The Role of Extended CFTR Gene Sequencing in Newborn Screening for Cystic Fibrosis
Abstract
1. Introduction
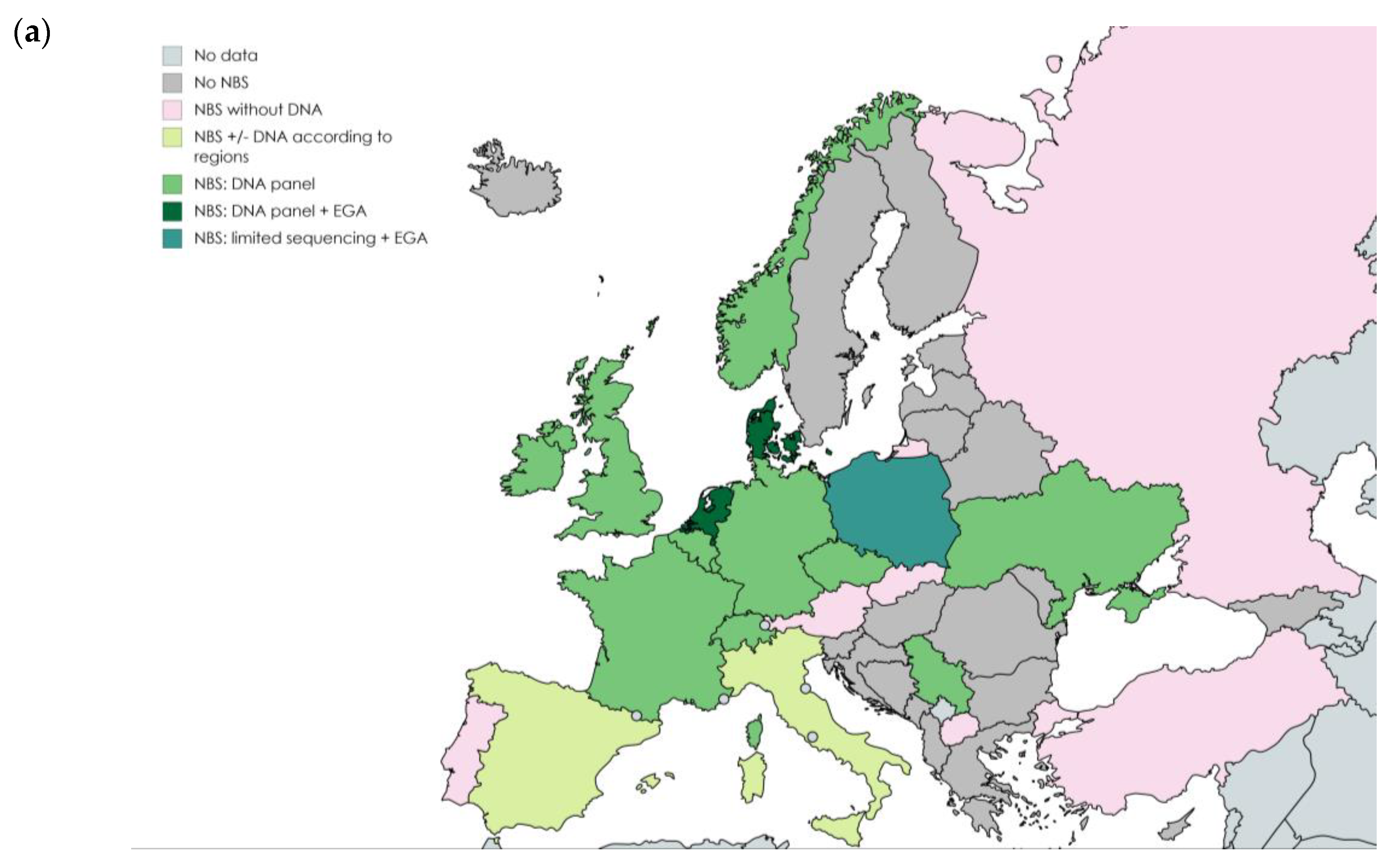
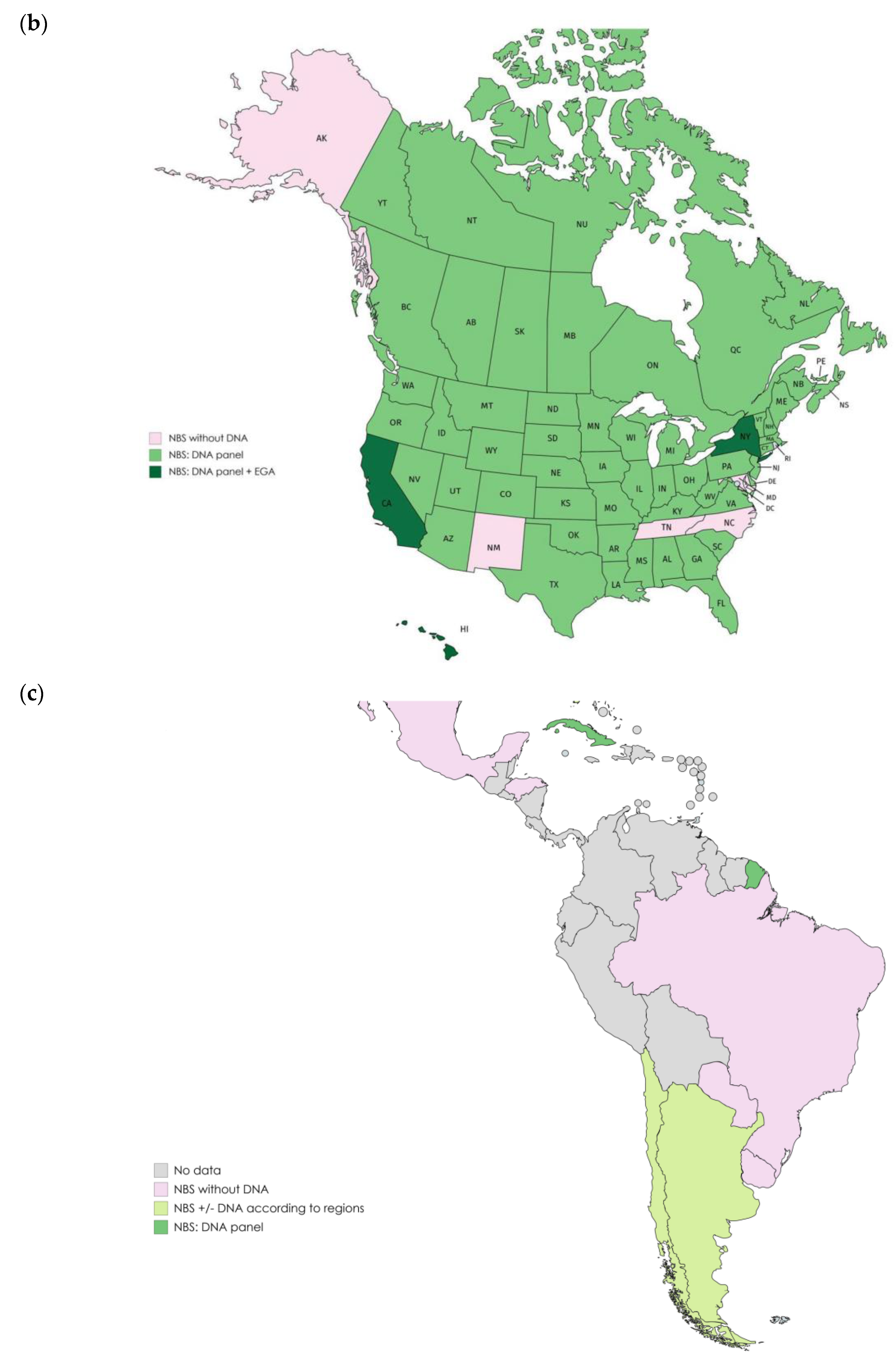
2. Overview of Newborn Screening Programs
2.1. Programs Including Variant Panels Only
2.2. Programs Including EGA
3. Values and Challenges of CFTR Extended Genetic Analysis
3.1. Values of CFTR Extended Genetic Analysis in Newborn Screening Programs for CF
3.1.1. Reduction of NBS False-Positive Results and Improvement of the PPV
3.1.2. Best Equity of CF Screening between Populations
3.1.3. Increased Knowledge of the Phenotypical Spectrum of CFTR Variants
3.1.4. Earlier Diagnosis and Access to Treatment
3.2. Challenges
3.2.1. Healthcare System Organization
3.2.2. Technical Issues
NBS Sequencing Platforms Development or Reorganization
Sequencing and Bioinformatics Limitations for Variant Detection
3.2.3. Byproducts of NBS for CF and Ethical Issues
Increased Detection of Inconclusive Diagnosis
Carrier Detection and Genetic Counseling
4. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barben, J.; Castellani, C.; Dankert-Roelse, J.; Gartner, S.; Kashirskaya, N.; Linnane, B.; Mayell, S.; Munck, A.; Sands, D.; Sommerburg, O.; et al. The expansion and performance of national newborn screening programmes for cystic fibrosis in Europe. J. Cyst. Fibros. 2017, 16, 207–213. [Google Scholar] [CrossRef] [PubMed]
- NSWG Annual Report 2018. Available online: https://www.ecfs.eu/sites/default/files/general-content-files/working-groups/NSWG%20annual%20report%202018v2.pdf (accessed on 21 February 2020).
