Abstract
Speech and language offer a rich, non-invasive window into brain health. Advances in machine learning (ML) have enabled increasingly accurate detection of neurodevelopmental and neurodegenerative disorders through these modalities. This paper envisions the future of ML in the early detection of neurodevelopmental disorders like autism spectrum disorder and attention-deficit/hyperactivity disorder, and neurodegenerative disorders, such as Parkinson’s disease and Alzheimer’s disease, through speech and language biomarkers. We explore the current landscape of ML techniques, including deep learning and multimodal approaches, and review their applications across various conditions, highlighting both successes and inherent limitations. Our core contribution lies in outlining future trends across several critical dimensions. These include the enhancement of data availability and quality, the evolution of models, the development of multilingual and cross-cultural models, the establishment of regulatory and clinical translation frameworks, and the creation of hybrid systems enabling human–artificial intelligence (AI) collaboration. Finally, we conclude with a vision for future directions, emphasizing the potential integration of ML-driven speech diagnostics into public health infrastructure, the development of patient-specific explainable AI, and its synergistic combination with genomics and brain imaging for holistic brain health assessment. Overcoming substantial hurdles in validation, generalization, and clinical adoption, the field is poised to shift toward ubiquitous, accessible, and highly personalized tools for early diagnosis.
1. Introduction
The global burden of neurodevelopmental and neurodegenerative disorders presents an escalating public health challenge. Conditions, such as autism spectrum disorder (ASD), attention-deficit/hyperactivity disorder (ADHD), Parkinson’s disease (PD), and Alzheimer’s disease (AD), significantly impact quality of life, necessitate extensive care, and impose immense socio-economic costs [1]. Globally, ASD affects roughly 1 in 100 children and about 1 in 127 individuals across the lifespan [2,3]. AD affects >55 million people worldwide and is projected to reach ~78 million by 2030 and ~139–153 million by 2050 [4,5]. PD affects ~8.5 million people and is projected to more than double by 2040–2050 [6,7].
It is important to mention that neurodevelopmental and neurodegenerative disorders differ fundamentally in etiology, diagnostic frameworks, and developmental trajectories. Neurodevelopmental disorders (e.g., ASD, ADHD) arise in the developmental period and are defined behaviorally in Diagnostic and Statistical Manual of Mental Disorders (DSM)/International Classification of Diseases (ICD) as disturbances in the acquisition of cognitive, language, motor, or social functions, with multifactorial genetic and environmental risk architecture and no validated biological diagnostic biomarkers to date [8,9,10]. By contrast, neurodegenerative diseases reflect progressive proteinopathies with characteristic pathobiology—β-amyloid and tau for AD within the AT(N) framework, and α-synuclein pathology for PD—supported by evolving fluid and imaging biomarkers that increasingly anchor diagnosis [11,12,13]. Diagnostic criteria also diverge: ADHD/ASD require developmental history and symptom-based thresholds in DSM/ICD, whereas AD now emphasizes biological definition and staged criteria, and PD employs Movement Disorder Society clinical diagnostic criteria with specified supportive and exclusion features [14,15].
A critical unmet need across these diverse conditions is the capacity for early and accurate diagnosis. Early detection is paramount, as it enables timely intervention, potentially slowing disease progression, improving developmental outcomes, and facilitating access to support services, thereby enhancing patient and caregiver well-being [16,17]. However, current diagnostic pathways often involve subjective clinical assessments, lengthy observation periods, and expensive, invasive neuroimaging or biochemical tests, leading to diagnostic delays and disparities in access [18,19].
In this context, speech and language emerge as exceptionally promising biomarkers. They are ubiquitous, non-invasive, cost-effective, and highly accessible, requiring only a microphone-equipped device for data collection [20]. Speech production and language processing are complex cognitive functions that rely on intricate neural networks. Consequently, disruptions in these networks, whether due to atypical development or neurodegeneration, often manifest as subtle yet detectable changes in vocal characteristics, prosody, articulation, fluency, syntax, semantics, and pragmatics [21]. These changes can precede overt clinical symptoms by years, offering a unique window for proactive intervention.
The advent of machine learning (ML) has revolutionized the ability to identify subtle patterns within complex data, making it an ideal candidate for extracting meaningful diagnostic signals from speech and language. ML algorithms can analyze vast quantities of acoustic (e.g., formant frequencies, pitch), linguistic (e.g., syntactic complexity, lexical choice), and paralinguistic (e.g., speech rate, intonation) features, identifying intricate relationships that are imperceptible to the human ear or traditional statistical methods [22]. This capability has propelled ML to the forefront of research in speech-based diagnostics, transforming raw vocalizations into actionable diagnostic insights.
This paper aims to discuss the future trajectories of ML-driven, speech- and language-based diagnostics for neurodevelopmental and neurodegenerative disorders. It provides a narrative review that synthesizes representative findings from current research, analyzes emerging technological and societal trends, and examines how these developments are shaping the landscape of early detection. Rather than offering a systematic evidence synthesis, the paper takes a conceptual and integrative approach, connecting advances across linguistics, ML, and clinical neuroscience to provide a forward-looking outlook on the transformative potential of ML in brain health diagnostics.
2. Background and Current Landscape
The application of ML to speech and language biomarkers for neurodevelopmental and neurodegenerative disorders has seen significant progress in recent years. Researchers have leveraged a diverse array of ML techniques to extract meaningful patterns from complex vocal and linguistic data, aiming to identify subtle deviations indicative of neurological conditions.
2.1. Overview of Current Machine Learning Techniques
The bedrock of most current diagnostic applications is supervised learning, a paradigm where models learn from labeled data, such as speech samples explicitly marked as healthy or disordered. Historically, Support Vector Machines have been prominent, proving effective for classification tasks by identifying an optimal hyperplane to separate different classes in a high-dimensional feature space. These have been widely applied with hand-crafted acoustic features, including Mel-frequency cepstral coefficients, jitter, and shimmer, particularly for conditions like PD [23]. Ensemble learning methods, such as Random Forests, also offer robustness and strong performance by building multiple decision trees and aggregating their predictions, especially when dealing with high-dimensional feature sets and complex interactions [24].
