Abstract
The appropriate categorisation of written information by health professionals is very important to guarantee its accessibility. Unfortunately, the information technology tools that support professionals on that task imply a heavy workload, so that the responsibility for categorising the written content is often delegated to administrative staff. Well-known health ontologies such as SNOMED-CT or MeSH provide a representation of the clinical contents to be used by the information systems. This research proposes a computer based method to automatically extract and code the diagnostics, procedures and treatments according to health ontologies. A Knowledge Management System based on an extended version of Drupal is used to implement and evaluate this proposal. Results provide a positive evidence on the application of the method to support medical professionals.
1. Introduction
Ontologies in medicine have the potential to improve data quality and patient safety, facilitating semantic interoperability by capturing clinical data in a standardised, unambiguous and granular manner [1]. SNOMED Clinical Terms (SNOMED-CT) [2] and Medical Subject Headings (MeSH) [3] are the most widely used medical ontologies. By using a medical ontology, health professionals can categorise their clinical documents with a recognised source of terms. However, these ontologies contains a large number of terms. For instance, SNOMED-CT contains more than 340,000 classified terms (https://www.snomed.org/snomed-ct/snomed-ct-worldwide). It is not possible for any person to be able of managing all these terms, but ontologies should be adopted without implying workload problems for health professionals.
Text categorisation is necessary to facilitate access to health professionals to the amount of information stored in Electronic Health Records (EHRs) [4]. EHRs are collections of electronic health information about patients for integrating health information to improve quality of care [5]. The constant increase in the number of EHRs, makes it essential the existence of mechanisms for the extraction of information to facilitate its use [6].
Knowledge Management Systems (KMS) can be used to effectively manage EHR systems, capture all the relevant information and make it available to health professionals. A KMS is a software designed to collect the relevant information within an organisation, making it explicit for their users to query and update. Recent case studies in hospitals demonstrated that using a KMS to manage their EHR improves their performance and service quality [7,8].
This paper describes a solution for gathering useful information from medical texts stored in the KMS records of medical institutions in order to automatically categorise their content and ensure the quality of the content published in an EHR. The rest of the paper is structured as follows: in the second section, the background is presented and the need for this research is justified; the third section presents the method proposed; the fourth section presents the evaluation of the method; the conclusions of this work are presented in the last section.
2. Background and Related Works
During routine patient care activities, health professionals describe the reasons for consultation, personal background, test results, clinical trials and treatments, among others, usually through natural language written reports, which are partially structured. As a result, the information stored in a EHR contains a high redundancy of terminology. To solve this issue, a recent paper proposes a system of patients’ records implemented with QuickView. This solution is based on a clustering approach to support health professionals with a navigable overview of the most important categories of patients’ medical history [4].
To categorise health documents, the administrative staff of the clinical centres are usually in charge of manually coding all that written information using the proper terms. Using ontologies enables to improve the quality of data, to support further assistance statistics and the financial management of the centres, to promote research, etc. [9]. However, this is a time-consuming task. Would it be possible to automate this process, thus reducing costs and any transcription errors?
A solution adopted to address this issue is the use of subsets of SNOMED-CT terms to facilitate clinical interaction of every specific field [10]. For instance, an ontology-based classification method to automatically categorise epilepsy types was developed using machine learning [11]. By using oncology-related SNOMED-CT terms, a system for automatically identifying cancer from large collections of free-text death certificates were developed to accurately report on cancer mortality. It was based on both a natural processing language and a supervised Support Vector Machines-based approach [12].
Dione is a Web Ontology Language (OWL) representation for the automatic classification of patients’ diseases by using SNOMED-CT annotations embedded in EHRs [13]. It is obtained by mapping SNOMED-CT with the ICD-10-CM diseases (International Classification of Diseases, Tenth Revision, Clinical Modification). Dione is an initial step towards the automatic classification, requiring the use of natural language processing techniques or text mining.
This research aims to provide a method for the automatic processing and standardisation of medical text in any speciality. This solution will reduce the effort of health professionals while categorising medical texts.
3. Method
The aim of this method is to extract the medical terms used by health professionals in their documents, analysing and categorising them following medical ontologies. This method has been devised through a design and creation research strategy, which focuses on developing an artefact based on IT applications [14]. The method comprises the following steps:
- Recording information generated by health professionals: First, the information generated by health professionals is collected in order to process it.
- Text analysis: Second, the medical text is analysed by splitting it into tokens.