- Massie, J.; Clements, B.; The Australian Paediatric Respiratory Group. Diagnosis of cystic fibrosis after newborn screening: The Australasian experience—twenty years and five million babies later: A consensus statement from the Australasian paediatric respiratory group. Pediatr. Pulmonol. 2005, 39, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Byrnes, C.A.; Vidmar, S.; Cheney, J.L.; Carlin, J.B.; Armstrong, D.S.; Cooper, P.J.; Grimwood, K.; Moodie, M.; Robertson, C.F.; Rosenfeld, M.; et al. Prospective evaluation of respiratory exacerbations in children with cystic fibrosis from newborn screening to 5 years of age. Thorax 2013, 68, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Pique, L.; Graham, S.; Pearl, M.; Kharrazi, M.; Schrijver, I. Cystic fibrosis newborn screening programs: Implications of the CFTR variant spectrum in nonwhite patients. Genet. Med. 2017, 19, 36–44. [Google Scholar] [CrossRef]
- Mak, D.Y.F.; Sykes, J.; Stephenson, A.L.; Lands, L.C. The benefits of newborn screening for cystic fibrosis: The Canadian experience. J. Cyst. Fibros. 2016, 15, 302–308. [Google Scholar] [CrossRef]
- Kharrazi, M.; Yang, J.; Bishop, T.; Lessing, S.; Young, S.; Graham, S.; Pearl, M.; Chow, H.; Ho, T.; Currier, R.; et al. Newborn Screening for Cystic Fibrosis in California. Pediatrics 2015, 136, 1062–1072. [Google Scholar] [CrossRef]
- Farrell, P.M.; White, T.B.; Howenstine, M.S.; Munck, A.; Parad, R.B.; Rosenfeld, M.; Sommerburg, O.; Accurso, F.J.; Davies, J.C.; Rock, M.J.; et al. Diagnosis of Cystic Fibrosis in Screened Populations. J. Pediatr. 2017, 181, S33–S44. [Google Scholar] [CrossRef]
- Southern, K.W.; Barben, J.; Gartner, S.; Munck, A.; Castellani, C.; Mayell, S.J.; Davies, J.C.; Winters, V.; Murphy, J.; Salinas, D.; et al. Inconclusive diagnosis after a positive newborn bloodspot screening result for cystic fibrosis; clarification of the harmonised international definition. J. Cyst. Fibros. 2019, 18, 778–780. [Google Scholar] [CrossRef]
- Castellani, C.; Southern, K.W.; Brownlee, K.; Dankert Roelse, J.; Duff, A.; Farrell, M.; Mehta, A.; Munck, A.; Pollitt, R.; Sermet-Gaudelus, I.; et al. European best practice guidelines for cystic fibrosis neonatal screening. J. Cyst. Fibros. 2009, 8, 153–173. [Google Scholar] [CrossRef]
- Castellani, C.; Massie, J.; Sontag, M.; Southern, K.W. Newborn screening for cystic fibrosis. Lancet Respir. Med. 2016, 4, 653–661. [Google Scholar] [CrossRef]
- Castellani, C.; Linnane, B.; Pranke, I.; Cresta, F.; Sermet-Gaudelus, I.; Peckham, D. Cystic Fibrosis Diagnosis in Newborns, Children, and Adults. Semin. Respir. Crit. Care Med. 2019, 40, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Werbrouck, A.; Verhaeghe, N.; de Wachter, E.; Simoens, S.; Annemans, L.; Putman, K. A model-based economic evaluation of four newborn screening strategies for cystic fibrosis in Flanders, Belgium. Acta Clin. Belg. 2019, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Castellani, C.; Duff, A.J.A.; Bell, S.C.; Heijerman, H.G.M.; Munck, A.; Ratjen, F.; Sermet-Gaudelus, I.; Southern, K.W.; Barben, J.; Flume, P.A.; et al. ECFS best practice guidelines: The 2018 revision. J. Cyst. Fibros. 2018, 17, 153–178. [Google Scholar] [CrossRef] [PubMed]
- Caggana, M.; (New York State Department of Health, Albany, NY 12208, USA). Personal communication, 2020.
