deamidation rate [26,27]. The species of the (N + 1) residue are also the important factor for the deamidation rate because the main chain on the C-terminal side is involved in the reaction. Experimental data of pentapeptides have indicated that the Gln deamidation rate is highest in a Gln-Gly sequence [25]. Gln deamidation in a Gln-Gly sequence is approximately 16 times faster than that in a Gln-Trp sequence, which has the lowest deamidation rate. Glutarimide formation is assumed not to occur in sequences with slower deamidation rate than the Gln-Pro sequence. On the other hand, the (N − 1) residues have little effect on the deamidation rate. In a previous study, we have supposed the two-step glutarimide formation (cyclization and deammoniation steps) from Gln (Scheme 2) [28]. A gem-hydroxylamine intermediate is generated in the cyclization step, and then the deammoniation step proceeds via proton transfers in the gem-hydroxylamine moiety. The detail effects of the (N + 1) residue on the mechanism of deamidation have not been reported.
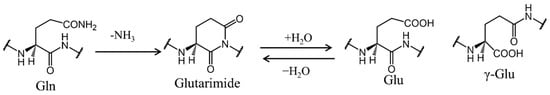
Scheme 1.
Deamidation pathway of glutamine residues via glutarimide.
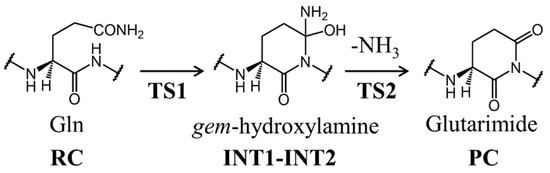
Scheme 2.
Proposed pathway of glutarimide formation from glutamine residues. The names of obtained geometries were represented in this figure (RC: reactant complex, TS: transition state, INT: intermediate, PC: product complex).
Quantum chemical calculation methods have been used to investigate the reaction mechanisms of various nonenzymatic post-translational modifications of proteins [28,29,30,31,32,33,34,35]. In these studies, the proton transfers required for the reactions were indicated, and reasonable activation barriers were determined. In addition, Takahashi et al. reported the effects of (N + 1) histidine residue on the Asn deamidation [30]. Therefore, quantum chemical calculations are available for the investigation of the detailed reaction mechanism of the nonenzymatic post-translational modifications that occur in proteins. In the present study, to reveal the effects of an (N + 1) Gly residue on the mechanism and reaction rate of deamidation, the Gln deamidation in Gln-Gly sequence was investigated using quantum chemical calculations.
2. Computational Methods
Gaussian 16 software was used for all calculations [36]. For the model compound, a glutaminyl glycine capped with acetyl and methylamino groups on the N-terminus and C-terminus, respectively (Ac-Gln-Gly-NMe) (Figure 1). The dihedral angles of the main chains were characterized as φ (C−N−Cα−C) and ψ (N−Cα−C−N). The dihedral angles of the side chain were represented as χ1 (N−Cα−Cβ−Cγ) and χ2 (Cα−Cβ−Cγ−Cδ). The initial structure of the reactant complex (RC) was constructed in reference to the optimized geometry in a previous study [28]. However, the interaction between Gly on the C-terminal side and the catalyst ion is unknown. We also analyzed the cyclization step from the initial structure as different in the catalyst ion and Gly binding. A dihydrogen phosphate (H2PO4−) ion was included in calculations as a catalyst. An H2PO4− ion has been suggested to act as the catalyst for the nonenzymatic reactions in proteins, as this ion has the properties of both a Lewis acid and a base. The optimization of all geometries for energy-minimum and transition state (TS) were performed without any constraints at the B3LYP/6-31+G(d,p) level. This level of theory was used in the previous study for Gln deamidation [28]. The connections of the energy minima and each TS were confirmed by intrinsic reaction coordinate (IRC) calculations. Vibrational frequency calculations were performed for calculations of the zero-point energies for all optimized geometries. We also confirmed that the energy-minimum geometries had no imaginary frequency, and the TS geometries had a single imaginary frequency. The single-point energies were calculated for all the optimized geometries at the MP2/6-311+G(2d,2p) level. The same energy calculation was performed for the optimized geometries of Ac-Gln-NMe obtained in the previous study [28]. The polarizable continuum model was used for all calculations. Coordinates of all optimized geometries are shown in Supplementary Materials (Tables S1−S12).
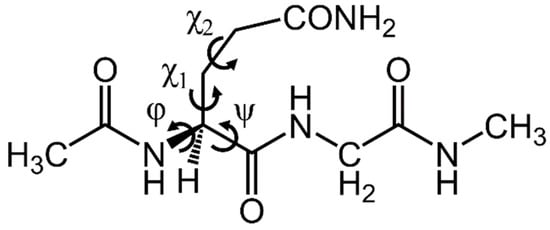
Figure 1.
Structures of Ac-Gln-Gly-NMe and the dihedral angles.
