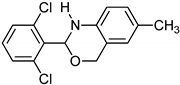
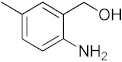
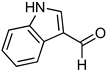
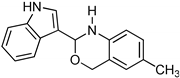
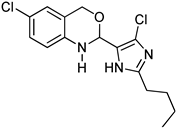
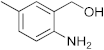
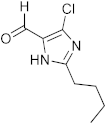
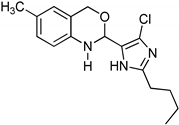
Abstract
Novel benzoxazines were synthesized by microwave irradiation and tested for their potential binding affinity towards receptors of advanced glycation end products (RAGE). We found that the compound (2-(2-bromophenyl)-6-methyl-2,4-dihydro-1H-benzo[d][1,3]oxazine) (3i) is a lead inhibitor of RAGE. Further, our in silico prediction that benzoxazines dock towards the AGE binding region of RAGE suggests that these ligands could bind effectively at the hydrophobic pocket of the receptor and additionally form key interactions with Arg48 and Arg104, revealing its diversity in developing anti-RAGE drugs to treat AGE–RAGE-dominant disease conditions. Functionally, we herein report the anti-tuberculosis activity of small molecules which could be bioactive in the culture of mycobacterium tuberculosis.
1. Introduction
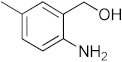
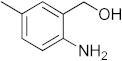
The recurrence of tuberculosis (TB) and the prevalence of diabetes mellitus (DM) is increasing in today’s world, which makes eliminating TB nearly impossible [1]. Diabetes mellitus is a disease categorized by the presence of elevated blood glucose levels due to the body’s inability to use or produce the insulin hormone. Type II diabetes is caused by the dysfunction of insulin receptors in which insulin receptor signaling is uncoupled from glucose uptake. Type II diabetes has been linked to vascular calcification through several different mechanisms such as hyperkalemia, hyperglycemia, oxidative stress, and hypercalcemia [2]. A major killer of mankind worldwide is diabetes and tuberculosis. The coexistence of DM and latent tuberculosis (LTBI) burdens global health, which is why routine bidirectional screenings are recommended [3]. Due to the impact of DM on both innate and adaptive immunity, patients with TB are prone to develop untreatable conditions with poor clinical outcomes and early death [4,5]. Inflammation, DM, and complications associated with it are caused by receptor for advanced glycation end product (RAGE), a multiligand receptor of the immunoglobulin superfamily [6]. Leukocytes, immune function, and infection response have been most extensively studied in relation to RAGE receptor signaling, one of the pathways implicated in diabetic complications [7]. RAGE is an immunoglobulin-like receptor that exists in numerous isoforms and binds to a variety of endogenous extracellular ligands and intracellular effectors. RAGE binds to multiple ligands such as AGE [8], phosphatidylserine [9], S100 proteins [10], lipopolysaccharides [11], high-mobility group box (HMGB)-1 [12], and amyloid-β (Aβ) [13]. Upon exposure to reducing sugars, advanced glycation end products (AGEs) are produced by the non-enzymatic glycation of proteins. Glycation mainly leads to loss of enzymatic function, aggregation, and protein cross-linking. The accumulation of AGEs plays a very important role in health disorders such as immune inflammation, diabetes mellitus, tuberculosis, and cardiovascular and neurodegenerative diseases [14]. RAGE mainly interacts with a variety of ligands because of its recognition of the 3D structures rather than the key amino acids of ligands. Therefore, RAGE was first known to act as a receptor for advanced glycation end products (AGEs). Ali Hafez Ali Mohammed El-Far et al. reported that papaverine works as a RAGE inhibitor in in vitro cell culture systems of RAGE-expressing HT1080 fibrosarcoma cells [15]. Azeliragon et al. also reported PF-04494700 or TTP488 will inhibit the interaction between RAGE and Aβ1−42, S100B, HMGB1, and CML [16,17]. Han et al. discovered pyrimidines as potential inhibitors of RAGE [18,19] and Choi et al. reported that benzoxazole derivatives inhibited the RAGE–Aβ interaction in vitro [20]. We reported oxazine as a potential anti-tumour agent that potentially mimics the pyranoside of sugar and binds VEGF, HB-EGF, and TNF-α [21] and also acted as a methionyl-t-RNA synthetase inhibitor [22]. We also reported the synthesis of benzoxazines and evaluated their anti-diabetic activities [23]. Previously reported methods of synthesis of oxazines require strong basic or acidic conditions, heating, and prolonged reaction times, herein we report the design and synthesis of benzoxazines under mild reaction conditions [24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41]. Further, our in vitro RAGE-binding studies, computational docking studies towards RAGE binding, and anti-tuberculosis evaluation of benzoxazines have revealed a newly discovered structure in RAGE-related pathways of many diseases, including DM and TB.
2. Materials and Methods
Chemicals
All chemicals used were of analytical grade and purchased from Sigma Aldrich, and SRL, Mumbai (India). 1H NMR spectra were recorded on an Agilent (400 MHz) spectrometer in CDCl3 solvent using TMS as an internal standard. Chemical shifts were expressed as δ ppm and abbreviations were assigned as follows: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, and J values are given in Hz. Mass spectra were determined on a Shimadzu LC-MS and ESI-MS. Elemental analyses were carried out using an Elemental Vario Cube CHNS Rapid Analyzer. Progress of the reaction was monitored by TLC pre-coated silica gel plates.