- Rodrigues, R.; Magalhaes, P.K.R.; Fernandes, M.I.M.; Gabetta, C.S.; Ribeiro, A.F.; Pedro, K.P.; Valdetaro, F.; Santos, J.L.F.; de Souza, R.M.; Pazin Filho, A.; et al. Neonatal screening for cystic fibrosis in São Paulo State, Brazil: A pilot study. Braz. J. Med. Biol. Res. 2009, 42, 973–978. [Google Scholar] [CrossRef] [PubMed]
- XI Congreso Latinoamericano de Errores Inatos del Metabolismo y Pesquisa Neonatal. 2019. Available online: http://www.sleimpn2019.org/index.php/trabajos/posters-pesquisa-neonatal (accessed on 21 February 2020).
- Massie, R.J.H.; Curnow, L.; Glazner, J.; Armstrong, D.S.; Francis, I. Lessons learned from 20 years of newborn screening for cystic fibrosis. Med. J. Aust. 2012, 196, 67–70. [Google Scholar] [CrossRef]
- Ministry of Health. Newborn Metabolic Screening Programme: Annual Report 2017; Ministry of Health: Wellington, New Zealand, 2018.
- Ross, L.F. Newborn Screening for Cystic Fibrosis: A Lesson in Public Health Disparities. J. Pediatr. 2008, 153, 308–313. [Google Scholar] [CrossRef]
- Skov, M.; Bækvad-Hansen, M.; Hougaard, D.M.; Skogstrand, K.; Lund, A.M.; Pressler, T.; Olesen, H.V.; Duno, M. Cystic fibrosis newborn screening in Denmark: Experience from the first 2 years. Pediatr. Pulmonol. 2020, 55, 549–555. [Google Scholar] [CrossRef]
- Tolstova, V.D.; Kashirskaya, N.Y.; Kapranov, N.I.; Khodunova, A.A.; Denisenkova, E.V.; Smazhil, E.V. First results of newborn screening program for CF in Russia. J. Cyst. Fibros. 2008, 7, S13. [Google Scholar] [CrossRef][Green Version]
- Başaran, A.E.; Başaran, A.; Uygun, D.F.K.; Alper, Ö.; Acican, D.; BïNgöl, A. Initial regional evaluation of the Cystic Fibrosis Newborn Screening Program: Data from the Mediterranean coast of Turkey. Turk. J. Med. Sci. 2019, 49, 1655–1661. [Google Scholar]
- Delgado Pecellín, I.; Pérez Ruiz, E.; Álvarez Ríos, A.I.; Delgado Pecellín, C.; Yahyaoui Macías, R.; Carrasco Hernández, L.; Marcos Luque, I.; Caro Aguilera, P.; Moreno Valera, M.J.; Quintana Gallego, M.E. Results of the Andalusian Cystic Fibrosis Neonatal Screening Program, 5 Years After Implementation. Arch. Bronconeumol. 2018, 54, 551–558. [Google Scholar] [CrossRef]
- Renner, S.; Schanzer, A.; Metz, T.; Szépfalusi, Z.; Zeyda, M. CF newborn screening in Austria: After 20 years changing the algorithm from IRT/IRT to IRT/PAP/IRT. J. Cyst. Fibros. 2018, 17, S18. [Google Scholar] [CrossRef]
- Marcão, A.; Barreto, C.; Pereira, L.; Vaz, L.; Cavaco, J.; Casimiro, A.; Félix, M.; Silva, T.; Barbosa, T.; Freitas, C.; et al. Cystic Fibrosis Newborn Screening in Portugal: PAP Value in Populations with Stringent Rules for Genetic Studies. Int. J. Neonatal Screen. 2018, 4, 22. [Google Scholar] [CrossRef]
- Sommerburg, O.; Stahl, M.; Hammermann, J.; Okun, J.; Kulozik, A.; Hoffmann, G.; Mall, M. Neugeborenenscreening auf Mukoviszidose in Deutschland: Vergleich des neuen Screening-Protokolls mit einem Alternativprotokoll. Klin. Padiatr. 2017, 229, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Brockow, I.; Nennstiel, U. Parents’ experience with positive newborn screening results for cystic fibrosis. Eur. J. Pediatr. 2019, 178, 803–809. [Google Scholar] [CrossRef]