3. Results and Discussion
To evaluate the effects of the (N + 1) Gly residue on the reaction mechanism and the activation barrier to Gln deamidation, the two-step pathway for the formation of glutarimide, cyclization and deammoniation steps was investigated and compared with the previously reported results in Ac-Gln-NMe [28]. The optimized geometries of reactant complex (RC), transition state 1 (TS1), and intermediate 1(INT1) for the cyclization step were obtained, and the optimized geometries of intermediate 2 (INT2), transition state 2 (TS2), and product complex (PC) for deammoniation step were obtained.
3.1. Comparison of the Optimized Geometries in the Cyclization Step
The cyclization step was investigated in three conformations (pathway A, B, and C) with different binding modes between the H2PO4− ion and Ac-Gln-Gly-NMe (Figure 2, Figure 3 and Figure 4). RC-A, -B, and -C were constructed based on the optimized geometry of RC in Ac-Gln-NMe [28]. When the main-chain conformation is almost not changed, RC is stabilized by the increased hydrogen bonds between the catalytic ions and peptides [35]. We examined positions of the H2PO4− ion, which can form two hydrogen bonds, involved in the proton transfer and one with the main chain. In RC of pathway A, the H2PO4− ion formed three hydrogen bonds bridging the main and side chains: with an oxygen atom in the Gly main chain, with an oxygen atom in the Gln side chain, and with the nitrogen in the Gln main chain (1.76, 1.74, and 1.76 Å, respectively) (Figure 2). The H2PO4− ion also formed a hydrogen bond with the oxygen atom of the Gly main chain in RC-C, while it formed a hydrogen bond with the nitrogen atom of the Gly in RC-B (Figure 3 and Figure 4). All proton transfers in the cyclization step proceeded via the H2PO4− ion. A proton of the Gly main chain was transferred to the H2PO4− ion at the early stage of the reaction, and this proton transfer was completed in TS1-A–C (Figure 2, Figure 3 and Figure 4). This proton abstraction by the H2PO4− ion was considered to enhance the nucleophilicity of the nitrogen atom in the Gly main chain. The other proton was transferred from the H2PO4− ion to the oxygen atom of the Gln side chain with a delay. No differences in the proton transfers for the TS1 formation were observed among the pathway A, B, and C. The distance between the nitrogen atom of the Gly main chain and the carbon atom of the Gln side chain (C−N distance) changed from 3.30 to 1.96 Å in pathway A, from 3.25 to 1.99 Å in pathway B, and from 3.25 to 1.93 Å in pathway C, while the C−N distance in INT1 was 1.53 Å in all pathways. The C−N distance in TS1-A, TS1-B, and TS1-C of Ac-Gln-Gly-NMe was shorter than that of Ac-Gln-NMe (2.07 Å), of which the optimized geometries obtained in ref. [28] are shown in Figure 5. The cyclization mechanism in Ac-Gln-Gly-NMe was similar to the reaction in Ac-Gln-NMe. However, a new hydrogen bond between the H2PO4− ion and the Gly main chain was observed in each TS1 of Ac-Gln-Gly-NMe, and these hydrogen bonds were maintained throughout the cyclization step.
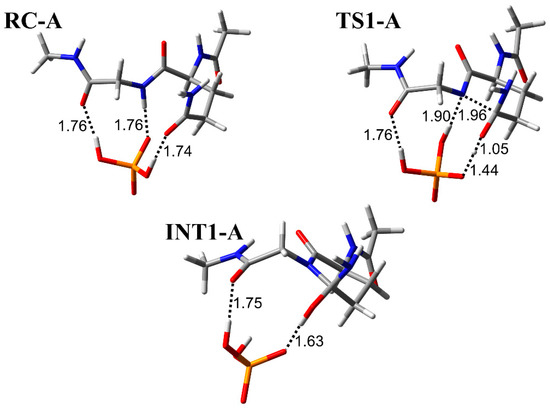
Figure 2.
The geometries in pathway A for the cyclization step in Ac-Gln-Gly-NMe. Selected interatomic distances are in units of Å. Atoms are represented as a stick model with typical colors (hydrogen: white, carbon: grey, nitrogen: blue, oxygen: red, phosphorus: orange).
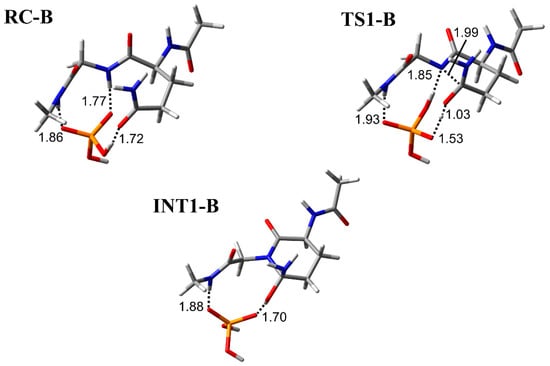
Figure 3.