The most transformative development in recent years has been the rise of deep neural networks. Convolutional Neural Networks (CNNs) are primarily utilized for analyzing spectrograms of speech, which are visual representations of sound frequencies over time. CNNs excel at capturing local patterns and hierarchical features, much like their application in image recognition [25], and have shown promise in detecting vocal tremors or atypical prosody [26]. Recurrent Neural Networks and Long Short-Term Memory networks are specifically designed to process sequential data, making them well-suited for capturing temporal dependencies in speech and language, such as fluency patterns, pauses, or grammatical structures [27]. These are particularly relevant for analyzing conversational dynamics or narrative coherence in conditions like AD. More recently, the Transformer architecture, with its self-attention mechanism, has become dominant, especially in natural language processing. While initially applied to textual data, the Transformer’s ability to model long-range dependencies and contextual relationships makes it increasingly relevant for speech analysis, particularly in conjunction with large pre-trained models [28]. Figure 1 illustrates a general ML pipeline for detecting speech or language disorders.
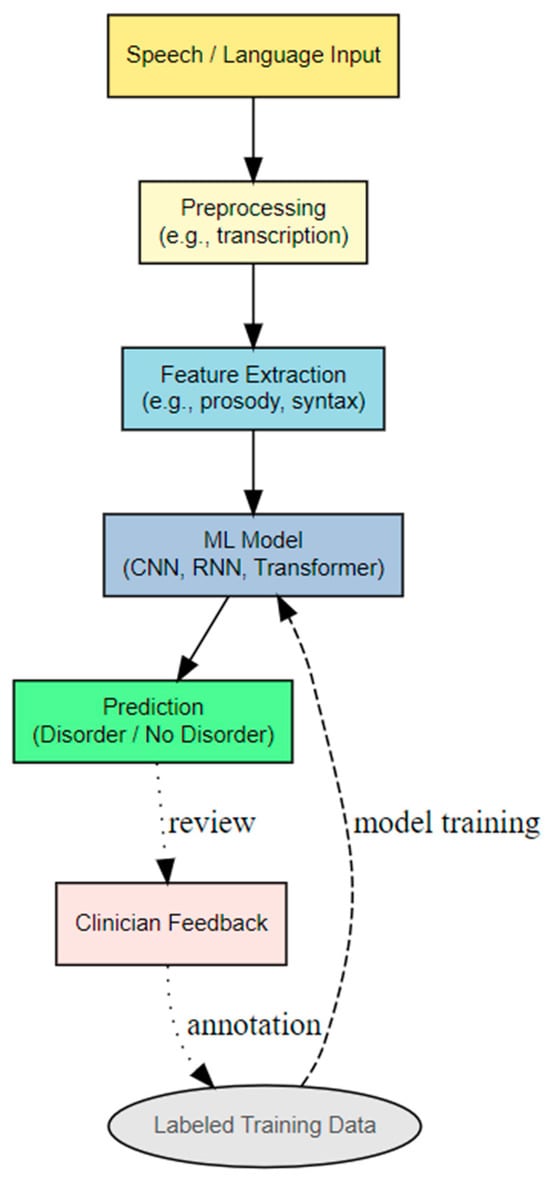
Figure 1.
General ML pipeline for detecting speech or language disorders.
Beyond supervised learning, self-supervised and unsupervised models address the challenge of limited labeled data. Self-supervised learning involves models learning representations from unlabeled data by solving pretext tasks, such as predicting masked words or future frames in a sequence. Models like Wav2Vec 2.0 and HuBERT, pre-trained on vast amounts of raw speech, learn rich, transferable representations that can then be fine-tuned on smaller, labeled datasets for specific diagnostic tasks [29], significantly reducing the need for extensive manual annotation. Unsupervised learning techniques, including clustering or anomaly detection, can identify unusual patterns in speech without prior labels [30], potentially flagging individuals who deviate significantly from a “normative” speech profile. For example, Bi et al. [31] introduced a fully unsupervised deep learning approach for diagnosing AD using magnetic resonance imaging (MRI) scans. The method employed unsupervised networks to automatically extract features from the MRI data, followed by an unsupervised prediction model to classify disease states. The model achieved high accuracy in distinguishing AD from mild cognitive impairment (MCI) and in differentiating MCI from normal controls. Parlett-Pelleriti et al. [32] presented a review of 43 studies that applied unsupervised ML techniques to ASD research. The authors concluded that unsupervised methods are particularly well-suited for ASD research due to the wide variability among individuals with the diagnosis, and both current applications and future developments in this area show significant potential.
Finally, multimodal learning recognizes that speech and language are often accompanied by other behavioral cues, integrating information from various sources. This approach might combine speech features with facial expressions, gaze patterns, body movements, or even physiological signals like heart rate variability, to provide a more comprehensive diagnostic picture [33]. For instance, in autism detection, combining vocalizations with eye-tracking data can enhance diagnostic accuracy [34].
2.2. Review of Current Applications
ML-driven speech and language diagnostics are actively being explored across a spectrum of neurodevelopmental and neurodegenerative disorders. For ASD, early detection efforts often focus on vocalizations in infants and toddlers, such as atypical cry patterns or reduced babbling complexity, and later, on prosodic abnormalities, repetitive language, or pragmatic difficulties in older children [35]. ML models analyze acoustic features, turn-taking patterns, and semantic content in these cases. Research into ADHD explores speech rate, variability, and disfluencies, as well as the coherence and organization of narratives, which can reflect executive function deficits [36].
In PD, dysarthria, affecting voice quality, articulation, and prosody, is a hallmark. ML models extensively analyze features like fundamental frequency (pitch) variations (jitter, shimmer), voice intensity, speech rate, and pauses to detect early signs of vocal impairment [37]. For AD and MCI, language decline is a prominent feature. ML models assess lexical diversity, semantic coherence, grammatical complexity, increased pauses, and repetitions in spontaneous speech or narrative tasks, such as picture description [38].
Notwithstanding these research advances, the vast majority of speech- and language-based ML applications remain experimental. To date, no model has completed prospective, multi-site clinical trials or obtained regulatory clearance for standalone diagnosis in these conditions.