- Diagnosis extraction: Third, medical vocabulary concepts included in the processed text are extracted.
- Coding by medical vocabularies: Fourth, the text is encoded by relating it to the list of tags proposed by the medical vocabularies.
- Returning resulted tags: Finally, the tags are returned so that external systems can use them to support health professionals tagging.
The method proposed is part of emPhasys (http://emphasys.uca.es/en/), an ICT instrument for the empowerment of users/patients, supported in the new paradigms of the Personalised Health Care or Customised Health Care [15]. Within emPhasys, the method will be implemented by a Knowledge Management Systems (KMS) module that will collect the medical information provided by the operation of other modules. This information will be transformed semantically to be available so that it can be exploited with data mining techniques.
4. Evaluation
The evaluation is divided into three subsections. Firstly, the deployment performed for the evaluation is described. Secondly, the results obtained are analysed. And thirdly, a discussion between the method and related works is presented.
4.1. Method Deployment
To carry out the implementation of the proposed method we used Apache Stanbol (https://stanbol.apache.org) and the MeSH and SNOMED-CT medical vocabularies. Apache Stanbol is a platform with a set of software components for semantic content management. Such components provide the tools to include semantic services in traditional content systems. The semantic services are provided using REST APIs. In this case, an Apache Stanbol instance was deployed on a server in which the MeSH and SNOMED-CT ontologies were configured and loaded.
The following steps have been taken to configure Apache Stanbol with MeSH and SNOMED-CT. First, the ontologies were loaded in RDF format. Second, the ontologies were indexed by the Stanbol EntityHub skipping the empty nodes. Finally, a Stanbol Keyword Linking has been created and the search engine options for the accuracy of results were configured. In addition, it has been included in an Apache Stanbol List Chain, thus enabling it to be used with other search engines.
4.2. Analysis of Results
For privacy reasons, actual EHR medical records were not accessed to evaluate the proposed system. Instead, an instance of Drupal Content Management System (CMS) was deployed to emulate the EHR KMS. CMSs and KMSs are similar tools to managing information, with differences related to the treatment of this information and the objective of its management (https://sixfeetup.com/blog/kms-vs-CMS-what-differences). We randomly chose a set of articles about health topics and loaded their abstracts in Drupal. With the Auto Recommend Content Tags (https://www.drupal.org/project/auto_recommended_tags) plug-in configured, Drupal can invoke the semantic service provided by Apache Stanbol and thus, supporting users to visualise terms related to the text they are typing.
The following steps were taken to configure the Drupal instance to collect the terms returned by Apache Stanbol: Firstly, the Auto Recommend Content Tags plug-in was installed in Drupal; secondly, a NodeJS service was required. Hence, it was installed in the server and launched it; finally, the Auto Recommend Content Tags plug-in to connect with the Apache Stanbol service was configured using the appropriate URL and port.
Then, Drupal instance was tested with the aforementioned abstracts by using both Mesh and SNOMED-CT ontologies. Firstly, Table 1 includes the relation between the terms provided by Apache Stanbol using the MeSH ontology and the keywords proposed by the authors. Second, Table 2 includes the same relation with the terms provided by Apache Stanbol but in this case, using the SNOMED-CT ontology.

Table 1.
Table showing the keywords of the articles and the keywords proposed by Apache Stanbol using the MeSH ontology.

Table 2.
Table showing the keywords of the articles and the keywords proposed by Apache Stanbol using SNOMED-CT ontology.
To calculate recall and precision metrics, we checked if the keywords proposed by the authors coincided with the keywords proposed by Apache Stanbol, as follows:
- Total match: The keyword proposed by the author appears in the result provided by Apache Stanbol.
- Partial match: The keyword proposed by the author is a compound word and it partially appears in the result provided by Apache Stanbol.
- No match: The keyword proposed by the author does not appear in the result provided by Apache Stanbol.
Firstly, the values for precision and recall metrics obtained using the MeSH ontology are 0.2 and 0.38. Secondly, the obtained values for these metrics using SNOMED-CT ontology are 0.45 and 0.0873, respectively. The analysed data can be publicly viewed in a Google Sheet (https://goo.gl/hvPTyL). The main reasons for these low-obtained values are the following:
- The keywords choice of an article is a subjective task. Different authors can choose different keywords for the same article.
- Apache Stanbol returns the related keywords to the words that appear in the abstract of each article only if they are also part of the ontology.
