Abstract
Peptic ulcer disease, affecting almost 20% of the worldwide population, depicts an urgent need for effective treatment due to the limited therapeutic options available and the side effects associated with current drugs. The disease is often linked with Helicobacter pylori infection and NSAID usage, both of which compromise the mucosal lining of the stomach. There is growing evidence that dietary polyphenols can contribute to the prevention and management of various chronic diseases, including cancer and gastrointestinal disorders. Among these, green tea has garnered significant attention due to its rich polyphenolic content and associated health benefits. The abundance of green tea polyphenols (GTPs) exhibits chemoprotective, antimicrobial, and antioxidant properties. This study explores a set of 65 GTPs against penicillin-binding proteins (PBPs) as a molecular target to prevent peptic ulceration. Our molecular docking analysis revealed that the polyphenol ‘Epigallocatechin gallate’ (EGCG) exhibited effective binding affinity towards PBPs (PDB code: 1QMF), with a docking score of (−17.23 kcal/mol), followed then by Theaflavin-3-gallate (−16.57 kcal/mol) and Epigallocatechin (−15.91 kcal/mol). In silico ADME profiling indicated favorable pharmacokinetics for EGCG, including no AMES toxicity, low hERG inhibition, and good intestinal absorption. Our study highlights EGCG as a potential inhibitor of H. pylori, providing a promising natural therapeutic candidate for the management of peptic ulcer disease.
1. Introduction
Helicobacter pylori infect up to 80% of children in developing countries, spreading via fecal–oral or oral–oral routes. Prevalence is higher with poor sanitation, aging, and among African Americans and Hispanics in the U.S., with no sex difference. Infection persists lifelong if untreated [1]. About 10–15% develop peptic ulcers, while gastric cancer is rarer and influenced by additional factors. H. pylori is also linked to gastric non-Hodgkin’s lymphomas, though these constitute under 3% of malignancies [2]. This bacillus is spiral-shaped and known as a major etiological cause of chronic gastritis. Even though we have conventional therapy, there are many severe side effects associated, raising concerns regarding drug toxicity. This conventional therapy would also have a major issue of drug resistance [3]; henceforth, there is an urgent need to develop alternative strategies comprising use of plant-derived extracts and phytochemicals, which recently have gained interest [4]. Flavonoid-rich extracts are of special focus due to their multiple mechanisms of action against H. pylori. Besides antibacterial activity, flavonoids enhance mucosal defense by exerting cytoprotective, antioxidative, and anti-inflammatory effects. Individual flavonoids often display multi-target anti-ulcer activity, including protection of intestinal barrier integrity, modulation of gastric secretions, regulation of enzymatic activity, immune modulation, and interference with microbial colonization [5]. Penicillin-binding proteins (PBPs), essential for bacterial cell wall maintenance, provide an additional therapeutic target [6]. Their allosteric binding sites facilitate conformational changes that increase substrate accessibility. Identifying inhibitors capable of binding both active and allosteric sites offer a promising therapeutic strategy. Green tea extracts, particularly catechins, exhibit inhibitory effects on H. pylori growth in vitro. Studies suggest that green tea intake, either prior to or following infection, can prevent or reduce gastric mucosal inflammation [7,8,9]. Its polyphenols suppress H. pylori-induced proliferation of gastric epithelial cells and inhibit urease activity, an enzyme critical for bacterial colonization and survival [1]. In this study, we evaluated phytomolecules from Green Tea against PBPs, exploring dual-site binding interactions to reveal novel inhibitory profiles. Further, in silico ADME analyses were conducted to assess pharmacokinetic properties, supporting the potential of flavonoids as adjuncts to conventional therapy.