- Sontag, M.K.; Lee, R.; Wright, D.; Freedenberg, D.; Sagel, S.D. Improving the Sensitivity and Positive Predictive Value in a Cystic Fibrosis Newborn Screening Program Using a Repeat Immunoreactive Trypsinogen and Genetic Analysis. J. Pediatr. 2016, 175, 150–158. [Google Scholar] [CrossRef]
- Rock, M.J.; Hoffman, G.; Laessig, R.H.; Kopish, G.J.; Litsheim, T.J.; Farrell, P.M. Newborn screening for cystic fibrosis in Wisconsin: Nine-year experience with routine trypsinogen/DNA testing. J. Pediatr. 2005, 147, S73–S77. [Google Scholar] [CrossRef]
- Terlizzi, V.; Mergni, G.; Buzzetti, R.; Centrone, C.; Zavataro, L.; Braggion, C. Cystic fibrosis screen positive inconclusive diagnosis (CFSPID): Experience in Tuscany, Italy. J. Cyst. Fibros. 2019, 18, 484–490. [Google Scholar] [CrossRef]
- Munck, A.; Delmas, D.; Audrézet, M.-P.; Lemonnier, L.; Cheillan, D.; Roussey, M. Optimization of the French cystic fibrosis newborn screening programme by a centralized tracking process. J. Med. Screen. 2018, 25, 6–12. [Google Scholar] [CrossRef]
- Rueegg, C.S.; Kuehni, C.E.; Gallati, S.; Baumgartner, M.; Torresani, T.; Barben, J. One-Year Evaluation of a Neonatal Screening Program for Cystic Fibrosis in Switzerland. Dtsch. Arztebl. Int. 2013, 110, 356–363. [Google Scholar] [CrossRef][Green Version]
- Krulišová, V.; Balaščaková, M.; Skalická, V.; Piskáčková, T.; Holubová, A.; Paděrová, J.; Křenková, P.; Dvořáková, L.; Zemková, D.; Kračmar, P.; et al. Prospective and parallel assessments of cystic fibrosis newborn screening protocols in the Czech Republic: IRT/DNA/IRT versus IRT/PAP and IRT/PAP/DNA. Eur. J. Pediatr. 2012, 171, 1223–1229. [Google Scholar] [CrossRef]
- Lundman, E.; Gaup, H.J.; Bakkeheim, E.; Olafsdottir, E.J.; Rootwelt, T.; Storrøsten, O.T.; Pettersen, R.D. Implementation of newborn screening for cystic fibrosis in Norway. Results from the first three years. J. Cyst. Fibros. 2016, 15, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Schlüter, D.K.; Southern, K.W.; Dryden, C.; Diggle, P.; Taylor-Robinson, D. Impact of newborn screening on outcomes and social inequalities in cystic fibrosis: A UK CF registry-based study. Thorax 2020, 75, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Dankert-Roelse, J.E.; Bouva, M.J.; Jakobs, B.S.; Janssens, H.M.; de Winter-de Groot, K.M.; Schönbeck, Y.; Gille, J.J.P.; Gulmans, V.A.M.; Verschoof-Puite, R.K.; Schielen, P.C.J.I.; et al. Newborn blood spot screening for cystic fibrosis with a four-step screening strategy in the Netherlands. J. Cyst. Fibros. 2019, 18, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Sands, D.; Zybert, K.; Mierzejewska, E.; Oltarzewski, M. Diagnosing cystic fibrosis in newborn screening in Poland—15 years of experience. Dev. Period Med. 2015, 19, 16–24. [Google Scholar]
- Sands, D.; (Zaklad Mukowiscydozy, Instytut Matki i Dziecka, Kasprzaka 17a, 01-211 Warsow, Poland). Personal communication, 2020.