However, these results show positive evidence about the possibilities of the method to support health professionals to choose existing keywords in medical ontologies. In this way, health professionals can categorise their work in a simpler and more validated way by medical vocabularies. Finally, Figure 1 and Figure 2 show the list of terms provided by Apache Stanbol using both ontologies displayed from the Drupal website.
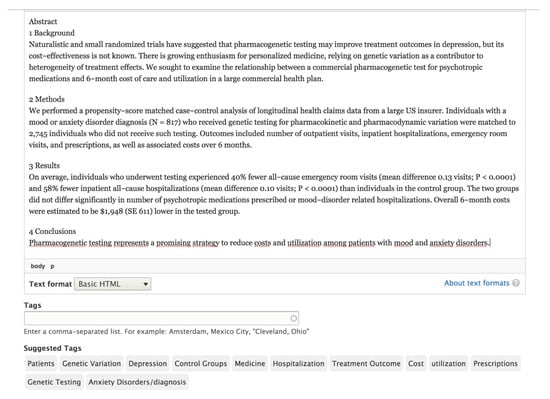
Figure 1.
List of terms proposed by Apache Stanbol using the MeSH ontology.
Figure 2.
List of terms proposed by Apache Stanbol using SNOMED-CT ontology.
4.3. Discussion
This subsection compares the ontology-based method proposed in this paper with several works that tackle the same problem.
QuickView is a system based on a clustering approach presented by Kreuzthaler et al. [4] to support health professionals categorising patients’ medical history. The authors pointed out that an important issue when clustering was that usually, the terms used to classify several documents were not the right terms. Thus, an automatic ontology-based method to classify health documents would solve this issue.
Several ontology-based methods by natural processing language and machine learning approach were found in the literature [11,12]. However, these methods addressed the same problem only for some specific field of medicine. Our ontology-based method uses a complete ontology to categorise medical texts regardless of the area to which they belong.
The summary of issues reduced by an automated method are shown in Table 3. Although first results are promising, further research is needed to draw stronger conclusions on the validity of our ontology-based method.

Table 3.
Table showing the issues found in the state of art.
5. Conclusions and Future Work
The use of medical ontologies is widespread in all areas of Health Sciences. Ontologies are used to categorise medical texts, a task that usually involves a workload for their users. This work presents a method for the automatic categorisation of medical texts through a specific software module, loaded with medical ontologies. The module has been tested with SNOMED-CT and MeSH vocabularies and checked against terms provided by the users. The results are promising, so additional experiments will be carried out.
As future work, this method will be integrated in the emPhasys platform and tested with actual EHRs. Then, the usability of the implementation will be assessed with the support of more professionals in the Health Sciences.
Funding
This work has been developed and funded by the EMPHASYS and VISAIGLE projects, funded by the Spanish National Research Agency (AEI) with ERDF funds under grants with ref. RTC-2016-5095-1 and TIN2017-85797-R.
References
- Lee, D.; de Keizer, N.; Lau, F.; Cornet, R. Literature review of SNOMED CT use. J. Am. Med. Inform. Assoc. 2013, 21, e11–e19. [Google Scholar] [CrossRef] [PubMed]
- Benson, T. Principles of Health Interoperability HL7 and SNOMED; Springer: London, UK, 2010. [Google Scholar]
- Lipscomb, C.E. Medical subject headings (MeSH). Bull. Med. Libr. Assoc. 2000, 88, 265. [Google Scholar] [PubMed]
- Kreuzthaler, M.; Pfeifer, B.; Vera, J.R.; Kramer, D.; Grogger, V.; Bredenfeldt, S.; Pedevilla, M.; Krisper, P.; Schulz, S. EHR Text Categorization for Enhanced Patient-Based Document Navigation. Stud. Health Technol. Inform. 2018, 248, 100–107. [Google Scholar] [PubMed]
- Gunter, T.D.; Terry, N.P. The emergence of national electronic health record architectures in the United States and Australia: models, costs, and questions. J. Med. Internet Res. 2005, 7, e3. [Google Scholar] [CrossRef] [PubMed]
- Kreuzthaler, M.; Schulz, S.; Berghold, A. Secondary use of electronic health records for building cohort studies through top-down information extraction. J. Biomed. Inform. 2015, 53, 188–195. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McCracken, S.S.; Edwards, J.S. Implementing a knowledge management system within an NHS hospital: a case study exploring the roll-out of an electronic patient record (EPR). Knowl. Manag. Res. Pract. 2017, 15, 1–11. [Google Scholar] [CrossRef]