2. Materials and Methods
2.1. Molecular Docking
For the current work, we docked 65 Green Tea phytomolecules (GTPs) from the Camellia sinensis plant, reported in various studies. These phytomolecules would then be drawn for their 2D structures in ‘ChemDraw 12.0 V’, subsequently converted to their 3D structures using ‘Chem3D’, and optimized further using ‘Avogadro software’. For the 3D crystal structure of protein, we downloaded the penicillin-binding protein (PDB code: 1QMF) from the Protein Database Bank (https://www.rcsb.org/structure/1QMF; accessed on 20 August 2025). The processing for the protein was performed using the ‘automated functionality’ in ‘MzDock V.2’, 2025 tool (https://sourceforge.net/projects/mzdock/; accessed on 20 August 2025), which is ‘a GUI-based pipeline for simulations’ [10]. The visualization for docking interactions were performed through ‘BIOVIA Discovery Studio 2025’ Visualizer.
2.2. In Silico Drug-Likeness and ADMET Analysis
We have screened the top three best-docked phytomolecule candidates from C. sinensis for their theoretical/predicted ADME properties using the ‘SwissADME’ (http://www.swissadme.ch, accessed on 20 August 2025). For toxicity assessments, we used ‘admetSAR’ (https://lmmd.ecust.edu.cn/admetsar3/predict.php accessed date on 20 August 2025) [11].
2.3. BOILED-Egg Model Analysis
The BOILED-Egg (Brain Or IntestinaL EstimateD permeation method) model analysis (http://www.swissadme.ch, accessed on 20 August 2025) serves as a theoretical estimation of brain access and gastrointestinal absorption profiles for the molecule. Using these two physicochemical parameters, it simultaneously predicts intestinal absorption and brain penetration. Owing to its speed, accuracy, conceptual simplicity, and intuitive graphical output, the model readily supports molecular design. The BOILED-Egg approach is widely applicable, including in drug candidate evaluation and chemical library screening during early drug discovery.
3. Results and Discussion
3.1. Docking Interaction Analyses
Our molecular docking analysis of 65 GTPs on the penicillin-binding protein target suggested that epigallocatechin gallate (EGCG) had highest affinity (−17.23 kcal/mol), followed by Theaflavin-3-gallate (−16.57 kcal/mol) and Epigallocatechin (−15.91 kcal/mol) (Table 1 and Table 2).
Docking studies revealed a range of binding affinities (Table 1), with several polyphenolic compounds showing strong interactions. Isotheaflavin (−14.1 kcal/mol), Oolonghomobisflavan B (−12.5 kcal/mol), and Epigallocatechin-3,4-di-O-gallate (−12.0 kcal/mol) exhibited the highest binding scores, stabilized by extensive hydrogen bonding (Thr550, Ser337, Asn397) and hydrophobic contacts (Trp374, Tyr568, Ala551).
Other promising candidates included Myricetin (−11.4 kcal/mol), Epicatechin-3,5-di-O-gallate (−11.4 kcal/mol), epigallocatechin gallate (−17.23 kcal/mol), and Epiafzelechin-3-O-gallate (−11.0 kcal/mol), all of which engaged in multiple H-bonds with key residues (Gln452, Ser337, His394) along with π–π stacking and van der Waals stabilization. Moderate affinity compounds, such as Barringtogenol (−9.3 kcal/mol), Epicatechin gallate (−9.1 kcal/mol), and Isoquercetin (−9.6 kcal/mol), also demonstrated favorable binding. In contrast, smaller molecules like benzaldehyde (−4.8 kcal/mol), cis-3-hexanol (−4.1 kcal/mol), and diphenylamine (−3.9 kcal/mol) showed weak interactions with minimal stabilizing contacts. Docking-interaction diagrams (2D and 3D) for top three hits have been shown in Figure 1 and Figure 2.

Table 1.
Docking-interaction energies of 65 selected bio-active molecules and 1 FDA-approved drug for target penicillin-binding proteins (PBPs).
Table 1.