The geometries in pathway B for the cyclization step in Ac-Gln-Gly-NMe. Selected interatomic distances are in units of Å. Atoms are represented as a stick model with typical colors (hydrogen: white, carbon: grey, nitrogen: blue, oxygen: red, phosphorus: orange).
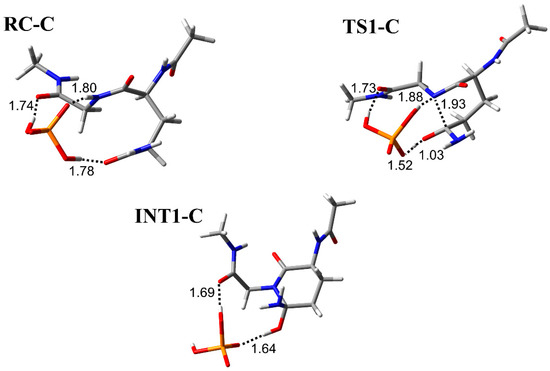
Figure 4.
The geometries in pathway C for the cyclization step in Ac-Gln-Gly-NMe. Selected interatomic distances are in units of Å. Atoms are represented as a stick model with typical colors (hydrogen: white, carbon: grey, nitrogen: blue, oxygen: red, phosphorus: orange).
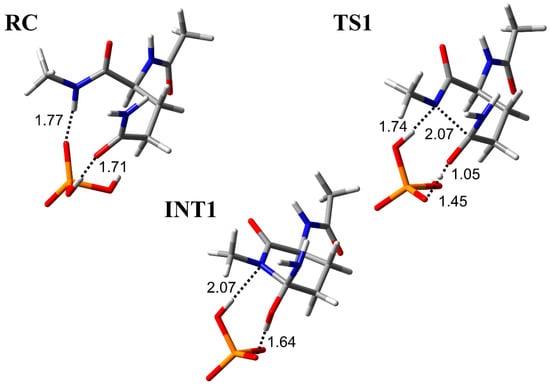
Figure 5.
The geometries for the cyclization step in Ac-Gln-NMe [28]. Selected interatomic distances are in units of Å. Atoms are represented as a stick model with typical colors (hydrogen: white, carbon: grey, nitrogen: blue, oxygen: red, phosphorus: orange).
To investigate the conformational change in cyclization step, the dihedral angles of Gln in pathway A, B, and C are shown in Table 1. The dihedral angle change that was the largest in the cyclization step in pathway A was the change in ψ (37°) in the TS1-A formation, and the change in ψ was relatively large in pathway B (47°). These changes in dihedral angle ψ of Ac-Gln-Gly-NMe were similar to the dihedral angle ψ of Ac-Gln-NMe (45°). Therefore, the (N + 1) Gly residue had almost no effect on the structural changes required for the reaction to proceed. In contrast, the changes of ψ in pathway C were relatively small (28°). In pathway C, the change of ψ with INT1-C formation was the largest (49°). Therefore, the binding mode of the catalytic ions was suggested to affect the conformational changes with the reaction. The dihedral angles of the optimized geometries were compared with those of the Gln residues in the crystal structure obtained from the protein data bank (PDB). The deamidation ratios of Gln16, Gln63, Gln92, and Gln120 were reported to be relatively high in the γS-crystallin obtained from cataractous lenses [8,37]. Because the crystal structure of γS-crystallin (PDB ID: 6FD8) forms dimer, the dihedral angles for chains A and B were extracted (Table 2). The dihedral angles ψ of Gln16, Gln63, and Gln92 were 128°–168°. These dihedral angles resemble the ψ of the optimized geometry for the RC. Therefore, Gln deamidation is more likely to occur in such structures.

Table 1.
Dihedral angles φ, ψ, χ1, and χ2 in pathway A, B, and C of the cyclization step.

Table 2.
Dihedral angles φ, ψ, χ1, and χ2 of frequently deamidated Gln in the crystal structures (PDB ID: 6FD8).
To identify the main pathway of Gln deamidation in a Gln-Gly sequence catalyzed by an H2PO4− ion, the relative energies for pathway A–C were calculated (Figure 6). Since the most stable geometry was RC-A, the relative energies were referred to RC-A. Although the number of hydrogen bonds in RC-B and RC-C was same as that of RC-A, the main chain conformations of RC-B and RC-C were unstable. The relative energy of TS1-A (111 kJ mol−1) was lower than those of TS1-B (141 kJ mol−1) and TS1-C (175 kJ mol−1). The activation barriers, which were relative energies of TS1 referred to each RC, of pathway B and C were 133 and 147 kJ mol−1. Therefore, pathway A was determined as the main pathway of Gln deamidation in Gln-Gly sequence. The difference between pathway A and other pathways was the binding mode of the catalyst ion, whereas the reaction mechanisms were similar. In addition, the dihedral angles were also similar except for φ of INT1-C. The reason for the higher activation energies in pathway B and C is considered to be unstable binding of the H2PO4− ion.