2.3. Datasets Used in Previous Work
The development of these ML models heavily relies on specialized datasets. The ADReSS (Alzheimer’s Dementia Recognition through Speech) is a benchmark dataset for AD detection from speech, containing audio recordings and transcripts from individuals with AD and healthy controls [39]. The DAIC-WOZ (Distress Analysis Interview Corpus—Wizard of Oz), while primarily for depression detection, contains conversational speech data relevant for analyzing psychiatric and neurological conditions, including features like prosody and turn-taking [40]. Coswara is a large crowdsourced dataset of respiratory sounds and speech, initially for COVID-19 detection, but valuable for exploring vocal biomarkers across various health conditions due to its scale and diversity [41]. The Child Language Data Exchange System (CHILDES) is a vast repository of transcribed child language data, crucial for studying typical and atypical language development [42]. Many studies also rely on smaller, institution-specific private clinical datasets collected from clinical populations, though these often limit generalizability due to their size and specific demographics.
2.4. Advantages and Limitations
The advantages of ML in this field include its non-invasiveness and accessibility, offering a low-cost alternative to traditional diagnostics and enabling broader access, especially in underserved areas. ML models have demonstrated significant potential for early detection, identifying subtle speech and language changes that precede overt clinical symptoms, thus offering a crucial window for early intervention [43]. Furthermore, these models offer scalability, capable of processing large volumes of data efficiently [44], making them suitable for population-level screening initiatives, and provide objective quantification of speech and language features, reducing subjectivity inherent in clinical assessments.
However, significant limitations persist. Generalizability is a major hurdle, as models trained on specific datasets often perform poorly when applied to new populations, different recording conditions, or diverse linguistic backgrounds, a phenomenon known as “dataset shift” [45]. Although group-level studies frequently report high accuracy, these findings rarely generalize across datasets, institutions, or populations, a limitation widely attributed to dataset shift and the absence of standardized data collection protocols [46,47]. Diagnostic reliability at the individual level remains elusive, and many well-known neurophysiological or behavioral biomarkers, such as the Mismatch Negativity (MMN) component, illustrate this gap. While MMN reliably differentiates groups with ASD from neurotypical controls, meta-analyses have concluded that it lacks the precision and reproducibility necessary for clinical implementation [48,49]. Similarly, although numerous ML studies report promising results in detecting ASD, no clinical SLP service currently employs ML tools for routine assessment or diagnosis, underscoring the persistent translational divide between algorithmic performance in research settings and real-world clinical utility. Data scarcity is another challenge; high-quality, clinically annotated speech datasets for rare disorders or specific disease stages are often small, imbalanced, and difficult to acquire due to privacy concerns and the cost of expert annotation, which limits the training of robust deep learning models [50]. Interpretability, often referred to as the “black-box problem”, means that many advanced deep learning models make it difficult to understand why a particular diagnosis was made. This lack of transparency is a significant barrier to clinical adoption, as clinicians require insights into the decision-making process for trust and validation [51]. Confounding factors also complicate analysis, as speech patterns are influenced by numerous variables beyond neurological conditions, including age, gender, medication, emotional state, education level, and cultural background, making it challenging to isolate disease-specific biomarkers [52]. Finally, no ML system has yet been approved by any regulatory agencies as a standalone diagnostic tool for autism or related neurodevelopmental conditions. Existing digital platforms, such as Canvas Dx, have only been authorized as diagnostic aids rather than definitive instruments, reflecting the early, investigational status of ML-based tools in this domain [53,54].
Despite these limitations, the rapid advancements in ML, particularly in deep learning and self-supervised learning, suggest a promising future for speech and language biomarkers in neurological diagnostics once key challenges are addressed. The following sections outline how these emerging trends may evolve and address current limitations in the use of ML for detection through speech and language biomarkers. Figure 2 illustrates an overview of the advantages and limitations of ML models in clinical practice.
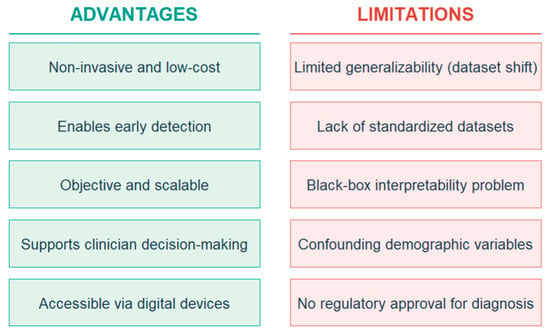
Figure 2.
Overview of the advantages and limitations of ML models in clinical practice.
3. Future Trends and Scenarios
The future of ML in the early detection of neurodevelopmental and neurodegenerative disorders via speech and language biomarkers is poised for transformative growth. This section discusses key trends and scenarios across several critical dimensions.
3.1. Data Availability and Quality
Major problems for current ML models in this domain are often data scarcity and heterogeneity. We foresee a significant shift towards increased data availability and improved quality, driven by several factors. Crowdsourcing and citizen science initiatives, exemplified by projects like Coswara, demonstrate the power of collective data contribution. Future efforts will see more sophisticated platforms enabling individuals to voluntarily contribute speech data, potentially through dedicated mobile applications. These platforms will incorporate gamification, clear consent mechanisms, and robust data anonymization to encourage participation and protect privacy, thereby significantly augmenting dataset size and diversity, particularly for rare disorders or specific demographic groups that are hard to reach through traditional clinical recruitment [55].
The proliferation of wearable technology and continuous monitoring devices, such as smartwatches and hearables equipped with high-quality microphones, will enable continuous, passive monitoring of speech and vocalizations in naturalistic environments. This shifts data collection from a single clinical snapshot to longitudinal, real-world data streams, capturing subtle, day-to-day fluctuations indicative of early disease onset or progression [56]. Imagine smart speakers or personal assistants subtly analyzing changes in vocal tremor or speech rate over months. To overcome privacy concerns and data silos, federated learning paradigms will become more prevalent. Instead of centralizing raw data, models will be trained locally on decentralized datasets, for example, in hospitals or on personal devices, with only model updates or aggregated insights being shared. This will enable collaborative model development across institutions without compromising patient privacy [57]. Large-scale data sharing consortia, adhering to strict ethical guidelines, will also emerge, pooling anonymized or synthetic speech data from diverse clinical cohorts. Furthermore, advances in synthetic data generation, utilizing generative adversarial networks (GANs) and diffusion models, will enable the creation of high-fidelity synthetic speech data that mimics real patient populations. This can augment scarce real datasets, especially for minority classes or specific pathological speech characteristics, helping to balance datasets and improve model robustness without exposing sensitive patient information [58].