3. General Procedure for the Synthesis of 1,3-Benzoxazines (3a–3j)
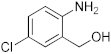
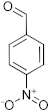
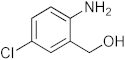
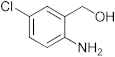
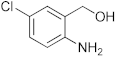
To a microwave tube equipped with a magnetic stir bar was added an equimolar mixture of 2-aminobenzylalcohol (1 mmol) and aromatic aldehyde (1 mmol) in acetic acid (5 mL) and irradiated in a microwave reactor at a 40 °C for 5 min at a maximum power of 320 W, and the reaction was monitored by using TLC. Then, after completion of the reaction the water was added to the reaction mixture and extracted with ethyl acetate and dried anhydrous sodium sulfate and the solvent was removed under reduced pressure, purified using column chromatography, and recrystallized from ethanol.
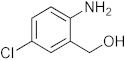
3.1. Synthesis of 6-Chloro-2-(4-nitrophenyl)-2,4-dihydro-1H-benzo[d][1,3]oxazine(3a)
The compound 3a was prepared from 2-amino-5-chlorobenzylalcohol and 4-nitrobenzaldehyde, yield 96%, melting point 75–77 °C. Elemental analysis calculated for C14H11ClN2O3: C, 57.83; H, 3.82; N, 9.65; found C, 57.99; H, 3.96; N, 9.51%, IR νmax (KBR,cm−1): 3260, 2895, 1485, 1264 cm−1; 1H NMR (CDCl3, 400 MHz): δ 8.08–8.06(d, 2H), 7.10(s, 1H), 7.08–6.98(d, 2H), 6.71–6.70(d, 2H), 5.28 (s, 1H), 5.06–5.02(d, 1H), 4.90–4.86(d, 1H), 4.13 (s, 1H); mass: m/z found for C14H11ClN2O3 was 291.0 (M+H)+, 293(M+2)+.
3.2. Synthesis of 6-Chloro-2-(2-methyl-1H-indol-3-yl)-2,4-dihydro-1H-benzo[d][1,3]oxazine(3b)
The compound 3b was prepared from 2-amino-5-chlorobenzylalcohol and 2-methylindole-3-carbaldehyde, yield 95%, melting point 96–98 °C. Elemental analysis calculated for C17H15ClN2O: C, 68.34; H, 5.06; N, 9.38; found C, 68.18; H, 4.91; N, 9.24%, IR νmax (KBR, cm−1): 3272, 2889, 1479, 1258 cm−1; 1H NMR (CDCl3, 400 MHz): δ 11.80 (s, 1H), 7.5–7.07(m, 7H), 5.15 (s, 1H), 4.98–4.92(m, 2H), 4.24 (s, 1H), 2.31(s, 3H); mass: m/z found for C17H15ClN2O was 299.0 (M+H)+.
3.3. Synthesis of 2-(2,6-Dichlorophenyl)-6-methyl-2,4-dihydro-1H-benzo[d][1,3]oxazine(3c)
The compound 3c was prepared from 2-amino-5-methylbenzylalcohol and 2,6-dichlorobenzaldehyde, yield 97%, melting point 61-63 °C. Elemental analysis calculated for C15H13Cl2NO: C, 61.25; H, 4.44; N, 4.75; found C, 61.12; H, 4.36; N, 4.61%, IR νmax (KBR, cm−1): 3266, 2881, 1471, 1264 cm−1; 1H NMR (CDCl3, 400 MHz): δ 7.50–6.41(m, 6H), 5.30 (s, 1H), 4.90–4.80(m, 2H), 4.12 (s, 1H), 2.32(s, 3H); mass: m/z found for C15H13Cl2NO was 294.0 (M+H)+.
3.4. Synthesis of 2-(1H-Indol-3-yl)-6-methyl-2,4-dihydro-1H-benzo[d][1,3]oxazine (3d)
The compound 3d was prepared from 2-amino-5-methylbenzylalcohol and indole-3-carbaldehyde, yield 94%, melting point 78–80 °C. Elemental analysis calculated for C17H16N2O: C, 77.24; H, 6.11; N, 10.61; found C, 77.09; H, 5.96; N, 10.45%, IR νmax (KBR, cm−1): 3260, 2875, 1467, 1265 cm−1; 1H NMR (CDCl3, 400 MHz): δ 10.15 (s,1H), 7.3–6.6(m, 8H), 5.25 (s, 1H), 4.84–4.74 (m, 2H), 4.10 (s, 1H), 2.33 (s, 3H); mass: m/z found for C17H16N2O was 265.0 (M+H)+.
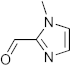
3.5. Synthesis of 6-Methyl-2-(1-methyl-1H-imidazol-5-yl)-2,4-dihydro-1H-benzo[d][1,3]oxazine(3e)
The compound 3e was prepared from 2-amino-5-methylbenzylalcohol and 1-methyl-1H-imidazole-5-carbaldehyde, yield 90%, melting point 63–64 °C. Elemental analysis calculated for C13H15N3O: C, 68.11; H, 6.60; N, 18.34; found C, 67.97; H, 6.47; N, 18.19%, IR νmax (KBR, cm−1): 3262, 2870, 1463, 1267 cm−1; 1H NMR (CDCl3, 400 MHz): δ 7.54 (s,1H), 7.20–6.55(m, 4H), 5.26 (s, 1H), 4.80–4.70 (m, 2H), 4.35 (s, 1H), 3.74(s, 3H), 2.34 (s, 3H); mass: m/z found for C13H15N3O was 230.0 (M+H)+.
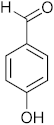
3.6. Synthesis of 4-(6-Methyl-2,4-dihydro-1H-benzo[d][1,3]oxazin-2-yl)phenol (3f)
The compound 3f was prepared from 2-amino-5-methylbenzylalcohol, 4-hydroxybenzaldehyde, yield 94%, melting point 78–80 °C. Elemental analysis calculated for C15H15NO2: C, 74.66; H, 6.26; N, 5.82; found C, 74.55; H, 6.35; N, 5.69%, IR νmax (KBR,cm−1): 3270, 2866, 1472, 1261 cm−1; 1H NMR (CDCl3, 400 MHz): δ 7.1–6.7 (m, 7H), 5.28(s, 1H), 5.02(s, 1H), 4.9–4.8(m, 2H), 4.13(s,1H), 2.3(s, 3H); mass: m/z found for C15H15NO2 was 242.0(M+H)+.