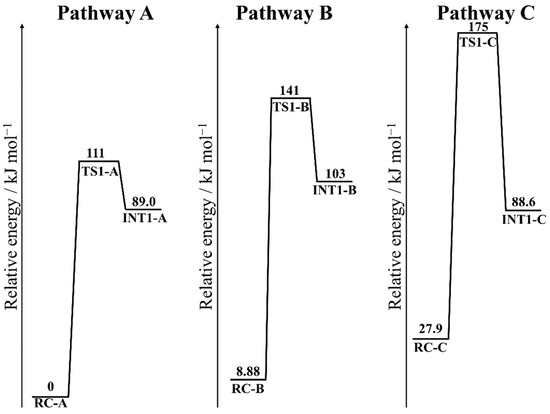
Figure 6.
Energy profiles for the cyclization step. The relative energies are referred to RC-A at B3LYP/6-31+G(d,p).
3.2. Comparison of the Optimized Geometries in the Deammoniation Step
The results of the cyclization step indicated that the main chain conformation needed to be stable. Therefore, the conformation of INT2, which is the reactant of the deammoniation step, was constructed using the previously reported stable conformer [28]. Although the repositioning of the H2PO4− ion are required for the conversion form INT1-A to INT2 (Figure 7), this alteration is thought to be energetically accessible because of the abundance of H2PO4− ions in vivo (Figure 8). In INT2, the H2PO4− ion formed two hydrogen bonds with the nitrogen atom and the hydroxyl group of the gem-hydroxylamine moiety (1.80 and 1.62 Å, respectively). Furthermore, a hydrogen bond between the oxygen atom of the Gly main chain and the H2PO4− ion was observed (1.84 Å). From INT2, two proton transfers occurred between the H2PO4− ion and the gem-hydroxylamine at initiation of the deammoniation step and resulted in TS2 formation (Figure 7). In TS2, the proton between the H2PO4− ion and the nitrogen atom of gem-hydroxylamine was close to this nitrogen atom, but the proton transfers did not complete. Therefore, a proton from the H2PO4− ion to the nitrogen was firstly transferred, and the other proton was transferred from the oxygen of gem-hydroxylamine to the H2PO4− ion with delay. The representative geometries in IRC also showed this order of proton transfers (Figure 9). These proton transfers occurred in the same order as in the case of Ac-Gln-NMe. The PC formation was accomplished after the two proton transfers and release of an ammonia molecule. There were almost no changes in all dihedral angles of the main chain and the side chain of throughout the deammoniation step (Table 3). In the presence or absence of the (N + 1) glycine, no large conformational change was required in the deammoniation step. In the crystal structure of γS-crystallin (PDB ID: 6FD8), the dihedral angles φ of relatively frequently deamidated Gln residues are from approximately −80° to −120° and are similar to the φ of INT2, TS2, and the PC (Table 2). Gln deamidation may occur easily in such conformations, which have a similar structure to a succinimide ring.
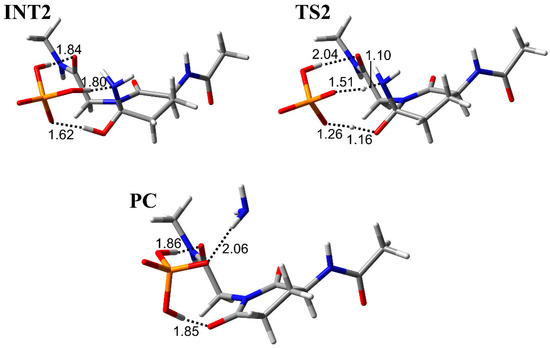
Figure 7.
The geometries for the deammoniation step in Ac-Gln-Gly-NMe. Selected interatomic distances are in units of Å. Atoms are represented as a stick model with typical colors (hydrogen: white, carbon: grey, nitrogen: blue, oxygen: red, phosphorus: orange).
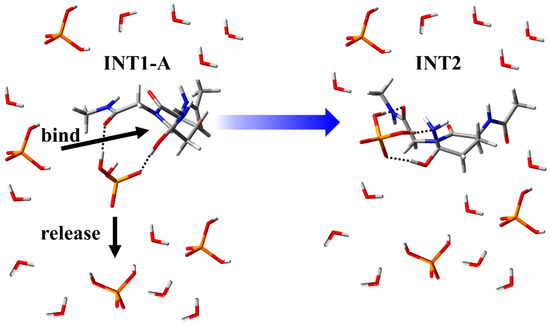
Figure 8.
Schematic diagram of the conversion from INT1-A to INT2 by the repositioning of the H2PO4− ion. Bulk water molecules and dihydrogen phosphate ions are illustrated.