However, these advancements will intensify ethical challenges with sensitive speech data. The continuous collection and analysis of highly personal vocal biomarkers raises profound questions about consent, data ownership, algorithmic bias, and potential misuse. Robust regulatory frameworks, explainable artificial intelligence (AI), and transparent data governance will be critical to building public trust and ensuring responsible innovation.
3.2. Model Evolution
The next generation of ML models will move beyond specialized, task-specific architectures towards more generalized and powerful paradigms. The rise of foundation models and large language models (LLMs) for health prediction will be central to this evolution. Pre-trained on massive, diverse datasets, foundation models, such as Whisper for speech recognition or large-scale LLMs for language understanding, will become the backbone for diagnostic applications. These models learn rich, transferable representations of speech and language that capture nuanced acoustic and linguistic patterns [59]. For instance, a Whisper-like model, pre-trained on millions of hours of speech, could be fine-tuned with a relatively small dataset of neurodevelopmental speech to achieve high diagnostic accuracy, leveraging its vast pre-existing knowledge of human vocalization. LLMs will not only transcribe but also analyze semantic content, coherence, and pragmatic aspects of speech, offering deeper linguistic insights.
Building on foundation models, transfer learning and domain adaptation will become standard. Models pre-trained on general speech or language tasks will be fine-tuned on smaller, disease-specific datasets. Furthermore, advanced domain adaptation techniques will enable models trained on one population or language to generalize more effectively to others, even with limited target domain data [60]. This will be crucial for addressing data scarcity and improving cross-cultural applicability. The fusion of speech with other modalities, such as video or physiological sensors, will be facilitated by multimodal foundation models that can jointly learn representations from different data streams. These models will capture the intricate interplay between vocal, facial, and bodily cues, leading to more robust and accurate diagnoses [61]. For example, a model could analyze speech prosody alongside subtle facial micro-expressions to detect early signs of emotional dysregulation in neurodevelopmental disorders or motor deficits in neurodegeneration. As data becomes more abundant and models more sophisticated, there will be a shift towards highly personalized diagnostic models. Few-shot learning techniques will allow models to adapt rapidly to individual patient characteristics with minimal new data, enabling tailored diagnostic pathways and monitoring [62].
3.3. Multilingual and Cross-Cultural Models
Currently, most research and datasets are biased towards English and Western populations, whereas languages such as African, Indigenous American, and Australian Indigenous languages remain underrepresented [63]. The future will see a concerted effort towards truly inclusive, global diagnostic systems. Addressing bias in training data will be paramount; acknowledging the inherent biases in current datasets, future efforts will prioritize the collection of diverse, representative speech data across languages, dialects, socio-economic backgrounds, and cultural contexts. Techniques for bias detection and mitigation within ML models will become standard practice, ensuring equitable performance across different groups [64].
While some acoustic biomarkers might be universal, such as vocal tremor, many linguistic and prosodic features are language-specific. Future models will need to effectively learn both universal and language-specific biomarkers. This will involve developing language-agnostic feature extraction methods or employing multilingual foundation models that can adapt to the details of different linguistic structures and cultural communication norms [65]. The ultimate forecast is for inclusive, global diagnostic systems, where diagnostic tools are equally effective for a patient in Tokyo as for one in rural Africa. This necessitates collaborative international research, open-source multilingual datasets, and the development of ML models that are inherently robust to linguistic and cultural variations. We project the emergence of “language-of-the-world” speech AI models capable of processing and analyzing speech in hundreds of languages, democratizing access to early detection globally [66].
3.4. Regulatory and Clinical Translation
The journey from research prototype to clinical utility is fraught with challenges, particularly regarding trust and validation. Explainability will be a key factor for clinical trust; black-box models will not gain widespread clinical adoption. Future regulatory bodies will demand high levels of explainability for ML-driven diagnostic tools. Clinicians need to understand why a model made a particular prediction, identify the most salient speech features contributing to the diagnosis, and assess the model’s confidence [67]. This will foster trust, enable clinical oversight, and facilitate integration into existing diagnostic workflows. Techniques like SHapley Additive exPlanations (SHAP) and Local Interpretable Model-agnostic Explanations (LIME) will become standard components of diagnostic ML systems [68,69].
Beyond technical explainability, ML interpretability and validation will occur in real-world trials. Models will undergo rigorous validation in large-scale, multi-center clinical trials. These trials will assess not only diagnostic accuracy (sensitivity, specificity) but also clinical utility, impact on patient outcomes, and cost-effectiveness in real-world settings. Emphasis will be placed on prospective studies that demonstrate the long-term benefits of early detection facilitated by ML [70]. Regulatory bodies, such as the Food and Drug Administration and European Medicines Agency, are expected to establish clear guidelines for the approval and deployment of AI-powered medical devices, specifically addressing speech-based diagnostics, building on proposed regulatory frameworks governing medical device use [71,72]. As ML models are deployed, they will require continuous monitoring and adaptive regulation for performance drift and bias. Regulatory frameworks will evolve to support adaptive regulation, allowing for model updates and re-validation based on real-world performance data, ensuring ongoing safety and efficacy.
3.5. Hybrid Systems and Human-AI Collaboration
The future is not about replacing clinicians but empowering them through intelligent tools. AI will primarily function as a second-opinion tool; ML systems will serve as sophisticated diagnostic aids, providing clinicians with a second opinion or flagging individuals requiring further assessment. They will enhance the efficiency and accuracy of human experts, particularly in screening large populations or identifying subtle patterns that might be missed during routine examinations [73]. Diagnostic platforms will offer real-time feedback to clinicians, providing immediate analysis of speech during patient interactions [74]. This could include visualizations of vocal abnormalities, linguistic complexity metrics, or risk scores, enabling more informed decision-making during consultations.
Adaptive diagnostic systems might emerge, which will learn from clinical outcomes and clinician feedback, continuously improving their performance. These systems will be able to adapt to new disease presentations, evolving diagnostic criteria, and individual patient responses to interventions, creating a virtuous cycle where clinical practice informs AI development, and AI insights enhance clinical care [75]. For example, if a clinician overrides an AI’s initial assessment, the system could learn from this discrepancy to refine its future predictions. Beyond clinical use, simplified, privacy-preserving patient-facing tools might be developed for self-monitoring, allowing individuals to track their own speech patterns over time and share data with their healthcare providers, fostering proactive health management.