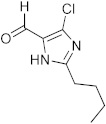
3.7. Synthesis of 2-(2-Butyl-4-chloro-1H-imidazol-5-yl)-6-chloro-2,4-dihydro-1H-benzo[d][1,3]oxazine (3g)
The compound 3g was prepared from 2-amino-5-chlorobenzylalcohol, 2-butyl-4-chloro-1H-imidazole-5-carbaldehyde, yield 90%, melting point 68–69 °C. Elemental analysis calculated for C15H17Cl2N3O: C, 55.23; H, 5.25; N, 12.88; found C, 55.16; H, 5.35; N, 5.09%, IR νmax (KBR, cm−1): 3274, 2860, 1470, 1268 cm−1; 1H NMR (CDCl3, 400 MHz): δ 10.1 (s, 1H), 7.2–7.1(m, 3H), 5.5(s, 1H), 4.7–4.6(m, 2H), 4.1(s, 1H), 2.61 (t, 2H), 1.60(m, 2H), 1.31(m, 2H), 0.81(t, 3H); mass: m/z found for C15H17Cl2N3O was 326.0 (M+H)+, 328.0 (M+2)+.
3.8. Synthesis of 2-(2-Butyl-4-chloro-1H-imidazol-5-yl)-6-methyl-2,4-dihydro-1H-benzo[d][1,3]oxazine (3h)
The compound 3 h was prepared from 2-amino-5-methylbenzylalcohol, 2-butyl-4-chloro-1H-imidazole-5-carbaldehyde, yield 94%, melting point 70–72 °C. Elemental analysis calculated for C16H20ClN3O: C, 62.84; H, 6.59; N, 13.74; found C, 62.75; H, 6.51; N, 13.60%, IR νmax (KBR, cm−1): 3276, 2872, 1462, 1270 cm−1; 1H NMR (CDCl3, 400 MHz): δ 12.60 (s, 1H), 7.31–7.05(m, 3H), 5.27(s, 1H), 4.70–4.60(m, 2H), 4.15(s, 1H),2.82(t, 2H), 2.34(s, 3H),1.66(m, 2H), 1.31(m, 2H), 0.99 (t, 3H); mass: m/z found for C16H20ClN3O was 306.0 (M+H)+, 308.0 (M+2)+.
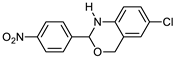
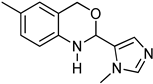
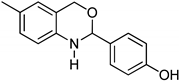
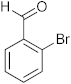
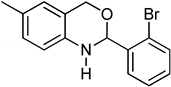
3.9. Synthesis of 2-(2-Bromophenyl)-6-methyl-2,4-dihydro-1H-benzo[d][1,3]oxazine (3i)
The compound 3i was prepared from 2-amino-5-methylbenzylalcohol, 2-bromobenzaldehyde, yield 93%, melting point 69–70 °C. Elemental analysis calculated for C15H14BrNO: C, 59.23; H, 4.64; N, 4.60; found C, 59.11; H, 4.45; N, 4.52%, IR νmax (KBR, cm−1): 3270, 2875, 1468, 1275 cm−1; 1H NMR (DMSO, 400 MHz): δ 7.5–7.0(m, 7H), 5.2(s, 1H), 4.8–4.6(m, 2H), 4.20(s, 1H), 2.30(s, 3H); mass: m/z found for C15H14BrNO was 304.0(M+H)+ and 306.0(M+2)+.
3.10. Synthesis of 6-Chloro-2-(1-methyl-1H-imidazol-5-yl)-2,4-dihydro-1H-benzo[d][1,3]oxazine (3j)
The compound 3j was prepared from 2-amino-5-chlorobenzylalcohol and 1-methyl-1H-imidazole-5-carbaldehyde, yield 90%, melting point 60–61 °C. Elemental analysis calculated for C12H12N3O: C, 57.72; H, 4.84; N, 16.83; found C, 57.82; H, 4.79; N, 16.60%, IR νmax (KBR, cm−1): 3265, 2868, 1459, 1263 cm−1; 1H NMR (DMSO, 400 MHz): δ 8.0–7.5 (m, 6H), 5.7 (s, 1H), 4.7–4.5 (m, 2H), 3.7(s, 3H); mass: m/z found for C12H12N3O was 250.0 (M+1)+.
3.11. AGE–RAGE Binding Assay
Inhibition of AGE–RAGE binding by small molecules was performed using enzyme-linked immunosorbent assay (ELISA). AGE was immobilized on 96-microtiter plates at room temperature. The wells were blocked with 3% bovine serum albumin (BSA) and/or 3% blocking reagent (Roche Applied Science) in PBS for 1 h at room temperature. During this time, RAGE was incubated with a small molecule inhibitor for 30 min. After washing with PBS containing 0.005% Tween 20 (PBS-T), the mixture of RAGE and small molecules was added, and the plate was incubated for another 1 h at room temperature. The reaction was carried out in PBS. Small molecules that inhibited the binding of RAGE to AGE through electrostatic or hydrophobic interactions were detected using antisera raised in rabbits against RAGE, followed by incubation with alkaline phosphatase-conjugated goat anti-rabbit IgG/IgM. Alkaline phosphatase activity was detected using p-nitrophenyl phosphate as a substrate, and the absorbance was measured at 415 nm.