Docking-interaction energies of 65 selected bio-active molecules and 1 FDA-approved drug for target penicillin-binding proteins (PBPs).
| Molecules | Binding Affinity (kcal/mol) | Molecules | Binding Affinity (kcal/mol) |
|---|---|---|---|
| Oolonghomobisflavan A | Theaflavic Acid | −7.21 | |
| Theasinensin D | −9.74 | Barrigenol R1 | |
| Theaflavin-3-gallate | −16.57 | Barringtogenol | −9.3 |
| Isotheaflavin | −14.1 | Camelliagenin | −6.41 |
| Epigallocatechin-3,5-Di-O-Gallate | −6.67 | Gallocatechin | −6.14 |
| Oolonghomobisflavan B | −12.5 | Catechin | −6.01 |
| Cis-3-Hexenol | −4.1 | Epicatechin | −5.28 |
| Epigallocatechin-3,4-Di-O-Gallate | −12.0 | Epiafzelechin | −4.07 |
| Vicenin 2 | −5.33 | Quercetin | −9.02 |
| Epicatechin-3,5-Di-O-Gallate | −11.4 | Cryptoxanthin | −9.11 |
| Rutin | −5.98 | Myricetin | −11.4 |
| Proanthocyanidin | −5.33 | Apigenin | −4.99 |
| Pheophytin | −4.22 | Nerolidol | −4.36 |
| Benzaldehyde | −4.8 | Kaempferol | −3.10 |
| Epitheaflavic Acid 3′-Gallate | −4.19 | Theanine | −2.90 |
| Epigallocatechin Gallate | −17.23 | Ascorbic Acid | −2.11 |
| Theasinensin E | −7.32 | Quinic Acid | −1.09 |
| Myricitrin | −11.4 | Succinic Acid | −1.7 |
| Theaflavin | −2.23 | Methyl Salicylate | −5.37 |
| Epicatechin Gallate | −9.1 | Theobromine | −5.21 |
| Kaempferitrin | −4.73 | Caffeine | −5.78 |
| Isoquercetin | −9.6 | Xanthine | −5.59 |
| Epiafzelechin 3-O-Gallate | −11.0 | Linalool Oxide | −5.88 |
| Pheophorbide | −7.34 | Phenylacetaldehyde | −5.71 |
| Epigallocatechin 3-O-P-Coumarate | −7.25 | Methylxanthine | −5.66 |
| Pheophorbide | −5.55 | Theophylline | −5.69 |
| Oxalic Acid | −5.03 | Geraniol | −5.31 |
| Cryptoxanthin | −5.21 | Hexanal | −5.36 |
| Isovitexin | −5.19 | Diphenylamine | −3.9 |
| Vitexin | −5.01 | Trans-2-Hexenal | −5.99 |
| Chlorogenic Acid | −4.09 | Linalool | −6.03 |
| Coumaroyl Quinic Acid | −7.02 | Phenylethanol | −6.07 |
| Epigallocatechin | −15.91 | Amoxicillin (Std.) | −10.93 |

Table 2.
Energy contribution of the key residues computed by docking methodology.
Table 2.
Energy contribution of the key residues computed by docking methodology.
| Sr. No. | Molecules | Docking Score (kcal/mol) | Residues with Contribution Energy |
|---|---|---|---|
| 1. | Amoxicillin (Std.) | −10.93 | Thr 526, Trp 374, Ser 337, Ser 395, Thr 550, Met 527, and Tyr 595 |
| 2. | Theaflavin-3-gallate | −16.57 | Trp374; Arg372; Phe570; Thr550; Ser548; Ser337; Asp373 |
| 3. | Epigallocatechin | −15.91 | Asn377; Trp374; Thr550; Phe450 |
| 4 | Epigallocatechin Gallate (EGCG) (Best docked) | −17.23 | Trp374; Gln552;Phe450;Gln452;Ser337; Phe570;Tyr568;Asn377;Arg372;Gly451;Gly336;Glu334;Ala551;Lys340;Ser395;Thr526;Phe570 |
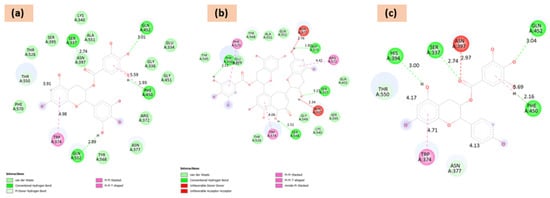
Figure 1.