In summary, the future of ML in speech-based diagnostics is characterized by abundant, diverse data, powerful and generalizable models, global inclusivity, stringent regulatory oversight, and a collaborative synergy between human expertise and AI. A summary of the forecasts is shown in Table 1.

Table 1.
Forecasts about ML diagnostics in clinical practice.
4. Conclusions and Future Directions
Despite major research advances, current ML systems have not yet achieved the reliability or regulatory readiness required for routine clinical use; their role remains exploratory and adjunctive rather than diagnostic. Nevertheless, as evidence and oversight frameworks mature, we anticipate integration into public health infrastructure. The non-invasive and scalable nature of speech-based diagnostics makes them potential candidates for integration into broader public health initiatives. We foresee a future where these tools are deployed for population-level screening, particularly in primary care settings or even through consumer-facing applications. This integration would significantly reduce diagnostic delays, improve access to care, and enable earlier interventions, leading to better long-term outcomes and reduced healthcare costs. This requires robust, validated, and user-friendly systems that can be operated by non-specialist healthcare providers or even directly by individuals in their homes, with results securely transmitted to clinicians for review.
Explainable, patient-specific ML systems will overcome the black-box problem through a strong emphasis on AI that provides actionable insights to clinicians and patients. Future systems will not just provide a diagnostic label but will highlight the specific acoustic, prosodic, or linguistic features that contributed to the prediction. Furthermore, these systems might become highly patient-specific, adapting to individual baseline speech patterns and tracking longitudinal changes. This personalized approach has the opportunity to move beyond population-level averages, enabling more precise monitoring of disease progression or response to therapy for each individual. The development of “digital twins” (i.e., virtual models that mirror a patient’s speech profile and are continuously updated with new data of a patient’s speech profile, continuously updated with new data) could allow for highly individualized risk assessments and treatment recommendations.
A powerful future direction lies in the synergistic integration with genomics and brain imaging, combining speech and language biomarkers with other omics data and neuroimaging modalities. Speech provides a functional, behavioral phenotype, while genomics offers insights into genetic predispositions, and brain imaging, such as functional MRI (fMRI) and positron emission tomography (PET), reveals structural and functional brain changes. Multi-omics integration, combining speech data with genomic information, could identify individuals at high genetic risk who also exhibit subtle speech atypicalities, allowing for ultra-early identification and preventive strategies [76]. Neuroimaging correlation, linking specific speech biomarkers with changes observed in brain imaging, could deepen our understanding of the neural correlates of speech production and language processing in disease. This could lead to the discovery of novel, more specific biomarkers and provide biological validation for speech-based diagnostic findings. The ultimate vision is a holistic brain health assessment platform that integrates diverse data streams—speech, genomics, neuroimaging, cognitive test results, and even wearable sensor data—to provide a comprehensive, multi-dimensional view of an individual’s neurological health. ML models will be crucial for fusing these disparate data types, identifying complex interactions, and providing a unified risk profile or diagnostic assessment. This integrated approach will move beyond single-modality diagnostics to a more subtle and accurate understanding of neurodevelopmental and neurodegenerative conditions, paving the way for truly personalized and preventive neurological care [77].
The present paper offers a conceptual and forward-looking synthesis of emerging trends for the use of ML in clinical translation. Despite rapid research progress, it is important to recognize that ML models in speech and language remain at the proof-of-concept stage. Bridging the gap between promising research and validated, generalizable clinical tools will require large-scale, longitudinal validation and appropriate regulatory oversight. Looking ahead, these developments point to a paradigm shift—from reactive diagnosis to proactive health management—where accessible, intelligent, and integrated technologies empower both individuals and clinicians in the continuous pursuit of brain health. Another important point lies in the joint discussion of neurodevelopmental (e.g., ASD, ADHD) and neurodegenerative (e.g., PD, AD) disorders within a shared analytical framework. While this comparative approach emphasizes the common value of speech and language as non-invasive biomarkers across the lifespan, these groups differ substantially in etiology, biomarker expression, and methodological requirements. These distinctions entail different data collection paradigms, ML architectures, and clinical validation pathways. Therefore, the integrated treatment adopted here should be viewed as a conceptual synthesis rather than a disorder-specific analytical framework.
Funding
This research received no external funding.
Data Availability Statement
No data are available.
Acknowledgments
This study was supported by the Phonetic Lab of the University of Nicosia.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ding, C.; Wu, Y.; Chen, X.; Chen, Y.; Wu, Z.; Lin, Z.; Kang, D.; Fang, W.; Chen, F. Global, regional, and national burden and attributable risk factors of neurological disorders: The Global Burden of Disease Study 1990–2019. Front. Public Health 2022, 10, 952161. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Autism Spectrum Disorders (Fact Sheet). 2025. Available online: https://www.who.int/news-room/fact-sheets/detail/autism-spectrum-disorders (accessed on 9 October 2025).
- Santomauro, D.F.; Erskine, H.E.; Herrera, A.M.M.; Miller, P.A.; Shadid, J.; Hagins, H.; Addo, I.Y.; Adnani, Q.E.S.; Ahinkorah, B.O.; Ahmed, A.; et al. The Global Epidemiology and Health Burden of the Autism Spectrum: Findings from the Global Burden of Disease Study 2021. Lancet Psychiatry 2025, 12, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Disease International. Dementia Statistics (Facts & Figures); Alzheimer’s Disease International: London, UK, 2020; Available online: https://www.alzint.org/about/dementia-facts-figures/dementia-statistics/ (accessed on 9 October 2025).
- Alzheimer’s Disease International. Numbers of People with Dementia Worldwide: An Update to the Estimates in the World Alzheimer Report 2015; Alzheimer’s Disease International: London, UK, 2020; Available online: https://www.alzint.org/resource/numbers-of-people-with-dementia-worldwide/ (accessed on 9 October 2025).
- World Health Organization (WHO). Parkinson Disease—Fact Sheet; World Health Organization: Geneva, Switzerland, 2023; Available online: https://www.who.int/news-room/fact-sheets/detail/parkinson-disease (accessed on 9 October 2025).