3.12. Microplate Alamar Blue Assay
We screened a large number of benzoxazines for their effectiveness against the growth of M. tuberculosis H37Rv strain (ATCC 27294), as reported previously from our lab [42]. A volume of 200 mL of deionized water was added to sterile 96-well plates in order to humidify the medium and minimize evaporation. Middlebrook 7H9 broth was added to some wells, as well as 2X benzoxazine solutions, at varying concentrations. Incubation was performed at 37 °C for 5 days after sealing the plate with parafilm. Each well was then filled with a mixture of 10% Tween-80 and alamarBlue reagent (50 mL) for 24 h. Bacterial growth was characterized by a pink color, while compound inhibition of bacterial growth was characterized by a blue color. The inhibitory activity was expressed as percentage of minimum inhibitory concentrations (MICs) of benzoxazines against M. tuberculosis. Positive controls included streptomycin, pyrazinamide, and ciprofloxacin.
3.13. Molecular Docking
All of the compounds were docked to the crystal structure of human RAGE receptor (PDB: 3O3U) [18] using the MOE program (MOE) [43]. Protonation states of amino acids were assigned using protonate3D from MOE [44]. Ligands were ionized at physiological pH using MOE. The binding site was defined by the amino acids Lys43, Lys44, Arg48, and Arg104. These amino acids were shown to be in the active site of the receptor [45]. Docking calculations were carried out using MOE’s induced-fit protocol with default parameters.
4. Results and Discussion
4.1. Chemical Synthesis of Newer Benzoxazines
1,3-benzoxazines were synthesized from substituted 2-aminobenzyl alcohol and aromatic aldehydes in acetic acid under microwave irradiation (Scheme 1). The previously reported procedures have a longer reaction duration and require the presence of solvent (Scheme 1). A series of novel 1, 3 benzoxazines were synthesized as shown in Table 1. The compounds obtained were characterized by melting point, IR, 1H NMR, elemental analysis, and mass spectral analysis (Supplementary Information).
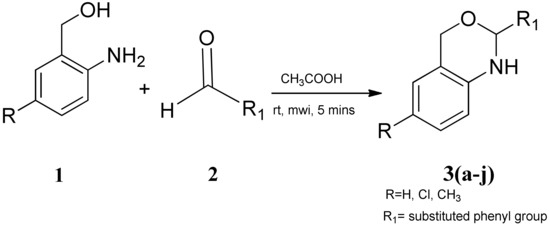
Scheme 1.
Synthesis of novel substituted benzoxazines from 3a–j.

Table 1.
List of newly synthesized benzoxazines.
4.2. Benzoxazines Inhibited the AGE–RAGE Binding in ELISA Method
An analysis of the 3D crystal structure of RAGE showed that it contains the positively charged surface present in the binding regions of ligands [46]. Further, Mizumoto et al. effectively determined the interaction of RAGE with the chondroitin/heparin sulphate glycosaminoglycans [47]. As a consequence of the above results, we speculate that the electron-rich nuclei containing small molecules can bind to RAGE and thus act as co-receptors. As a result, ELISA experiments were conducted with benzoxazines, which contain nitrogen and other heteroatoms utilizing the method established at Sugahara’s laboratory [48]. As demonstrated in Figure 1, compound 3i showed significant inhibition of the AGE–RAGE binding pattern, compared to the solvent (DMSO) control (Figure 1). Benzoxazines bearing a 2-bromo-phenyl group were found to be effective in disrupting the AGE–RAGE binding in vitro.
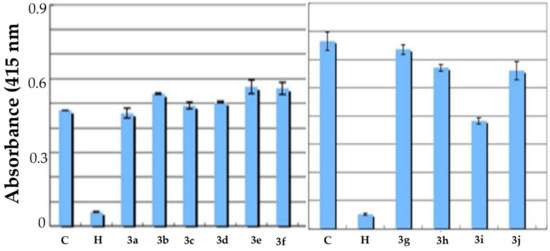
Figure 1.
Inhibitory effect of benzoxazines (compounds 3a-3j, 10 mg) on AGE binding to RAGE by the ELIZA method. C, DMSO control; H, heparin (0.5 mg).
Furthermore, our in vitro analysis found that compound 3i with 10–20 mg completely inhibited the binding of RAGE to AGE, indicating that the compound was targeting RAGE in a cell-free system (Figure 2). Hydrophobic and electronegative functional groups may be the reason for the inhibition of AGE–RAGE binding by benzoxazines. Heparin was used as a positive control.
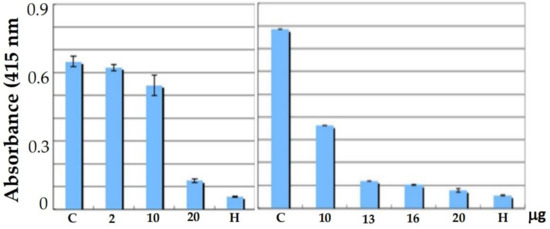
Figure 2.
Concentration-dependent inhibitory effect of benzoxazine 3i on AGE binding to RAGE by the ELIZA method. C, DMSO control; H, heparin (0.5 mg).
The highly sulfated glycosaminoglycan called chondroitin sulfate E is a well-known RAGE binding biomolecule, for which there are a few reports available in which the small molecules bind towards RAGE in vitro. In vitro RAGE inhibitory activity of pyrazole-5-carboxamides bearing 4-fluorophenoxy group showed the highest binding, when compared to other similar structures, as determined by the structure–activity relationship and surface plasmon resonance techniques [48]. We herein demonstrated the RAGE binding benzoxazine small molecule 3i by the in vitro ELISA method.
