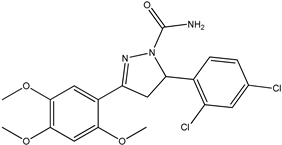
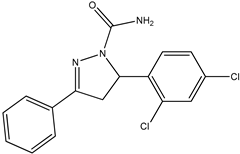
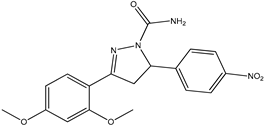
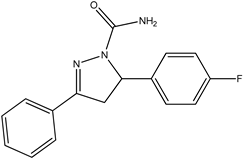
Abstract
Malaria, a devastating disease caused by Plasmodium parasites, continues to pose a significant threat to global health, with increasing resistance to current antimalarial drugs. In this study, we employed an in silico approach to design and evaluate novel 2-pyrazoline carboxamide derivatives as potential protease inhibitors against Plasmodium falciparum. Our results show that all the designed ligands exhibit good drug-like properties, satisfying Lipinski’s rule of five, and have low toxicity profiles. Molecular docking studies revealed that five newly designed ligands (P5, P6, P7, P11, and P13) exhibit promising binding affinities and interactions with key protease enzymes involved in the hemoglobin degradation pathway, including Falcipain-2, Falcipain-3, and Plasmepsin-2 with PDB (Protein Data Bank) codes of 6JW9, 3BWK, and 1LF3, respectively. Notably, ligand P13 showed the strongest binding affinity with Falcipain-2, forming an additional hydrogen bond with CYS42, an essential residue for the enzyme’s catalytic activity. The interactions between the ligands and the enzymes suggest a competitive inhibition mechanism, with the potential to disrupt the hemoglobin degradation pathway and halt the parasite’s lifecycle. The biological implications of these findings are significant, as they suggest that these novel ligands could be effective against Plasmodium parasites, particularly in the context of increasing resistance to current antimalarial drugs. Overall, this study provides valuable insights into the potential of novel 2-pyrazoline carboxamide derivatives to serve as protease inhibitors against Plasmodium parasites, highlights their potential as a promising strategy for antimalarial drug development, and demonstrates the importance of in silico approaches in the discovery of novel therapeutics.
1. Introduction
Malaria is caused by Plasmodium parasites and transmitted via Anopheles mosquitoes [1]. It remains a global health challenge, with 249 million cases and 608,000 fatalities in 2022 [2]. The emergence of drug-resistant strains of P. falciparum underscores the urgent requirement for novel antimalarial therapies [3]. Targeting parasite-specific pathways, such as protease enzymes, which are crucial for hemoglobin degradation and amino acid supply, has emerged as a promising strategy [4,5]. Pyrazoline derivatives, recognized for their broad biological activities, have shown potential in inhibiting FP-2, offering a pathway for the development of new antimalarial drugs [6,7].
2. Materials and Methods
2.1. In Silico Drug-likeness and Toxicity Predictions
The SwissADME platform (http://www.swissadme.ch/index.php accessed on 19 November 2023) was used to provide insights into ADME (Absorption, Distribution, Metabolism, and Excretion) parameters, pharmacokinetic properties, drug-likeness attributes, and the suitability of compounds from a medicinal chemistry perspective [8]. The assessment of toxicity was carried out using the ProTox 3.0 web tool (https://comptox.charite.de/protox3/ accessed on 19 November 2023), which identified potential toxicities by analyzing various components within the drug.
2.2. Molecular Docking Analysis
The ligand pyrazoline carboxamide was designed using Chem Draw Ultra 12.0, which was later subjected to Spartan14 for Energy Minimization and optimization and saved in mol2 format. These files were then processed with AutoDock tools to generate pdbqt files for molecular docking using AutoDock vina [9]. The crystal structures of the target proteins, Falcipain-2 (PDB ID: 6JW9) and Falcipain-3 (PDB ID: 3BWK), and Plasmepsin-2 (PDB ID: 1LF3) were obtained from the website www.rcsb.org. Water molecules were removed, and hydrogen atoms were added to the protein structure using UCSF Chimera. Molecular docking was performed using AutoDock Tools and Autodock Vina in the grid box size for falcipain-2 28Å × 24Å × 20Å centered at −8.889, 15.368, −38.694 (X, Y, Z coordinates), for falcipain-3 16Å × 16Å × 12Å centered at 5.96, −22.35, 50.07 (X, Y, Z coordinates), and for plasmepsin-2 12Å × 20Å × 18Å centered at 16.22, 6.85, 27.61 (X, Y, Z coordinates). BIOVIA Discovery studio 2020 Client was used to analyze the obtained conformations of the each docked complex ligand interaction.