- Bhidayasiri, R.; Sringean, J.; Phumphid, S.; Anan, C.; Thanawattano, C.; Deoisres, S.; Panyakaew, P.; Phokaewvarangkul, O.; Maytharakcheep, S.; Buranasrikul, V.; et al. The Rise of Parkinson’s Disease Is a Global Challenge, but Efforts to Tackle This Must Begin at a National Level: A Protocol for National Digital Screening and “Eat, Move, Sleep” Lifestyle Interventions to Prevent or Slow the Rise of Non-Communicable Diseases in Thailand. Front. Neurol. 2024, 15, 1386608. [Google Scholar]
- Reed, G.M.; First, M.B.; Kogan, C.S.; Hyman, S.E.; Gureje, O.; Gaebel, W.; Maj, M.; Stein, D.J.; Maercker, A.; Tyrer, P.; et al. Innovations and Changes in the ICD-11 Classification of Mental, Behavioural and Neurodevelopmental Disorders. World Psychiatry 2019, 18, 3–19. [Google Scholar] [CrossRef]
- World Health Organization. Clinical Descriptions and Diagnostic Requirements for ICD-11 Mental, Behavioural and Neurodevelopmental Disorders; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Sauer, A.K.; Stanton, J.; Hans, S.; Grabrucker, A. Autism Spectrum Disorders: Etiology and Pathology; Exon Publications: Brisbane, Australia, 2021; pp. 1–15. [Google Scholar]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a Biological Definition of Alzheimer’s Disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Andrews, J.S.; Beach, T.G.; Buracchio, T.; Dunn, B.; Graf, A.; Hansson, O.; Ho, C.; Jagust, W.; McDade, E.; et al. Revised Criteria for Diagnosis and Staging of Alzheimer’s Disease: Alzheimer’s Association Workgroup. Alzheimer’s Dement. 2024, 20, 5143–5169. [Google Scholar] [CrossRef]
- Gauthier, S.; Rosa-Neto, P. Alzheimer’s Disease Biomarkers: Amyloid, Tau, Neurodegeneration (ATN)—Where Do We Go from Here? Pract. Neurol. 2019, 88, 60–61. [Google Scholar]
- Postuma, R.B.; Berg, D.; Stern, M.; Poewe, W.; Olanow, C.W.; Oertel, W.; Obeso, J.; Marek, K.; Litvan, I.; Lang, A.E.; et al. MDS Clinical Diagnostic Criteria for Parkinson’s Disease. Mov. Disord. 2015, 30, 1591–1601. [Google Scholar] [CrossRef]
- Franke, B.; Michelini, G.; Asherson, P.; Banaschewski, T.; Bilbow, A.; Buitelaar, J.K.; Cormand, B.; Faraone, S.V.; Ginsberg, Y.; Haavik, J.; et al. Live Fast, Die Young? A Review on the Developmental Trajectories of ADHD across the Lifespan. Eur. Neuropsychopharmacol. 2018, 28, 1059–1088. [Google Scholar] [CrossRef]
- Zwaigenbaum, L.; Bauman, M.L.; Choueiri, R.; Kasari, C.; Carter, A.; Granpeesheh, D.; Mailloux, Z.; Smith Roley, S.; Wagner, S.; Fein, D.; et al. Early intervention for children with autism spectrum disorder under 3 years of age: Recommendations for practice and research. Pediatrics 2015, 136 (Suppl. 1), S60–S81. [Google Scholar] [CrossRef]
- Leibing, A. The Earlier the Better: Alzheimer’s Prevention, Early Detection, and the Quest for Pharmacological Interventions. Cult. Med. Psychiatry 2014, 38, 217–236. [Google Scholar] [CrossRef]
- McLoughlin, C.; Lee, W.H.; Carson, A.; Stone, J. Iatrogenic harm in functional neurological disorder. Brain 2025, 148, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.; Wittens, M.M.J.; Engelborghs, S.; van Herwijnen, M.H.; Tsamou, M.; Roggen, E.; Smeets, B.; Krauskopf, J.; Briedé, J.J. Beyond CSF and neuroimaging assessment: Evaluating plasma miR-145-5p as a potential biomarker for mild cognitive impairment and Alzheimer’s disease. ACS Chem. Neurosci. 2024, 15, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Vogel, A.P.; Gharahkhani, P.; Renteria, M.E. Speech and language biomarkers for Parkinson’s disease prediction, early diagnosis and progression. NPJ Parkinson’s Dis. 2025, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, G.P.; Theodorou, E. Abilities of children with developmental language disorders in perceiving phonological, grammatical, and semantic structures. J. Autism Dev. Disord. 2023, 53, 4483–4487. [Google Scholar] [CrossRef]
- Myszczynska, M.A.; Ojamies, P.N.; Lacoste, A.M.; Neil, D.; Saffari, A.; Mead, R.; Hautbergue, G.M.; Holbrook, J.D.; Ferraiuolo, L. Applications of machine learning to diagnosis and treatment of neurodegenerative diseases. Nat. Rev. Neurol. 2020, 16, 440–456. [Google Scholar] [CrossRef]
- Moro-Velazquez, L.; Gomez-Garcia, J.A.; Arias-Londoño, J.D.; Dehak, N.; Godino-Llorente, J.I. Advances in Parkinson’s disease detection and assessment using voice and speech: A review of the articulatory and phonatory aspects. Biomed. Signal Process. Control 2021, 66, 102418. [Google Scholar] [CrossRef]
- Quaye, G.E. Random Forest for High-Dimensional Data. Ph.D. Thesis, The University of Texas at El Paso, El Paso, TX, USA, 2024. [Google Scholar]
- Vásquez-Correa, J.C.; Arias-Vergara, T.; Orozco-Arroyave, J.R.; Eskofier, B.; Klucken, J.; Nöth, E. Multimodal assessment of Parkinson’s disease: A deep learning approach. IEEE J. Biomed. Health Inform. 2018, 23, 1618–1630. [Google Scholar] [CrossRef]
- Weede, P.; Smietana, P.D.; Kuhlenbäumer, G.; Deuschl, G.; Schmidt, G. Two-stage convolutional neural network for classification of movement patterns in tremor patients. Information 2024, 15, 231. [Google Scholar] [CrossRef]
- Graves, A. Supervised Sequence Labelling with Recurrent Neural Networks; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, N. Attention is all you need. In Advances in Neural Information Processing Systems; Curran Associates: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Baevski, A.; Zhou, Y.; Mohamed, A.; Auli, M. Wav2vec 2.0: A framework for self-supervised learning of speech representations. Adv. Neural Inf. Process. Syst. 2020, 33, 12449–12460. [Google Scholar]