4.3. In Silico Analysis of Predicted Binding Poses of Benzoxazine Series Compounds to the RAGE Receptor
Docking calculations predicted two common binding modes for the benzoxazine series compounds. In the first binding pose, ligands occupy hydrophobic pocket near Arg48 and Arg104 and have key interactions with these two arginines. Significant interactions are the cation–pi interaction between Arg104 and the aromatic ring of the benzoaxine fragment, and the hydrogen bond interaction between Arg48 and imidazole and phenol moieties of the ligands. In the second binding pose, ligands are interacting mostly with Arg104. The benzene ring of the benzoxazine fragment forms a cation–pi interaction with the Arg104 sidechain. Arg104 also forms a hydrogen bond with the oxygen of the benzoxazine fragment. Interestingly, the two binding modes described here are similar to the predicted binding modes of 4,6-diphenylpyrimidine analogs to the RAGE receptor by Han et al. [18]. In conclusion, in silico prediction suggests that these ligands bind to a hydrophobic pocket at the surface of the RAGE protein and form key interactions with Arg48 and Arg104 (Figure 3).
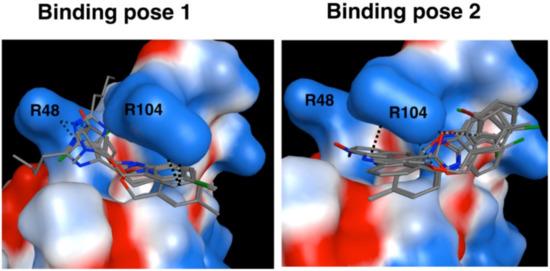
Figure 3.
Predicted binding poses of benzoxazine series compounds to the RAGE receptor. RAGE was represented by a molecular surface map based on electrostatic properties; from blue to red, electrostatic potential goes from positive to negative. Dashed green lines show representative H-bond and cation–pi interactions between RAGE and compounds.
4.4. Anti-Tuberculosis Activity of Small Molecules That Target RAGE
Previous research found that TB patients had higher levels of sRAGE when compared to controls. AGE expression was higher in the lungs of the TB infected patients. Therefore, we were interested in using RAGE inhibitors as anti-tuberculosis agents. In addition, some of the benzoxazines were reported as anti-TB agents. Therefore, we evaluated the anti-TB activity of all the benzoxazines against M. tuberculosis H37Rv strain and results are presented in Table 2. Some of the compounds showed growth-inhibitory activity against the tested mycobacterium at lower MIC values. Streptomycin and pyrazinamide were used as positive controls. The compound 3i was found to be a better anti-tuberculosis agent, capable of targeting RAGE in vitro.

Table 2.
Inhibition of H37Rv strain of M. tuberculosis by small molecules.
In clinical studies, DM patients who had TB and were treated with metformin showed somewhat reduced levels of AGE and sRAGE in comparison to those who were not treated with metformin, suggesting that RAGE inhibitors might be useful for treating DM patients with TB. TB patients with DM acquired previously might benefit from compound 3i-related drug development. As well, when compared with patients who survived, the deceased TB patients had lower levels of soluble RAGE and higher levels of leptin [49], indicating that this discovery could shed light on developing new drugs with anti-RAGE properties.
5. Conclusions
Consequently, we report the synthesis of novel benzoxazine compounds using a microwave irradiation method, which is valuable for preparing benzoxazine compound libraries and testing their role in inhibiting RAGE. As a lead inhibitor, we found compound 3i (2-(2-bromophenyl)-6-methyl-2,4-dihydro-1H-benzo[d][1,3]oxazine), and in-silico predictions suggested that these ligands bound to a hydrophobic pocket on the surface of the RAGE protein and interacted with Arg48 and Arg104. In addition, the anti-tuberculosis activity of the benzoxazines was also studied. Overall, this work may be useful for the identification of lead molecules for RAGE inhibition.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/applmicrobiol3010017/s1. Figure S1: 1H NMR of compound 3a. Figure S2: ESI-MS of compound 3a. Figure S3: 1H NMR of compound 3b. Figure S4: ESI-MS of compound 3b. Figure S5: ESI-MS of compound 3c. Figure S6: 1H NMR of compound 3d. Figure S7: ESI-MS of compound 3d. Figure S8: ESI-MS of compound 3e. Figure S9: 1H NMR of compound 3f. Figure S10: ESI-MS of compound 3f. Figure S11: 1H NMR of compound 3g. Figure S12: ESI-MS of compound 3g. Figure S13: ESI-MS of compound 3h. Figure S14: 1H NMR of compound 3i. Figure S15: ESI-MS of compound 3i. Figure S16: 1H NMR of compound 3j. Figure S17: ESI-MS of compound 3j.
Author Contributions
H.B., S.M. and S.B. performed the experiments. B.B. and P.E.L. designed the experiments, provided resources, and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
H.B. thanks UGC and CSIR for research fellowship. Basappa thanks DBT-NER and the Vision Group on Science and Technology (CESEM), Government of Karnataka for funding. The authors also acknowledge late Dr. Kazuyuki Sughahara and Prof. Yamada S. for providing support in evaluating the AGE–RAGE binding studies.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
The synthesized compounds are available at Basappa Laboratory.