3. Discussion
Table 1 shows that all the designed ligands satisfy Lipinski’s rule of five, indicating good drug-like properties. Their high gastrointestinal absorption (GI) potential suggests efficient oral bioavailability and their moderate lipophilicity (MLogP) suggests good solubility, while their hydrogen bond acceptors (HbA) may enhance their binding interactions. In the toxicity evaluation, the predicted LD50 values of all the designed ligands fell within the low-toxicity range.

Table 1.
Chemical descriptions of P5, P6, P7, P11, and P13.
Furthermore, the molecular docking results presented in this study offer valuable insights into the potential inhibitory effects of newly designed ligands (P5, P6, P7, P11, and P13) against key proteases of Plasmodium falciparum (Table 2). Falcipain-2 is a crucial cysteine protease involved in the hemoglobin degradation pathway of Plasmodium falciparum, the parasite responsible for malaria. By cleaving hemoglobin within the food vacuole, falcipain-2 provides the free amino acids necessary for parasite protein synthesis [10]. Falcipain-3 is also implicated in hemoglobin degradation. However, gene disruption studies suggest that falcipain-3 might have an indispensable function, possibly due to its role in erythrocyte invasion [11]. Plasmepsin-2 is another vital enzyme in the hemoglobin degradation pathway. It can be inhibited by preventing the supply of amino acids to the parasite, thereby halting its proliferation [12]. The inhibition of these enzymes is a promising strategy for antimalarial drug development, especially in the face of increasing drug resistance.

Table 2.
Analysis of the theoretical oral bioavailability of the designed compounds based on Lipinski’s rule of five, GI absorption, predicted LD50, and toxicity class.
The native ligands (E64, C1P, and EH5) serve as benchmarks for the docking study, providing a reference for the binding affinities and interactions expected of effective inhibitors. The newly designed ligands, P5, P6, P7, P11, and P13, were evaluated for their binding affinities and interactions with the active site residues of the target enzymes.
Falcipain-2 (PDB ID: 6JW9): The native ligand E64 showed moderate binding affinity, forming key hydrogen bonds with residues such as GLN36 and HIS174 (Table 3). These interactions are crucial to the stability of the enzyme–ligand complex and suggest a competitive inhibition mechanism. Among the newly designed ligands, P13 exhibited the strongest binding affinity (Table 3), which could be attributed to its additional hydrogen bond with CYS42, as presented in (Table 3), an essential residue for the catalytic activity of falcipain-2. This interaction not only enhances the binding affinity but also suggests a specificity that could translate to a potent inhibitory effect on the enzyme’s function in hemoglobin degradation.

Table 3.
Binding energies of designed pyrazolines carbaxomides and respective co-crystallized ligands.
Falcipain-3 (PDB ID: 3BWK): C1P, the native ligand, demonstrated a high binding affinity (Table 3), engaging in hydrogen bonds with residues like GLN45 and TRP215 (Table 3). These interactions are indicative of a natural regulatory mechanism of the enzyme’s activity. The newly designed ligands showed comparable binding affinities (Table 3), with interactions involving key residues such as TYR90 (Table 3). The consistent involvement of this residue across several ligands, as shown in Table 3, highlights its importance in the binding process and suggests its potential for effective inhibition, which could impact the parasite’s ability to invade red blood cells.
Plasmepsin-2 (PDB ID: 1LF3): EH5, the native ligand, exhibited a notably high binding affinity, forming extensive interactions with the enzyme (Table 3), including hydrogen bonds with VAL78 and SER79 (Table 3). Such interactions are indicative of a potent natural inhibition mechanism. The newly designed ligands, particularly P13, also demonstrated promising binding affinities and interactions. The hydrogen bond with ASP214, a residue that is critical to the enzyme’s activity, suggests that these ligands could effectively disrupt the hemoglobin degradation pathway, depriving the parasite of essential nutrients.
The biological implications of these findings are significant. By inhibiting the activity of these enzymes, the newly designed ligands could effectively halt the parasite’s lifecycle, leading to a cessation of disease progression. This is particularly crucial in the context of the increasing resistance to the current antimalarial drugs.