- Beke, A.; Szaszák, G. Unsupervised clustering of prosodic patterns in spontaneous speech. In Proceedings of the Text, Speech and Dialogue, TSD 2012, Brno, Czech Republic, 3–7 September 2012; Springer: Berlin/Heidelberg, Germany, 2012; pp. 648–655. [Google Scholar]
- Bi, X.; Li, S.; Xiao, B.; Li, Y.; Wang, G.; Ma, X. Computer aided Alzheimer’s disease diagnosis by an unsupervised deep learning technology. Neurocomputing 2020, 392, 296–304. [Google Scholar] [CrossRef]
- Parlett-Pelleriti, C.M.; Stevens, E.; Dixon, D.; Linstead, E.J. Applications of unsupervised machine learning in autism spectrum disorder research: A review. Rev. J. Autism Dev. Disord. 2023, 10, 406–421. [Google Scholar]
- Kumar, S.; Rani, S.; Sharma, S.; Min, H. Multimodality fusion aspects of medical diagnosis: A comprehensive review. Bioengineering 2024, 11, 1233. [Google Scholar] [CrossRef] [PubMed]
- Rashid, A.F.; Shaker, S.H. Review of autistic detection using eye tracking and vocalization based on deep learning. J. Algebr. Stat. 2022, 13, 286–297. [Google Scholar]
- Oller, D.K.; Niyogi, P.; Gray, S.; Richards, J.A.; Gilkerson, J.; Xu, D.; Yapanel, U.; Warren, S.F. Automated vocal analysis of naturalistic recordings from children with autism, language delay, and typical development. Proc. Natl. Acad. Sci. USA 2010, 107, 13354–13359. [Google Scholar] [CrossRef]
- Kuijper, S.J.; Hartman, C.A.; Bogaerds-Hazenberg, S.; Hendriks, P. Narrative production in children with autism spectrum disorder (ASD) and children with attention-deficit/hyperactivity disorder (ADHD): Similarities and differences. J. Abnorm. Psychol. 2017, 126, 63. [Google Scholar] [CrossRef]
- Tsanas, A.; Little, M.A.; McSharry, P.E.; Spielman, J.; Ramig, L.O. Novel speech signal processing algorithms for high-accuracy classification of Parkinson’s disease. IEEE Trans. Biomed. Eng. 2012, 59, 1264–1271. [Google Scholar] [CrossRef]
- Thaler, F.; Gewald, H. Language characteristics supporting early Alzheimer’s diagnosis through machine learning—A literature review. Health Inform. Int. J. 2021, 10, 10102. [Google Scholar] [CrossRef]
- Luz, S.; Haider, F.; de la Fuente Garcia, S.; Fromm, D.; MacWhinney, B. Alzheimer’s dementia recognition through spontaneous speech. Front. Comput. Sci. 2021, 3, 780169. [Google Scholar] [CrossRef]
- Gratch, J.; Artstein, R.; Lucas, G.M.; Stratou, G.; Scherer, S.; Nazarian, A.; Wood, R.; Boberg, J.; DeVault, D.; Marsella, S.; et al. The Distress Analysis Interview Corpus of human and computer interviews. In Proceedings of the LREC, Reykjavik, Iceland, 26–31 May 2014; pp. 3123–3128. [Google Scholar]
- Sharma, N.; Krishnan, P.; Kumar, R.; Ramoji, S.; Chetupalli, S.R.; Ghosh, P.K.; Ganapathy, S. Coswara—A database of breathing, cough, and voice sounds for COVID-19 diagnosis. arXiv 2020, arXiv:2005.10548. [Google Scholar]
- MacWhinney, B. The CHILDES Project: Tools for Analyzing Talk. Volume I: Transcription Format and Programs; Psychology Press: New York, NY, USA, 2014. [Google Scholar]
- Brahmi, Z.; Mahyoob, M.; Al-Sarem, M.; Algaraady, J.; Bousselmi, K.; Alblwi, A. Exploring the role of machine learning in diagnosing and treating speech disorders: A systematic literature review. Psychol. Res. Behav. Manag. 2024, 2205–2232. [Google Scholar] [CrossRef]
- Potla, R.T. Scalable Machine Learning Algorithms for Big Data Analytics: Challenges and Opportunities. J. Artif. Intell. Res. 2022, 2, 124–141. [Google Scholar]
- Quiñonero-Candela, J.; Sugiyama, M.; Schwaighofer, A.; Lawrence, N.D. Dataset Shift in Machine Learning; MIT Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Washington, P.; Wall, D.P. A review of and roadmap for data science and machine learning for the neuropsychiatric phenotype of autism. Annu. Rev. Biomed. Data Sci. 2023, 6, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Health Care Service Corporation (HCSC). Digital Health Technologies: Diagnostic Applications; Policy No. PSY301.024; Effective 15 December 2024; Health Care Service Corporation (HCSC): Chicago, IL, USA, 2024. [Google Scholar]
- Schwartz, S.; Shinn-Cunningham, B.; Tager-Flusberg, H. Meta-analysis and systematic review of the literature characterizing auditory mismatch negativity in individuals with autism. Neurosci. Biobehav. Rev. 2018, 87, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Lassen, J.; Oranje, B.; Vestergaard, M.; Foldager, M.; Kjær, T.W.; Arnfred, S.; Aggernæs, B. Reduced mismatch negativity in children and adolescents with autism spectrum disorder is associated with their impaired adaptive functioning. Autism Res. 2022, 15, 1469–1481. [Google Scholar] [CrossRef]
- Ghasemzadeh, H.; Hillman, R.E.; Mehta, D.D. Toward generalizable machine learning models in speech, language, and hearing sciences: Estimating sample size and reducing overfitting. J. Speech Lang. Hear. Res. 2024, 67, 753–781. [Google Scholar] [CrossRef]
- Jiang, X.; Hu, Z.; Wang, S.; Zhang, Y. Deep learning for medical image-based cancer diagnosis. Cancers 2023, 15, 3608. [Google Scholar] [CrossRef]