References
- Kumar, N.P.; Moideen, K.; Nancy, A.; Viswanathan, V.; Shruthi, B.S.; Sivakumar, S.; Hissar, S.; Kornfeld, H.; Babu, S. Systemic RAGE ligands are upregulated in tuberculosis individuals with diabetes co-morbidity and modulated by anti-tuberculosis treatment and metformin therapy. BMC Infect. Dis. 2019, 19, 1039. [Google Scholar] [CrossRef]
- Kay, A.M.; Simpson, C.L.; Stewart, J.A. The Role of AGE/RAGE Signaling in Diabetes-Mediated Vascular Calcification. J. Diabetes Res. 2016, 2016, 6809703. [Google Scholar] [CrossRef]
- Dooley, K.E.; Chaisson, R.E. Tuberculosis and diabetes mellitus: Convergence of two epidemics. Lancet Infect. Dis. 2009, 9, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.A.; Harries, A.D.; Jeon, C.Y.; Hart, J.E.; Kapur, A.; Lönnroth, K.; Ottmani, S.-E.; Goonesekera, S.D.; Murray, M.B. The impact of diabetes on tuberculosis treatment outcomes: A systematic review. BMC Med. 2011, 9, 81. [Google Scholar] [CrossRef]
- Martinez, N.; Kornfeld, H. Diabetes and immunity to tuberculosis. Eur. J. Immunol. 2014, 44, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.M.; Yan, S.D.; Yan, S.F.; Stern, D.M. The multiligand receptor RAGE as a progression factor amplifying immune and inflammatory responses. J. Clin. Investig. 2001, 108, 949–955. [Google Scholar] [CrossRef]
- Berbudi, A.; Rahmadika, N.; Tjahjadi, A.; Ruslami, R. Type 2 Diabetes and its Impact on the Immune System. Curr. Diabetes Rev. 2020, 16, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.M.; Vianna, M.; Gerlach, M.; Brett, J.; Ryan, J.; Kao, J.; Esposito, C.; Hegarty, H.; Hurley, W.; Clauss, M. Isolation and characterization of two binding proteins for advanced glycosylation end products from bovine lung which are present on the endothelial cell surface. J. Biol. Chem. 1992, 267, 14987–14997. [Google Scholar] [CrossRef]
- He, M.; Kubo, H.; Morimoto, K.; Fujino, N.; Suzuki, T.; Takahasi, T.; Yamada, M.; Yamaya, M.; Maekawa, T.; Yamamoto, Y.; et al. Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells. EMBO Rep. 2011, 12, 358–364. [Google Scholar] [CrossRef]
- Hofmann, M.A.; Drury, S.; Fu, C.; Qu, W.; Taguchi, A.; Lu, Y.; Avila, C.; Kambham, N.; Bierhaus, A.; Nawroth, P.; et al. RAGE mediates a novel proinflammatory axis: A central cell surface receptor for S100/calgranulin polypeptides. Cell 1999, 97, 889–901. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Harashima, A.; Saito, H.; Tsuneyama, K.; Munesue, S.; Motoyoshi, S.; Han, D.; Watanabe, T.; Asano, M.; Takasawa, S.; et al. Septic shock is associated with receptor for advanced glycation endproducts (RAGE) ligation of LPS. J. Immunol. 2011, 186, 3248–3257. [Google Scholar] [CrossRef] [PubMed]
- Hori, O.; Brett, J.; Slattery, T.; Cao, R.; Zhang, J.; Chen, J.; Nagashima, M.; Lundh, E.R.; Vijay, S.; Nitecki, D.; et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin: Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J. Biol. Chem. 1995, 270, 25752–25761. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.D.; Chen, X.; Fu, J.; Chen, M.; Zhu, H.; Roher, A.; Slattery, T.; Zhao, L.; Nagashima, M.; Morser, J.; et al. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer’s disease. Nature 1996, 382, 685–691. [Google Scholar] [CrossRef]
- Bongarzone, S.; Savickas, V.; Luzi, F.; Gee, A.D. Targeting the Receptor for Advanced Glycation Endproducts (RAGE): A Medicinal Chemistry Perspective. J. Med. Chem. 2017, 60, 7213–7232. [Google Scholar] [CrossRef]
- El-Far, A.H.A.M.; Munesue, S.; Harashima, A.; Sato, A.; Shindo, M.; Nakajima, S.; Inada, M.; Tanaka, M.; Takeuchi, A.; Tsuchiya, H.; et al. In vitro anticancer effects of a RAGE inhibitor discovered using a structure-based drug design system. Oncol. Lett. 2018, 15, 4627–4634. [Google Scholar] [CrossRef] [PubMed]
- Galasko, D.; Bell, J.; Mancuso, J.Y.; Kupiec, J.W.; Sabbagh, M.N.; van Dyck, C.; Thomas, R.G.; Aisen, P.S. Alzheimer’s Disease Cooperative, S. Clinical trial of an inhibitor of RAGE-Abeta interactions in Alzheimer disease. Neurology 2014, 82, 1536–1542. [Google Scholar] [CrossRef]
- Sabbagh, M.N.; Agro, A.; Bell, J.; Aisen, P.S.; Schweizer, E.; Galasko, D. PF-04494700, an oral inhibitor of receptor for ad-vanced glycation end products (RAGE), in Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2011, 25, 206–212. [Google Scholar] [CrossRef]
- Han, Y.T.; Choi, G.-I.; Son, D.; Kim, N.-J.; Yun, H.; Lee, S.; Chang, D.J.; Hong, H.-S.; Kim, H.; Ha, H.-J.; et al. Ligand-Based Design, Synthesis, and Biological Evaluation of 2-Aminopyrimidines, a Novel Series of Receptor for Advanced Glycation End Products (RAGE) Inhibitors. J. Med. Chem. 2012, 55, 9120–9135. [Google Scholar] [CrossRef]