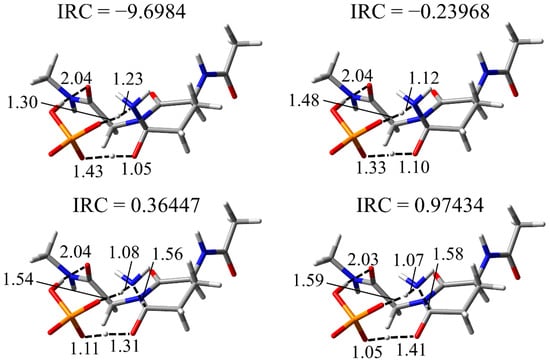
Figure 9.
Four representative geometries on IRC of the deammoniation step. Selected interatomic distances are shown in Å. Atoms are represented as a stick model with typical colors (hydrogen: white, carbon: grey, nitrogen: blue, oxygen: red, phosphorus: orange).

Table 3.
Dihedral angles in the deammoniation step.
3.3. Energy Profiles
To compare the activation barrier of the reaction in Ac-Gln-NMe and Ac-Gln-Gly-NMe more accurately, the activation barriers were calculated for both model compounds using the B3LYP/6-31;G(d,p), MP2/6-311+G(2d,2p), CAM-B3LYP/6-311+G(2d,2p), ωB97XD/6-311+G(2d,2p), and HF/6-311+G(2d,2p) levels (Table 4 and Table 5). The energy obtained by HF methods was too high for the reaction proceeding. MP2 methods yielded the lowest activation barrier, and the activation barriers in CAM-B3LYP/6-311+G(2d,2p) and ωB97XD/6-311+G(2d,2p) were similar to that in B3LYP/6-31+G(d,p). The MP2/6-311+G(2d,2p) level has been used to investigate the activation barrier of nonenzymatic posttranslational modifications of proteins, and the calculated values were reasonable in comparison with the experimental values [36,38]. Therefore, the energy diagram in this calculation level is shown in Figure 10, and electronic and zero-point energies in MP2/6-311+G(2d,2p)//B3LYP/6-31+G(d,p) are shown in Tables S13 and S14. For the calculation of the relative energies of Ac-Gln-Gly-NMe, the optimized geometries of pathway A were employed. In both reactions, the barrier heights of the cyclization steps were significantly high in comparison with the barrier of the deammoniation steps. Therefore, both in Ac-Gln-NMe and Ac-Gln-Gly-NMe, the cyclization step was the rate-limiting step. The activation barriers for Ac-Gln-NMe were 96.8 kJ mol−1, and for Ac-Gln-Gly-NMe were 85.4 kJ mol−1. In previous studies, the experimental data indicated that the deamidation rate of Gln residues was slower than that of Asn residues, and the activation energy of Asn deamidation was 80–100 kJ mol−1 [9,39]. Furthermore, the activation barriers of Asn deamidation reported in previous computational studies were 80–90 kJ mol−1 [29,30,31,38]. In addition, these activation energies include values obtained at the same calculation level as that in the present study. The calculated activation barrier of Gln deamidation was higher or slightly higher than that of Asn deamidation. Thus, both for Ac-Gln-NMe and Ac-Gln-Gly-NMe, the activation barrier calculated using MP2 or DFT methods in this study appears reasonable. The calculated activation barrier for Ac-Gln-Gly-NMe was 11.4 kJ mol−1 lower than that for Ac-Gln-NMe (Figure 10). The (N + 1) Gly residue is suggested to reduce the activation barrier. The differences in TS1 were the C–N distance and the hydrogen bond formation (Figure 2). Our results suggest that the stability of TS1 is enhanced by alteration of the distribution of electron density, which is caused by the hydrogen bond formation between catalytic ions and an (N + 1) residues.

Table 4.
Relative energy for Ac-Gln-NMe calculated by various calculation levels with a 6-311+G(2d,2p) basis set.

Table 5.
Relative energy for Ac-Gln-Gly-NMe obtained by various calculation levels with a 6-311+G(2d,2p) basis set.
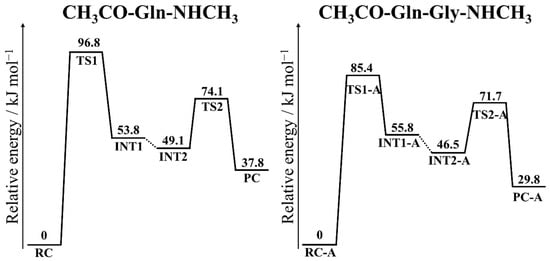
Figure 10.
Energy profiles for the two step glutarimide formation. The relative energies in Ac-Gln-NMe and Ac-Gln-Gly-Nme were referred to in each reactant at MP2/6-311+G(2d,2p).
The optimized geometries proposed the reason why the activation barrier of Gln deamidation was higher, or slightly higher, than that of Asn deamidation. In Asn deamidation, the structural changes of the main chain for the TS1 formation occurred on both the N- and C- terminal sides [38]. On the other hand, TS1 formation in Gln deamidation required the structural changes of the main chain only on the C-terminal side. Since the side chain of Gln is longer, the conformational change upon cyclization occurs in the side chain and does not affect the main chain on the N-terminal side. As the result, twisting of the main chain may occur, and TS1 may become unstable (Figure 11).