- Ziętkiewicz, E.; Rutkiewicz, E.; Pogorzelski, A.; Klimek, B.; Voelkel, K.; Witt, M. CFTR mutations spectrum and the efficiency of molecular diagnostics in Polish cystic fibrosis patients. PLoS ONE 2014, 9, e89094. [Google Scholar] [CrossRef]
- Currier, R.J.; Sciortino, S.; Liu, R.; Bishop, T.; Alikhani Koupaei, R.; Feuchtbaum, L. Genomic sequencing in cystic fibrosis newborn screening: What works best, two-tier predefined CFTR mutation panels or second-tier CFTR panel followed by third-tier sequencing? Genet. Med. 2017, 19, 1159–1163. [Google Scholar] [CrossRef]
- Cirilli, N.; Southern, K.W.; Buzzetti, R.; Barben, J.; Nährlich, L.; Munck, A.; Wilschanski, M.; De Boeck, K.; Derichs, N. Real life practice of sweat testing in Europe. J. Cyst. Fibros. 2018, 17, 325–332. [Google Scholar] [CrossRef]
- Schrijver, I.; Pique, L.; Graham, S.; Pearl, M.; Cherry, A.; Kharrazi, M. The Spectrum of CFTR Variants in Nonwhite Cystic Fibrosis Patients: Implications for Molecular Diagnostic Testing. J. Mol. Diagn. 2016, 18, 39–50. [Google Scholar] [CrossRef]
- Cystic Fibrosis Mutation Data Base (CFMDB). Available online: http://www.genet.sickkids.on.ca/ (accessed on 21 February 2020).
- Sosnay, P.R.; Siklosi, K.R.; Van Goor, F.; Kaniecki, K.; Yu, H.; Sharma, N.; Ramalho, A.S.; Amaral, M.D.; Dorfman, R.; Zielenski, J.; et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat. Genet. 2013, 45, 1160–1167. [Google Scholar] [CrossRef]
- Claustres, M.; Thèze, C.; des Georges, M.; Baux, D.; Girodon, E.; Bienvenu, T.; Audrezet, M.-P.; Dugueperoux, I.; Férec, C.; Lalau, G.; et al. CFTR-France, a national relational patient database for sharing genetic and phenotypic data associated with rare CFTR variants. Hum. Mutat. 2017, 38, 1297–1315. [Google Scholar] [CrossRef]
- Wells, J.; Rosenberg, M.; Hoffman, G.; Anstead, M.; Farrell, P.M. A Decision-Tree Approach to Cost Comparison of Newborn Screening Strategies for Cystic Fibrosis. Pediatrics 2012, 129, e339–e347. [Google Scholar] [CrossRef] [PubMed]
- Deeb, K.K.; Metcalf, J.D.; Sesock, K.M.; Shen, J.; Wensel, C.A.; Rippel, L.I.; Smith, M.; Chapman, M.S.; Zhang, S. The c.1364C>A (p.A455E) Mutation in the CFTR Pseudogene Results in an Incorrectly Assigned Carrier Status by a Commonly Used Screening Platform. J. Mol. Diagn. 2015, 17, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Kim, E.; Lee, I.; Marcotte, E.M. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci. Rep. 2015, 5, 17875. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Shen, P.; Odegaard, J.I.; Fung, E.; Chiang, T.; Peng, G.; Davis, R.W.; Wang, W.; Kharrazi, M.; Schrijver, I.; et al. Next-Generation Molecular Testing of Newborn Dried Blood Spots for Cystic Fibrosis. J. Mol. Diagn. 2016, 18, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Bergougnoux, A.; Délétang, K.; Pommier, A.; Varilh, J.; Houriez, F.; Altieri, J.P.; Koenig, M.; Férec, C.; Claustres, M.; Lalau, G.; et al. Functional characterization and phenotypic spectrum of three recurrent disease-causing deep intronic variants of the CFTR gene. J. Cyst. Fibros. 2019, 18, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Boemer, F.; Fasquelle, C.; d’Otreppe, S.; Josse, C.; Dideberg, V.; Segers, K.; Guissard, V.; Capraro, V.; Debray, F.G.; Bours, V. A next-generation newborn screening pilot study: NGS on dried blood spots detects causal mutations in patients with inherited metabolic diseases. Sci. Rep. 2017, 7, 17641. [Google Scholar] [CrossRef] [PubMed]