- Ramanarayanan, V.; Lammert, A.C.; Rowe, H.P.; Quatieri, T.F.; Green, J.R. Speech as a biomarker: Opportunities, interpretability, and challenges. Perspect. ASHA Spec. Interest Groups 2022, 7, 276–283. [Google Scholar] [CrossRef]
- Premera Blue Cross. Prescription Digital Health Diagnostic Aid for Autism Spectrum Disorder (Medical Policy 3.03.01); Premera Blue Cross: Spokane, WA, USA, 2024. [Google Scholar]
- Kleine, A.K.; Kokje, E.; Hummelsberger, P.; Lermer, E.; Gaube, S. AI-enabled clinical decision support tools for mental healthcare: A product review. OSF Preprints 2023. Available online: https://osf.io/ez43g (accessed on 3 November 2025). [CrossRef]
- Schmitz, H.; Howe, C.L.; Armstrong, D.G.; Subbian, V. Leveraging mobile health applications for biomedical research and citizen science: A scoping review. J. Am. Med. Inform. Assoc. 2018, 25, 1685–1698. [Google Scholar] [CrossRef]
- Majumder, S.; Mondal, T.; Deen, M.J. Wearable sensors for remote health monitoring. Sensors 2017, 17, 130. [Google Scholar] [CrossRef]
- Rieke, N.; Hancox, J.; Li, W.; Milletari, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The future of digital health with federated learning. NPJ Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Advances in Neural Information Processing Systems; Curran Associates: Red Hook, NY, USA, 2014; Volume 27. [Google Scholar]
- Radford, A.; Kim, J.W.; Xu, T.; Brockman, G.; McLeavey, C.; Sutskever, I. Robust speech recognition via large-scale weak supervision. In Proceedings of the International Conference on Machine Learning, Honolulu, HI, USA, 23–29 July 2023; PMLR: Honolulu, HI, USA, 2023; pp. 28492–28518. [Google Scholar]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Baltrušaitis, T.; Ahuja, C.; Morency, L.P. Multimodal machine learning: A survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 41, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wang, T.; Cai, P.; Mondal, S.K.; Sahoo, J.P. A comprehensive survey of few-shot learning: Evolution, applications, challenges, and opportunities. ACM Comput. Surv. 2023, 55, 1–40. [Google Scholar] [CrossRef]
- Larasati, R. Inclusivity of AI Speech in Healthcare: A Decade Look Back. arXiv 2025, arXiv:2505.10596. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, J.M.; Sarro, F.; Harman, M. A comprehensive empirical study of bias mitigation methods for machine learning classifiers. ACM Trans. Softw. Eng. Methodol. 2023, 32, 1–30. [Google Scholar] [CrossRef]
- Mohamed, A.; Lee, H.Y.; Borgholt, L.; Havtorn, J.D.; Edin, J.; Igel, C.; Kirchhoff, K.; Li, S.-W.; Livescu, K.; Maaløe, L.; et al. Self-supervised speech representation learning: A review. IEEE J. Sel. Top. Signal Process. 2022, 16, 1179–1217. [Google Scholar] [CrossRef]
- Babu, A.; Wang, C.; Tjandra, A.; Lakhotia, K.; Xu, Q.; Goyal, N.; Singh, K.; von Platen, P.; Saraf, Y.; Pino, J.; et al. XLS-R: Self-supervised cross-lingual speech representation learning at scale. arXiv 2021, arXiv:2111.09296. [Google Scholar]
- Gunning, D.; Stefik, M.; Choi, J.; Miller, T.; Stumpf, S.; Yang, G.Z. XAI—Explainable artificial intelligence. Sci. Robot. 2019, 4, eaay7120. [Google Scholar] [CrossRef]
- Nohara, Y.; Matsumoto, K.; Soejima, H.; Nakashima, N. Explanation of Machine Learning Models Using Shapley Additive Explanation and Application for Real Data in Hospital. Comput. Methods Programs Biomed. 2022, 214, 106584. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.R.; Khan, N. Deterministic Local Interpretable Model-Agnostic Explanations for Stable Explainability. Mach. Learn. Knowl. Extr. 2021, 3, 525–541. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–60. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration. Artificial Intelligence and Machine Learning in Software as a Medical Device. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device (accessed on 19 August 2025).
- European Medicines Agency. Reflection Paper on the Use of Artificial Intelligence (AI) in the Lifecycle of Medicinal Products; EMA: London, UK, 2024; Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-use-artificial-intelligence-ai-medicinal-product-lifecycle_en.pdf (accessed on 3 November 2025).
- Talitckii, A.; Kovalenko, E.; Anikina, A.; Zimniakova, O.; Semenov, M.; Bril, E.; Shcherbak, A.; Dylov, D.V.; Somov, A. Avoiding misdiagnosis of Parkinson’s disease with the use of wearable sensors and artificial intelligence. IEEE Sens. J. 2020, 21, 3738–3747. [Google Scholar] [CrossRef]
- Pham, T.D.; Holmes, S.B.; Zou, L.; Patel, M.; Coulthard, P. Diagnosis of pathological speech with streamlined features for long short-term memory learning. Comput. Biol. Med. 2024, 170, 107976. [Google Scholar] [CrossRef]
- Kumari, P.; Chauhan, J.; Bozorgpour, A.; Huang, B.; Azad, R.; Merhof, D. Continual learning in medical image analysis: A comprehensive review of recent advancements and future prospects. arXiv 2023, arXiv:2312.17004. [Google Scholar] [CrossRef]
- Hassan, S.; Akaila, D.; Arjemandi, M.; Papineni, V.; Yaqub, M. MINDSETS: Multi-omics integration with neuroimaging for dementia subtyping and effective temporal study. Sci. Rep. 2025, 15, 15835. [Google Scholar] [CrossRef]
- Bhonde, S.B.; Prasad, J.R. Machine learning approach to revolutionize use of holistic health records for personalized healthcare. Int. J. Adv. Sci. Technol. 2020, 29, 313–321. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).