Two-dimensional docking-interaction diagrams for (a) epigallocatechin gallate, (b) Theaflavin-3-gallate, and (c) epigallocatechin, respectively, against PBP.
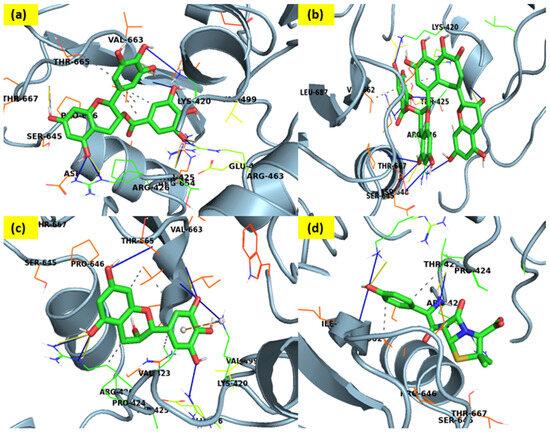
Figure 2.
Three-dimensional docking-interaction diagrams for (a) epigallocatechin gallate, (b) Theaflavin-3-gallate, (c) epigallocatechin, and (d) Amoxicillin, a standard drug, respectively, against PBP.
3.2. The ADME Analysis and BOILED-Egg
The in silico ADMET (absorption, distribution, metabolism, excretion, and toxicity) profiles of the top three docked hits are summarized in Table 3. The ADME analysis shows clear differences in drug-likeness among the compounds from Green Tea. Molecules with low TPSA (<90 Å2) and moderate lipophilicity (MLOGP ~1–3), such as apigenin, benzaldehyde, caffeine, geraniol, linalool, and methyl salicylate, exhibited high GI absorption and, in several cases, BBB permeability (apigenin, benzaldehyde, cis-3-hexenol, diphenylamine). Larger polyphenols (e.g., epigallocatechin gallates, rutin, proanthocyanidins) with high TPSA (>150 Å2) denoted poor GI absorption, no BBB penetration, multiple Lipinski violations, and reduced bioavailability scores (≤0.17), limiting oral drug potential (Supplementary file, Table S1).
Most small molecules are not P-gp substrates and generally non-inhibitors of major CYP isoforms, except for certain flavonoids (e.g., kaempferol, quercetin, myricetin) that inhibited CYP1A2, CYP2C9, or CYP3A4, suggesting possible drug–drug interaction liabilities. Compounds with good solubility and no Lipinski violations (e.g., ascorbic acid, theophylline, oxalic/succinic acid) demonstrated favorable oral bioavailability, though not all crossed the BBB. Theaflavin-3-gallate, epigallocatechin, and epigallocatechin gallate (EGCG) demonstrated favorable intestinal absorption (Figure 3), no Blood–Brain Barrier permeability, and were predicted to be non-carcinogenic, non-AMES toxic, with class IV acute oral toxicity.

Table 3.
Predictive ADMET analysis for top three best-docked hits.
Table 3.
Predictive ADMET analysis for top three best-docked hits.
| Properties | Theaflavin-3-Gallate | Epigallocatechin | Epigallocatechin Gallate (EGCG) * |
|---|---|---|---|
| CYP450 2C9 Substrate | Non-substrate | Non-substrate | Non-substrate |
| CYP450 2D6 Substrate | Non-substrate | Non-substrate | Non-substrate |
| CYP450 3A4 Substrate | Non-substrate | Non-substrate | Non-substrate |
| Human Ether-a-go-go-Related Gene Inhibition | Weak inhibitor | Weak inhibitor | Weak inhibitor |
| AMES Toxicity | Non-AMES toxic | Non-AMES toxic | Non-AMES toxic |
| Carcinogens | None | None | None |
| Acute Oral Toxicity | IV | IV | IV |
| P-glycoprotein Inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| Rat Acute Toxicity (LD50, mol/kg) | 2.6693 | 1.8700 | 2.6643 |
| Human Intestinal Absorption | + | + | + |
| AlogP | 3.19 | 1.25 | 2.23 |
| H-Bond Acceptor | 16 | 7 | 11 |
| H-Bond Donor | 11 | 6 | 8 |
| Tetrahymena pyriformis (pIGC50 (ug/L)) | 0.595 | 0.792 | 0.913 |
| Blood–Brain Barrier | - | - | - |
* Best docked.