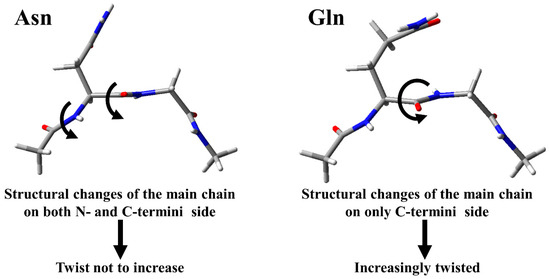
Figure 11.
Schematic diagram of the reason why the activation barrier of Gln deamidation is higher than Asn deamidation.
4. Conclusions
In the present study, to reveal the effects of an (N + 1) Gly residue on glutarimide formation, we investigated the reaction mechanism of the glutarimide formation from Gln residues through cyclization and deammoniation steps. Calculations for the cyclization step were performed in three pathways, and pathway A was determined as a main pathway. The reaction mechanism for Ac-Gln-Gly-NMe was similar to that for Ac-Gln-NMe. The conformational changes in the main pathway were observed at the C-terminus during TS1 formation and at the N-terminus during the INT2 formation. The characteristics of the dihedral angle changes in Ac-Gln-Gly-NMe were the same as those in Ac-Gln-NMe. Hydrogen bond formation was different between Ac-Gln-Gly-NMe and Ac-Gln-NMe. Ac-Gln-NMe formed two hydrogen bonds with the H2PO4− ion throughout the reaction; in contrast, Ac-Gln-Gly-NMe formed three hydrogen bonds. This increased number of hydrogen bonds may stabilize the complex formation of the catalytic ion and Gln. For the reaction to proceed, it is important that the side chain of peripheral residues does not obstruct the formation of hydrogen bond between catalytic ions and the main chain. In the main pathway, the hydrogen bond formation on the C-terminal side stabilized TS1 as well as RC. The hydrogen bond can mitigate the deviation of the charge distribution in TS1. As a result, the hydrogen bond on the C-terminal side decreased the activation barrier of the deamidation reaction in Ac-Gln-Gly-NMe. Our results suggest that the (N + 1) Gly residue is not a three-dimensional obstacle and, further, reduces the activation barrier of the Gln deamidation. Many mysteries in the slow deamidated sequences, such as Gln-Trp, will be solved in future studies. The findings in the present study help to clarify the deamidation reaction at Gln residues and to understand the pathogenesis of age-related and autoimmune diseases.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/appliedchem1020011/s1. Table S1: title, Coordinates of all atoms in RC-A at B3LYP/6-31+G(d,p), Table S2: Coordinates of all atoms in TS1-A at B3LYP/6-31+G(d,p), Table S3: Coordinates of all atoms in INT1-A at B3LYP/6-31+G(d,p), Table S4: Coordinates of all atoms in RC-B at B3LYP/6-31+G(d,p), Table S5: Coordinates of all atoms in TS1-B at B3LYP/6-31+G(d,p), Table S6: Coordinates of all atoms in INT1-B at B3LYP/6-31+G(d,p), Table S7: Coordinates of all atoms in RC-C at B3LYP/6-31+G(d,p), Table S8: Coordinates of all atoms in TS1-C at B3LYP/6-31+G(d,p), Table S9: Coordinates of all atoms in INT1-C at B3LYP/6-31+G(d,p), Table S10: Coordinates of all atoms in INT2 at B3LYP/6-31+G(d,p), Table S11: Coordinates of all atoms in TS2 at B3LYP/6-31+G(d,p), Table S12: Coordinates of all atoms in PC at B3LYP/6-31+G(d,p), Table S13: Electronic and zero-point energies for all structures in CH3COGlnNH3CH3 at MP2/6-311+G(2d,2p)//B3LYP/6-31+G(d,p), Table S14: Electronic and zero-point energies for all structures in CH3COGlnNH3CH3 at MP2/6-311+G(2d,2p)//B3LYP/6-31+G(d,p).