- Bergougnoux, A.; Boureau-Wirth, A.; Rouzier, C.; Altieri, J.-P.; Verneau, F.; Larrieu, L.; Koenig, M.; Claustres, M.; Raynal, C. A false positive newborn screening result due to a complex allele carrying two frequent CF-causing variants. J. Cyst. Fibros. 2016, 15, 309–312. [Google Scholar] [CrossRef][Green Version]
- Terlizzi, V.; Castaldo, G.; Salvatore, D.; Lucarelli, M.; Raia, V.; Angioni, A.; Carnovale, V.; Cirilli, N.; Casciaro, R.; Colombo, C.; et al. Genotype-phenotype correlation and functional studies in patients with cystic fibrosis bearing CFTR complex alleles. J. Med. Genet. 2017, 54, 224–235. [Google Scholar] [CrossRef]
- Salinas, D.B.; Sosnay, P.R.; Azen, C.; Young, S.; Raraigh, K.S.; Keens, T.G.; Kharrazi, M. Benign and Deleterious Cystic Fibrosis Transmembrane Conductance Regulator Mutations Identified by Sequencing in Positive Cystic Fibrosis Newborn Screen Children from California. PLoS ONE 2016, 11, e0155624. [Google Scholar] [CrossRef]
- Ren, C.L.; Borowitz, D.S.; Gonska, T.; Howenstine, M.S.; Levy, H.; Massie, J.; Milla, C.; Munck, A.; Southern, K.W. Cystic Fibrosis Transmembrane Conductance Regulator-Related Metabolic Syndrome and Cystic Fibrosis Screen Positive, Inconclusive Diagnosis. J. Pediatr. 2017, 181, S45–S51. [Google Scholar] [CrossRef]
- Ooi, C.Y.; Castellani, C.; Keenan, K.; Avolio, J.; Volpi, S.; Boland, M.; Kovesi, T.; Bjornson, C.; Chilvers, M.A.; Morgan, L.; et al. Inconclusive Diagnosis of Cystic Fibrosis After Newborn Screening. Pediatrics 2015, 135, e1377–e1385. [Google Scholar] [CrossRef] [PubMed]
- Munck, A.; Bourmaud, A.; Bellon, G.; Picq, P.; Farrell, P.M.; DPAM Study Group. Phenotype of children with inconclusive cystic fibrosis diagnosis after newborn screening. Pediatr. Pulmonol. 2020, 55, 918–928. [Google Scholar] [CrossRef] [PubMed]
- Thauvin-Robinet, C.; Munck, A.; Huet, F.; Génin, E.; Bellis, G.; Gautier, E.; Audrézet, M.-P.; Férec, C.; Lalau, G.; Georges, M.D.; et al. The very low penetrance of cystic fibrosis for the R117H mutation: A reappraisal for genetic counselling and newborn screening. J. Med. Genet. 2009, 46, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Boussaroque, A.; Audrézet, M.-P.; Raynal, C.; Sermet-Gaudelus, I.; Bienvenu, T.; Férec, C.; Bergougnoux, A.; Lopez, M.; Scotet, V.; Munck, A.; et al. Penetrance is a critical parameter for assessing the disease liability of CFTR variants. J. Cyst. Fibros. 2020, in press. [Google Scholar] [CrossRef]
- Hayeems, R.Z.; Miller, F.A.; Barg, C.J.; Bombard, Y.; Carroll, J.C.; Tam, K.; Kerr, E.; Chakraborty, P.; Potter, B.K.; Patton, S.; et al. Psychosocial Response to Uncertain Newborn Screening Results for Cystic Fibrosis. J. Pediatr. 2017, 184, 165–171. [Google Scholar] [CrossRef]
- McClaren, B.J.; Metcalfe, S.A.; Aitken, M.; Massie, R.J.; Ukoumunne, O.C.; Amor, D.J. Uptake of carrier testing in families after cystic fibrosis diagnosis through newborn screening. Eur. J. Hum. Genet. 2010, 18, 1084–1089. [Google Scholar] [CrossRef]
- Dequeker, E.; Stuhrmann, M.; Morris, M.A.; Casals, T.; Castellani, C.; Claustres, M.; Cuppens, H.; des Georges, M.; Ferec, C.; Macek, M.; et al. Best practice guidelines for molecular genetic diagnosis of cystic fibrosis and CFTR-related disorders—updated European recommendations. Eur. J. Hum. Genet. 2009, 17, 51–65. [Google Scholar] [CrossRef]