4. Conclusions
In conclusion, this molecular docking study presents compelling evidence that the newly designed ligands have the potential to serve as effective inhibitors of crucial malaria parasite proteases. Their strong binding affinities and specific interactions with key active site residues make them promising candidates for the development of new antimalarial drugs. Future work will involve the experimental validation of these in silico predictions to assess the therapeutic potential of these ligands.
Author Contributions
Conceptualization, Y.J., I.Y.A., A.N.H. and M.A.; methodology, Y.J., I.Y.A., A.N.H. and M.A.; software, Y.J., A.N.H., M.A., L.A.H., M.S.Y. and Z.S.; validation, Y.J., I.Y.A., A.N.H., M.A. and J.A.; formal analysis, Y.J., I.Y.A., A.N.H., M.A., J.A., L.A.H., M.S.Y. and Z.S.; investigation, Y.J., I.Y.A., A.N.H., M.A. and J.A.; resources, Y.J., A.N.H., L.A.H., M.S.Y. and Z.S.; writing original draft preparation, Y.J., I.Y.A., A.N.H. and M.A.; writing review and editing, Y.J., I.Y.A., A.N.H., M.A. and J.A.; supervision, I.Y.A., A.N.H. and M.A.; project administration, M.A, I.Y.A., A.N.H. and Y.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors are deeply grateful to the staff members of the Department of Pharmaceutical and Medicinal Chemistry, Ahmadu Bello University Zaria, Kaduna, Nigeria for the computational resources.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ravindar, L.; Hasbullah, S.A.; Rakesh, K.P.; Hassan, N.I. Pyrazole and pyrazoline derivatives as antimalarial agents: A key review. Eur. J. Pharm. Sci. 2023, 183, 106365. [Google Scholar] [CrossRef] [PubMed]
- WHO. The 2023 WHO World Malaria Report; WHO: Geneva, Switzerland, 2023. [Google Scholar]
- Adigun, R.A.; Malan, F.P.; Balogun, M.O.; October, N. Design, synthesis, and in silico-in vitro antimalarial evaluation of 1,2,3-triazole-linked dihydropyrimidinone quinoline hybrids. Struct. Chem. 2023, 34, 2065–2082. [Google Scholar] [CrossRef]
- Aggarwal, S.; Paliwal, D.; Kaushik, D.; Gupta, G.K.; Kumar, A. Synthesis, Antimalarial Evaluation and SAR Study of Some 1,3,5-Trisubstituted Pyrazoline Derivatives. Lett. Org. Chem. 2019, 16, 807–817. [Google Scholar] [CrossRef]
- Goud, P.M.; Sheri, A.; Desai, P.V.; Watkins, E.B.; Tekwani, B.; Sabnis, Y.; Avery, M.A. Design, synthesis and evaluation of trisubstituted thiazoles targeting Plasmodium falciparum cysteine proteases. Med. Chem. Res. 2005, 14, 74–105. [Google Scholar] [CrossRef]
- Himangini Pathak, D.P.; Sharma, V.; Kumar, S. Designing novel inhibitors against falcipain-2 of Plasmodium falciparum. Bioorganic Med. Chem. Lett. 2018, 28, 1566–1569. [Google Scholar] [CrossRef]
- Wiratama, M.; Satria, S.; Waskitha, W.; Haryadi, W.; Wahyuningsih, T.D. Synthesis, antimalarial activity assay and molecular docking study of N-substituted chloro-pyrazolines. Trop. J. Pharm. Res. 2022, 21, 1255–1261. [Google Scholar] [CrossRef]
- Boudou, F.; Sehmi, A.; Belakredar, A.; Zaoui, O. Synthesis, characterization, antimicrobial activity, and in silico assessment of a novel pyrazoline carboxamide heterocyclic compound. Bangladesh J. Pharmacol. 2023, 18, 152–161. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, P.J. Falcipain cysteine proteases of malaria parasites: An update. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140362. [Google Scholar] [CrossRef] [PubMed]
- Molina-Franky, J.; Patarroyo, M.E.; Kalkum, M.; Patarroyo, M.A. The Cellular and Molecular Interaction Between Erythrocytes and Plasmodium falciparum Merozoites. Front. Cell. Infect. Microbiol. 2022, 12, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.L.; Dans, M.G.; Balbin, J.M.; De, T.F.; Gilson, P.R.; Beeson, J.G.; Boyle, M.J.; Wilson, D.W. Targeting malaria parasite invasion of red blood cells as an antimalarial strategy. FEMS Microbiol. Rev. 2019, 43, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Avoiding Investment in Doomed Drugs. Curr. Drug Discov. 2001, 1, 17–19. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).