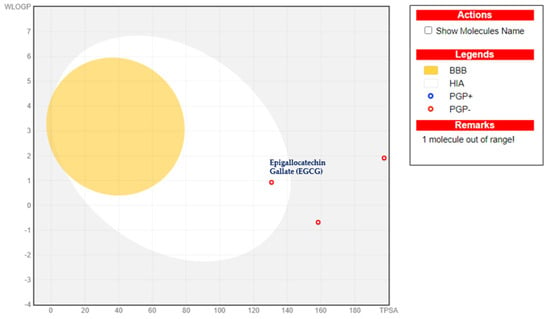
Figure 3.
BOILED−Egg model analysis for the best-docked epigallocatechin gallate (EGCG).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ecsoc-29-26744/s1, Table S1: ADMET Profile of phytochemicals.
Author Contributions
Conceptualization, P.A. and J.R.; methodology P.G. and S.N.M.; writing—original draft preparation, S.N.M. and R.S.; writing—review and editing S.N.M. and R.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data used in this work is already available in the manuscript.
Acknowledgments
Author thanks, ‘D. Y. Patil University, Navi Mumbai, India’ and School of Pharmacy, DYPU, Navi Mumbai, India for the provision of CADD facility.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yadav, S.; Pandey, A.; Mali, S.N. From Lab to Nature: Recent Advancements in the Journey of Gastroprotective Agents from Medicinal Chemistry to Phytotherapy. Eur. J. Med. Chem. 2024, 272, 116436. [Google Scholar] [CrossRef] [PubMed]
- Sugano, K.; Tack, J.; Kuipers, E.J.; Graham, D.Y.; El-Omar, E.M.; Miura, S.; Haruma, K.; Asaka, M.; Uemura, N.; Malfertheiner, P. Kyoto global consensus report on Helicobacter pylori gastritis. Gut 2015, 64, 1353–1367. [Google Scholar] [CrossRef] [PubMed]
- Dadgostar, P. Antimicrobial resistance: Implications and costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Valle, M.; Orellana-Palma, P.; Petzold, G. Plant-based polyphenols: Anti-Helicobacter pylori effect and improvement of gut microbiota. Antioxidants 2022, 11, 109. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Scortecci, K.C.; Boylan, F. A Review on Flavonoids as Anti-Helicobacter pylori Agents. Appl. Sci. 2025, 15, 3936. [Google Scholar] [CrossRef]
- Gerrits, M.M.; Schuijffel, D.; Van Zwet, A.A.; Kuipers, E.J.; Vandenbroucke-Grauls, C.M.J.E.; Kusters, J.G. Alterations in penicillin-binding protein 1A confer resistance to β-lactam antibiotics in Helicobacter pylori. Antimicrob. Agents Chemother. 2002, 46, 2229–2233. [Google Scholar] [CrossRef] [PubMed]
- Makarewicz, M.; Drożdż, I.; Tarko, T.; Duda-Chodak, A. The Interactions between polyphenols and microorganisms, especially gut microbiota. Antioxidants 2021, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Filosa, S.; di Meo, F.; Crispi, S. Polyphenols-gut microbiota interplay and brain neuromodulation. Neural Regen. Res. 2018, 13, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Mali, S.N.; Thorat, B.R.; Gupta, D.R.; Pandey, A. Mini-Review of the Importance of Hydrazides and Their Derivatives—Synthesis and Biological Activity. Eng. Proc. 2021, 11, 21. [Google Scholar] [CrossRef]
- Kabier, M.; Gambacorta, N.; Trisciuzzi, D.; Kumar, S.; Nicolotti, O.; Mathew, B. MzDOCK: A free ready-to-use GUI-based pipeline for molecular docking simulations. J. Comput. Chem. 2024, 45, 1980–1986. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).