- Han, Y.T.; Kim, K.; Son, D.; An, H.; Kim, H.; Lee, J.; Park, H.-J.; Lee, J.; Suh, Y.-G. Fine tuning of 4,6-bisphenyl-2-(3-alkoxyanilino)pyrimidine focusing on the activity-sensitive aminoalkoxy moiety for a therapeutically useful inhibitor of receptor for advanced glycation end products (RAGE). Bioorganic Med. Chem. 2015, 23, 579–587. [Google Scholar] [CrossRef]
- Choi, K.; Lim, K.S.; Shin, J.; Kim, S.H.; Suh, Y.-G.; Hong, H.-S.; Kim, H.; Ha, H.-J.; Kim, Y.-H.; Lee, J.; et al. 6-Phenoxy-2-phenylbenzoxazoles, novel inhibitors of receptor for advanced glycation end products (RAGE). Bioorganic Med. Chem. 2015, 23, 4919–4935. [Google Scholar] [CrossRef]
- Murugan, S.; Kavitha, C.V.; Purushothaman, A.; Nevin, K.G.; Sugahara, K.; Rangappa, K.S. A small Oxazine compound as an anti-tumor agent:A novel pyranoside mimetic that bind to VEGF, HB-EGF and TNF-α. Cancer Lett. 2010, 297, 231–243. [Google Scholar]
- Bharathkumar, H.; Mohan, C.D.; Rangappa, S.; Kang, T.; Keerthy, H.K.; Fuchs, J.E.; Kwon, N.H.; Bender, A.; Kim, S.; Rangappa, K.S. Screening of quinoline, 1,3-benzoxazine, and 1,3-oxazine-based small molecules against iso-lated methionyl-tRNA synthetase and A549 and HCT116 cancer cells including an in silico binding mode analysis. Org. Biomol. Chem. 2015, 13, 9381–9387. [Google Scholar] [CrossRef] [PubMed]
- Bharathkumar, H.; Sundaram, M.S.; Jagadish, S.; Paricharak, S.; Hemshekhar, M.; Mason, D.; Kemparaju, K.; Girish, K.S.; Bender, A.; Rangappa, K.S. Novel benzoxazines, Novel Benzoxazine-Based Aglycones Block Glucose Uptake In Vivo by Inhibiting Glycosidases. PLoS ONE 2014, 9, e102759. [Google Scholar] [CrossRef]
- Sukhorukov, A.Y.; Nirvanappa, A.C.; Swamy, J.; Ioffe, S.L.; Nanjunda Swamy, S.; Basappa Rangappa, K.S. Synthesis and characterization of novel 1,2-oxazine-based small molecules that targets acetylcholinesterase. Bioorg. Med. Chem. Lett 2014, 1–24, 3618–3621. [Google Scholar] [CrossRef]
- Nirvanappa, A.C.; Mohan, C.D.; Rangappa, S.; Ananda, H.; Sukhorukov, A.Y.; Shanmugam, M.K.; Sundaram, M.S.; Nayaka, S.C.; Girish, K.S.; Chinnathambi, A.; et al. Novel Synthetic Oxazines Target NF-κB in Colon Cancer In Vitro and Inflammatory Bowel Disease In Vivo. PLoS ONE 2017, 6–12, e0175659. [Google Scholar] [CrossRef]
- Subramanian, G.; Rajeev, C.P.B.; Mohan, C.D.; Sinha, A.; Chu, T.T.; Anusha, S.; Ximei, H.; Fuchs, J.E.; Bender, A.; Rangappa, K.S.; et al. Synthesis and in vitro evaluation of hydrazinyl phthalazines against malaria parasite, Plasmodium falciparum. Bioorg. Med. Chem. Lett. 2016, 26, 3300–3306. [Google Scholar] [CrossRef] [PubMed]
- Fongmoon, D.; Shetty, A.K.; Basappa; Yamada, S.; Sugiura, M.; Kongtawelert, P.; Sugahara, K. Chondroitinase-mediated Degradation of Rare 3-O-Sulfated Glucuronic Acid in Functional Oversulfated Chondroitin Sulfate K and E. J. Biol. Chem. 2007, 282, 36895–36904. [Google Scholar] [CrossRef] [PubMed]
- Baburajeev, C.P.; Mohan, C.D.; Ananda, H.; Rangappa, S.; Fuchs, J.E.; Jagadish, S.; Siveen, K.S.; Chinnathambi, A.; Alharbi, S.A.; Zayed, M.E.; et al. Development of Novel Triazolo-Thiadiazoles from Heterogeneous “Green” Catalysis as Protein Tyrosine Phosphatase 1B Inhibitors. Sci. Rep. 2015, 5, 14195. [Google Scholar] [CrossRef]
- Kanchugarakoppal, S.R.; Basappa. New cholinesterase inhibitors: Synthesis and structure-activity relationship studies of 1,2-benzisoxazole series and novel imidazolyl-d 2-isoxazolines. J. Phys. Organ. Chem. 2005, 18, 773–778. [Google Scholar] [CrossRef]
- Basappa; Kavitha, C.; Rangappa, K. Simple and an efficient method for the synthesis of 1-[2-dimethylamino-1-(4-methoxy-phenyl)-ethyl]-cyclohexanol hydrochloride: (±) venlafaxine racemic mixtures. Bioorganic Med. Chem. Lett. 2004, 14, 3279–3281. [Google Scholar] [CrossRef]
- Sadashiva, M.P.; Basappa, B.; NanjundaSwamy, S.; Li, F.; Manu, K.A.; Sengottuvelan, M.; Prasanna, D.S.; Anilkumar, N.C.; Sethi, G.; Sugahara, K.; et al. Anti-cancer activity of novel dibenzo[b,f]azepine tethered isoxazoline derivatives. BMC Chem. Biol. 2012, 12, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Pandey, V.; Wang, B.; Mohan, C.D.; Raquib, A.R.; Rangappa, S.; Srinivasa, V.; Fuchs, J.E.; Girish, K.S.; Zhu, T.; Bender, A.; et al. Discovery of a small-molecule inhibitor of specific serine residue BAD phosphorylation. Proc. Natl. Acad. Sci. USA 2018, 115, E10505–E10514. [Google Scholar] [CrossRef]
- Blanchard, V.; Chevalier, F.; Imberty, A.; Leeflang, B.R.; Basappa; Sugahara, K.; Kamerling, J.P. Conformational Studies on Five Octasaccharides Isolated from Chondroitin Sulfate Using NMR Spectroscopy and Molecular Modeling. Biochemistry 2007, 46, 1167–1175. [Google Scholar] [CrossRef]