Author Contributions
Conceptualization, K.K. and A.O.; methodology, K.K.; software, A.O.; validation, Y.I., E.K., T.N. and N.F.; formal analysis, H.A.; investigation, H.A.; resources, A.O.; data curation, K.K.; writing—original draft preparation, H.A.; writing—review and editing, K.K., T.N., A.O. and N.F.; visualization, H.A.; supervision, N.F.; project administration, K.K.; funding acquisition, K.K., A.O., Y.I. and N.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Grants-in-Aid for Scientific Research (17K08257, 19J23595, and 21K15244) from the Japan Society for the Promotion of Science.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Geiger, T.; Clarke, S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987, 262, 785–794. [Google Scholar] [CrossRef]
- Lindner, H.; Helliger, H. Age-dependent deamidation of asparagine residues in proteins. Exp. Gerontol. 2001, 36, 1551–1563. [Google Scholar] [CrossRef]
- Robinson, N.E.; Robinson, A.B. Molecular clocks. Proc. Natl. Acad. Sci. USA 2001, 98, 944–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, N.E. Protein deamidation. Proc. Natl. Acad. Sci. USA 2002, 99, 5283–5288. [Google Scholar] [CrossRef] [Green Version]
- Lampi, K.J.; Wilmarth, P.A.; Murray, M.R.; David, L.L. Lens β-crystallins: The role of deamidation and related modifications in aging and cataract. Prog. Biophys. Mol. Biol. 2014, 115, 21–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, W.G.; Papaconstantinou, J. Aging of α-crystallins during development of the lens. Proc. Natl. Acad. Sci. USA 1969, 64, 404–410. [Google Scholar] [CrossRef] [Green Version]
- Takemoto, L.; Boyle, D. Deamidation of specific glutamine residues from α-A crystallin during aging of the human lens. Biochemistry 1998, 37, 13681–13685. [Google Scholar] [CrossRef] [PubMed]
- Lapko, V.N.; Purkiss, A.G.; Smith, D.L.; Smith, J.B. Deamidation in human gamma S-crystallin from cataractous lenses is influenced by surface exposure. Biochemistry 2002, 41, 8638–8648. [Google Scholar] [CrossRef]
- Patel, K.; Borchardt, R.T. Chemical pathways of peptide degradation. II. Kinetics of deamidation of an asparaginyl residue in a model hexapeptide. Pharm. Res. 1990, 7, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, C.; O’Connor, P.B. Glutamine deamidation: Differentiation of glutamic acid and gamma-glutamic acid in peptides by electron capture dissociation. Anal. Chem. 2010, 82, 3606–3615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capasso, S.; Mazzarella, L.; Sica, F.; Zagari, A. First evidence of spontaneous deamidation of glutamine residue via cyclic imide to α- and γ-glutamic residue under physiological conditions. J. Chem. Soc. Chem. Commun. 1991, 23, 1667–1668. [Google Scholar] [CrossRef]
- Moss, C.X.; Matthews, S.P.; Lamont, D.J.; Watts, C. Asparagine deamidation perturbs antigen presentation on class II major histocompatibility complex molecules. J. Biol. Chem. 2005, 280, 18498–18503. [Google Scholar] [CrossRef] [Green Version]
- Doyle, H.A.; Gee, R.J.; Mamula, M.J. Altered immunogenicity of isoaspartate containing proteins. Autoimmunity 2007, 40, 131–137. [Google Scholar] [CrossRef]
- Doyle, H.A.; Aswad, D.W.; Mamula, M.J. Autoimmunity to isomerized histone H2B in systemic lupus erythematosus. Autoimmunity 2013, 46, 6–13. [Google Scholar] [CrossRef]
- Qin, Z.; Zhu, J.X.; Aswad, D.W. The d-isoAsp-25 variant of histone H2B is highly enriched in active chromatin: Potential role in the regulation of gene expression? Amino Acids 2016, 48, 599–603. [Google Scholar] [CrossRef] [Green Version]
- Diós, Á.; Elek, R.; Szabó, I.; Horváth, S.; Gyimesi, J.; Király, R.; Werkstetter, K.; Koletzko, S.; Fésüs, L.; Korponay-Szabó, I.R. Gamma-gliadin specific celiac disease antibodies recognize p31-43 and p57-68 alpha gliadin peptides in deamidation related manner as a result of cross-reaction. Amino Acids 2021, 53, 1051–1063. [Google Scholar] [CrossRef]
- Lerner, A.; Benzvi, C. Microbial transglutaminase is a very frequently used food additive and is a potential inducer of autoimmune/neurodegenerative diseases. Toxics 2021, 9, 233. [Google Scholar] [CrossRef] [PubMed]
- van Lummel, M.; Duinkerken, G.; van Veelen, P.A.; de Ru, A.; Cordfunke, R.; Zaldumbide, A.; Gomez-Touriño, I.; Arif, S.; Peakman, M.; Drijfhout, J.W.; et al. Posttranslational modification of HLA-DQ binding islet autoantigens in type 1 diabetes. Diabetes 2014, 63, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Boswell, C.A.; Tesar, D.B.; Mukhyala, K.; Theil, F.P.; Fielder, P.J.; Khawli, L.A. Effects of charge on antibody tissue distribution and pharmacokinetics. Bioconjug. Chem. 2010, 21, 2153–2163. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.D.; van Enk, J.Z.; Flynn, G.C. Human antibody Fc deamidation in vivo. Biologicals 2009, 37, 313–322. [Google Scholar] [CrossRef]