- Choy, K.L.T.; Siu, K.Y.P.; Ho, T.S.G.; Wu, C.; Lam, H.Y.; Tang, V.; Tsang, Y.P. An intelligent case-based knowledge management system for quality improvement in nursing homes. VINE J. Inf. Knowl. Manag. Syst. 2018, 48, 103–121. [Google Scholar] [CrossRef]
- Zeshan, F.; Mohamad, R. Medical ontology in the dynamic healthcare environment. Procedia Comput. Sci. 2012, 10, 340–348. [Google Scholar] [CrossRef]
- Rodríguez-Solano, C.; Cáceres, J.; Sicilia, M.Á. Generating SNOMED CT subsets from clinical glossaries: An exploration using clinical guidelines. In Proceedings of the International Conference on ENTERprise Information Systems, Vilamoura, Portugal, 5–7 October 2011; pp. 117–127. [Google Scholar]
- Kassahun, Y.; Perrone, R.; De Momi, E.; Berghöfer, E.; Tassi, L.; Canevini, M.P.; Spreafico, R.; Ferrigno, G.; Kirchner, F. Automatic classification of epilepsy types using ontology-based and genetics-based machine learning. Artif. Intell. Med. 2014, 61, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Koopman, B.; Zuccon, G.; Nguyen, A.; Bergheim, A.; Grayson, N. Automatic ICD-10 classification of cancers from free-text death certificates. Int. J. Med. Inform. 2015, 84, 956–965. [Google Scholar] [CrossRef] [PubMed]
- del Mar Roldán-García, M.; García-Godoy, M.J.; Aldana-Montes, J.F. Dione: An OWL representation of ICD-10-CM for classifying patients’ diseases. J. Biomed. Semant. 2016, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Oates, B. Design and Creation. Researching Information Systems and Computing; Sage: Newcastle upon Tyne, UK, 2005; pp. 108–124. [Google Scholar]
- Collins, F. Has the revolution arrived? Nature 2010, 464, 674. [Google Scholar] [CrossRef] [PubMed]
- Høst, A.; Halken, S. A prospective study of cow milk allergy in Danish infants during the first 3 years of life. Allergy 1990, 45, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Sylvia, L.; H., A.A. Posttraumatic Headache: Classification by Symptom—Based Clinical Profiles. Headache J. Head Face Pain 2018, 58, 873–882. [Google Scholar]
- Urke, H.B.; Mittelmark, M.B.; Amugsi, D.A.; Matanda, D.J. Resources for nurturing childcare practices in urban and rural settings: Findings from the Colombia 2010 Demographic and Health Survey. Child Care Health Dev. 2018, 44, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Perlis, R.H.; Mehta, R.; Edwards, A.M.; Tiwari, A.; Imbens, G.W. Pharmacogenetic testing among patients with mood and anxiety disorders is associated with decreased utilization and cost: A propensity—Score matched study. Depress. Anxiety 2018. [Google Scholar] [CrossRef] [PubMed]
- Hanna, G.L.; Liu, Y.; Isaacs, Y.E.; Ayoub, A.M.; Brosius, A.; Salander, Z.; Arnold, P.D. Error-related brain activity in adolescents with obsessive—Compulsive disorder and major depressive disorder. Depress. Anxiety 2018. [Google Scholar] [CrossRef] [PubMed]
- Lars, E. Future Preventive Therapy: Are There Promising Drug Targets? Headache Curr. 2006, 3, 101–107. [Google Scholar] [CrossRef]
- Acosta, S.A.; Tajiri, N.; Bickford, P.C.; Borlongan, C.V. Cell Proliferation in the Brains of Adult Rats Exposed to Traumatic Brain Injury. In Neurostereology; Wiley-Blackwell: Hoboken, NJ, USA, 2013; Chapter 2; pp. 27–38. [Google Scholar] [CrossRef]
- Maternal medication and the baby. In Neonatal Formulary 7; Wiley-Blackwell: Hoboken, NJ, USA, 2014; Chapter 18; pp. 560–607.
- Wildhaber, J.; Carroll, W.D.; Brand, P.L. Global impact of asthma on children and adolescents’ daily lives: The room to breathe survey. Pediatr. Pulmonol. 2012, 47, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Wanaporn, A.; Surachai, K.; Somchai, S. Natural history of snoring and obstructive sleep apnea in Thai school-age children. Pediatr. Pulmonol. 2005, 39, 415–420. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).