- Anusha, S.; Mohan, C.D.; Ananda, H.; Baburajeev, C.; Rangappa, S.; Mathai, J.; Fuchs, J.E.; Li, F.; Shanmugam, M.K.; Bender, A.; et al. Adamantyl-tethered-biphenylic compounds induce apoptosis in cancer cells by targeting Bcl homologs. Bioorganic Med. Chem. Lett. 2016, 26, 1056–1060. [Google Scholar] [CrossRef]
- Basappa; Sugahara, K.; Thimmaiah, K.N.; Bid, H.K.; Houghton, P.J.; Rangappa, K.S. Anti-Tumor Activity of a Novel HS-Mimetic-Vascular Endothelial Growth Factor Binding Small Molecule. PLoS ONE 2012, 7, e39444. [Google Scholar] [CrossRef]
- Sebastian, A.; Pandey, V.; Mohan, C.D.; Chia, Y.T.; Rangappa, S.; Mathai, J.; Baburajeev, C.P.; Paricharak, S.; Mervin, L.H.; Bulusu, K.C.; et al. Novel Adamantanyl-Based Thiadiazolyl Pyrazoles Targeting EGFR in Triple-Negative Breast Cancer. ACS Omega 2016, 1, 1412–1424. [Google Scholar] [CrossRef] [PubMed]
- Rakesh, K.S.; Jagadish, S.; Vinayaka, A.C.; Hemshekhar, M.; Paul, M.; Thushara, R.M.; Sundaram, M.S.; Swaroop, T.R.; Mohan, C.D.; Basappa; et al. A New Ibuprofen Derivative Inhibits Platelet Aggregation and ROS Mediated Platelet Apoptosis. PLoS ONE 2014, 9, e107182. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.A.; Jayarama, S.; Basappa; Salimath, B.P.; Rangappa, K.S. Pro-apoptotic activity of imidazole derivatives mediated by up-regulation of Bax and activation of CAD in Ehrlich Ascites Tumor cells. Investig. New Drugs 2007, 25, 343–350. [Google Scholar] [CrossRef]
- Mohan, C.D.; Bharathkumar, H.; Dukanya Rangappa, S.; Shanmugam, M.K.; Chinnathambi, A.; Alharbi, S.A.; Alahmadi, T.A.; Bhattacharjee, A.; Lobie, P.E.; Deivasigamani, A.; et al. N-Substituted Pyrido-1,4-Oxazin-3-Ones Induce Apoptosis of Hepatocellular Carcinoma Cells by Targeting NF-κB Signaling Pathway. Front. Pharmacol. 2018, 5, 1125. [Google Scholar] [CrossRef]
- Basappa Rangappa, K.S.; Sugahara, K. Roles of glycosaminoglycans and glycanmimetics in tumor progression and metastasis. Glycoconj J. 2014, 31, 461–467. [Google Scholar] [CrossRef]
- Basappa, B.; Mantelingu, K.; Sadashiva, M.; Rangappa, K. A Simple and Efficient Method for the Synthesis of 1,2-Benzisoxazoles: A Series of Its Potent Acetylcholinesterase Inhibitors. ChemInform 2005, 36. [Google Scholar] [CrossRef]
- Anusha, S.; Cp, B.; Mohan, C.D.; Mathai, J.; Rangappa, S.; Mohan, S.; Chandra Paricharak, S.; Mervin, L.; Fuchs, J.E.; Mahedra, M.; et al. A Nano-MgO and Ionic Liquid-Catalyzed ‘Green’ Synthesis Protocol for the Development of Adamantyl-Imidazolo-Thiadiazoles as Anti-Tuberculosis Agents Targeting Sterol 14α-Demethylase (CYP51). PLoS ONE 2015, 15–10, e0139798. [Google Scholar] [CrossRef] [PubMed]
- Chemical Computing Group Inc. Molecular Operating Environment (MOE) 2014.09; Chemical Computing Group Inc.: Montreal, QC, Canada, 2014. [Google Scholar]
- Labute, P. Protonate3D: Assignment of ionizationstates and hydrogen coordinates tomacromolecular structures. Proteins 2009, 75, 187–205. [Google Scholar] [CrossRef]
- Matsumoto, S.; Yoshida, T.; Murata, H.; Yamamoto, H.; Kobayashi, Y.; Ohkubo, T. Solution Structure of the Variable-Type Domain of the Receptor for Advanced Glycation End Products: New Insight into AGE−RAGE Interaction. Biochemistry 2008, 47, 12299–12311. [Google Scholar] [CrossRef]
- Xu, D.; Young, J.H.; Krahn, J.M.; Song, D.; Corbett, K.D.; Chazin, W.J.; Pedersen, L.C.; Esko, J.D. Stable RAGE-Heparan Sulfate Complexes Are Essential for Signal Transduction. ACS Chem. Biol. 2013, 8, 1611–1620. [Google Scholar] [CrossRef]
- Mizumoto, S.; Takahashi, J.; Sugahara, K. Receptor for Advanced Glycation End Products (RAGE) Functions as Receptor for Specific Sulfated Glycosaminoglycans, and Anti-RAGE Antibody or Sulfated Glycosaminoglycans Delivered in Vivo Inhibit Pulmonary Metastasis of Tumor Cells. J. Biol. Chem. 2012, 287, 18985–18994. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.T.; Kim, K.; Choi, G.-I.; An, H.; Son, D.; Kim, H.; Ha, H.-J.; Son, J.-H.; Chung, S.-J.; Park, H.-J.; et al. Pyrazole-5-carboxamides, novel inhibitors of receptor for advanced glycation end products (RAGE). Eur. J. Med. Chem. 2014, 79, 128–142. [Google Scholar] [CrossRef]
- da Silva, L.F.; Skupien, E.C.; Lazzari, T.K.; Holler, S.R.; de Almeida, E.G.C.; Zampieri, L.R.; Coutinho, S.E.; Andrades, M.; Silva, D.R. Advanced glycation end products (AGE) and receptor for AGE (RAGE) in patients with active tuberculosis, and their relationship between food intake and nutritional status. PLoS ONE 2019, 14, e0213991. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).