- Harris, R.J.; Kabakoff, B.; Macchi, F.D.; Shen, F.J.; Kwong, M.; Andya, J.D.; Shire, S.J.; Bjork, N.; Totpal, K.; Chen, A.B. Identification of multiple sources of charge heterogeneity in a recombinant antibody. J. Chromatogr. B Biomed. Sci. Appl. 2001, 752, 233–245. [Google Scholar] [CrossRef]
- Tran, J.C.; Tran, D.; Hilderbrand, A.; Andersen, N.; Huang, T.; Reif, K.; Hotzel, I.; Stefanich, E.G.; Liu, Y.; Wang, J. Automated affinity capture and on-tip digestion to accurately quantitate in vivo deamidation of therapeutic antibodies. Anal. Chem. 2016, 88, 11521–11526. [Google Scholar] [CrossRef]
- Du, Y.; Walsh, A.; Ehrick, R.; Xu, W.; May, K.; Liu, H. Chromatographic analysis of the acidic and basic species of recombinant monoclonal antibodies. MAbs 2012, 4, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.E.; Robinson, A.B. Molecular Clocks: Deamidation of Asparaginyl and Glutaminyl Residues in Peptides and Proteins; Althouse Press: Cave Junction, OR, USA, 2004. [Google Scholar]
- Robinson, N.E.; Robinson, Z.W.; Robinson, B.R.; Robinson, A.L.; Robinson, J.A.; Robinson, M.L.; Robinson, A.B. Structure-dependent non-enzymatic deamidation of glutaminyl and asparaginyl pentapeptides. J. Pept. Res. 2004, 63, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Scotchler, J.W.; Robinson, A.B. Deamidation of glutaminyl residues: Dependence on pH, temperature, and ionic strength. Anal. Biochem. 1974, 59, 319–322. [Google Scholar] [CrossRef]
- Pace, A.L.; Wong, R.L.; Zhang, Y.T.; Kao, Y.H.; Wang, Y.J. Asparagine deamidation dependence on buffer type, pH, and temperature. J. Pharm. Sci. 2013, 102, 1712–1723. [Google Scholar] [CrossRef]
- Kato, K.; Nakayoshi, T.; Kurimoto, E.; Oda, A. Computational studies on the non-enzymatic deamidation mechanisms of Glutamine residues. ACS Omega 2019, 4, 3508–3513. [Google Scholar] [CrossRef]
- Manabe, N.; Kirikoshi, R.; Takahashi, O. Glycolic acid-catalyzed deamidation of asparagine residues in degrading PLGA matrices: A computational study. Int. J. Mol. Sci. 2015, 16, 7261–7272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, O.; Manabe, N.; Kirikoshi, R. A computational study of the mechanism of succinimide formation in the Asn-His sequence: Intramolecular catalysis by the His side chain. Molecules 2016, 21, 327. [Google Scholar] [CrossRef] [Green Version]
- Kirikoshi, R.; Manabe, N.; Takahashi, O. Succinimide formation from an NGR-containing cyclic peptide: Computational evidence for catalytic roles of phosphate buffer and the arginine side chain. Int. J. Mol. Sci. 2017, 18, 429. [Google Scholar] [CrossRef] [Green Version]
- Nakayoshi, T.; Kato, K.; Kurimoto, E.; Oda, A. Possible mechanisms of non-enzymatic formation of dehydroalanine residue catalyzed by dihydrogen phosphate ion. J. Phys. Chem. B 2019, 123, 3147–3155. [Google Scholar] [CrossRef] [PubMed]
- Halim, M.A.; Almatarneh, M.H.; Poirier, R.A. Mechanistic study of the deamidation reaction of glutamine: A computational approach. J. Phys. Chem. B 2014, 118, 2316–2330. [Google Scholar] [CrossRef] [PubMed]
- Nakayoshi, T.; Kato, K.; Fukuyoshi, S.; Takahashi, O.; Kurimoto, E.; Oda, A. Molecular mechanisms of succinimide formation from aspartic acid residues catalyzed by two water molecules in the aqueous phase. Int. J. Mol. Sci. 2021, 22, 509. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Nakayoshi, T.; Ishikawa, Y.; Kurimoto, E.; Oda, A. Computational analysis of the mechanism of non-enzymatic peptide bond cleavage at the C-terminal side of an asparagine residue. ACS Omega 2021, 6, 30078–30084. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Revision A.03; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Hooi, M.Y.; Raftery, M.J.; Truscott, R.J. Age-dependent deamidation of glutamine residues in human γS crystallin: Deamidation and unstructured regions. Protein Sci. 2012, 21, 1074–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, K.; Nakayoshi, T.; Kurimoto, E.; Oda, A. Mechanisms of deamidation of asparagine residues and effects of main-chain conformation on activation energy. Int. J. Mol. Sci. 2020, 21, 7035. [Google Scholar] [CrossRef]
- Connolly, B.D.; Tran, B.; Moore, J.M.R.; Sharma, V.K.; Kosky, A. Specific catalysis of asparaginyl deamidation by carboxylic acids: Kinetic, thermodynamic, and quantitative structure-property relationship analyses. Mol. Pharm. 2014, 11, 1345–1358. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).