- Castellani, C.; Cuppens, H.; Macek, M.; Cassiman, J.J.; Kerem, E.; Durie, P.; Tullis, E.; Assael, B.M.; Bombieri, C.; Brown, A.; et al. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J. Cyst. Fibros. 2008, 7, 179–196. [Google Scholar] [CrossRef]
- Boussaroque, A.; Bergougnoux, A.; Raynal, C.; Audrézet, M.-P.; Sasorith, S.; Férec, C.; Bienvenu, T.; Girodon, E. Pitfalls in the interpretation of CFTR variants in the context of incidental findings. Hum. Mutat. 2019, 40, 2239–2246. [Google Scholar] [CrossRef]
- Ceyhan-Birsoy, O.; Murry, J.B.; Machini, K.; Lebo, M.S.; Yu, T.W.; Fayer, S.; Genetti, C.A.; Schwartz, T.S.; Agrawal, P.B.; Parad, R.B.; et al. Interpretation of Genomic Sequencing Results in Healthy and Ill Newborns: Results from the BabySeq Project. Am. J. Hum. Genet. 2019, 104, 76–93. [Google Scholar] [CrossRef]
- Kingsmore, S.F. Newborn testing and screening by whole-genome sequencing. Genet. Med. 2016, 18, 214–216. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wilcken, B.; Wiley, V. Fifty years of newborn screening. J. Paediatr. Child Health 2015, 51, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Nijmeijer, S.C.M.; Conijn, T.; Lakeman, P.; Henneman, L.; Wijburg, F.A.; Haverman, L. Attitudes of the general population towards preconception expanded carrier screening for autosomal recessive disorders including inborn errors of metabolism. Mol. Genet. Metab. 2019, 126, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Delatycki, M.B.; Alkuraya, F.; Archibald, A.; Castellani, C.; Cornel, M.; Grody, W.W.; Henneman, L.; Ioannides, A.S.; Kirk, E.; Laing, N.; et al. International perspectives on the implementation of reproductive carrier screening. Prenat. Diagn. 2020, 40, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Castellani, C.; Picci, L.; Tridello, G.; Casati, E.; Tamanini, A.; Bartoloni, L.; Scarpa, M.; Assael, B.M. Cystic fibrosis carrier screening effects on birth prevalence and newborn screening. Genet. Med. 2016, 18, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.C.; Mall, M.A.; Gutierrez, H.; Macek, M.; Madge, S.; Davies, J.C.; Burgel, P.-R.; Tullis, E.; Castaños, C.; Castellani, C.; et al. The future of cystic fibrosis care: A global perspective. Lancet Respir. Med. 2020, 8, 65–124. [Google Scholar] [CrossRef]
- Gruber, A.; Pacault, M.; El Khattabi, L.A.; Vaucouleur, N.; Orhant, L.; Bienvenu, T.; Girodon, E.; Vidaud, D.; Leturcq, F.; Costa, C.; et al. Non-invasive prenatal diagnosis of paternally inherited disorders from maternal plasma: Detection of NF1 and CFTR mutations using droplet digital PCR. Clin. Chem. Lab. Med. 2018, 56, 728–738. [Google Scholar] [CrossRef]
- Guissart, C.; Tran Mau Them, F.; Debant, V.; Viart, V.; Dubucs, C.; Pritchard, V.; Rouzier, C.; Boureau-Wirth, A.; Haquet, E.; Puechberty, J.; et al. A Broad Test Based on Fluorescent-Multiplex PCR for Noninvasive Prenatal Diagnosis of Cystic Fibrosis. Fetal. Diagn. Ther. 2019, 45, 403–412. [Google Scholar] [CrossRef]

| Countries/States | 1st Tier | 2nd Tier | 3rd Tier | 4th Tier | Safety Net | IRT1 > Cut Off | Sensitivity (wo MI) | PPV CF | Ratio CF:CFSPID | Carrier Frequency |
|---|---|---|---|---|---|---|---|---|---|---|
| Brazil (Sao Paulo) [16] | IRT | IRT | 0.73–1.67% | 86–100% | 3–19% | ND 1.1:1 (Turkey) | NA | |||
| Russia [22] | ||||||||||
| Slovakia [1] | ||||||||||
| Turkey [23] | ||||||||||
| Spain (Andalusia) [24] | ||||||||||
| Austria [25] | PAP | IRT | 0.97% | 23% | 25:1 * | NA | ||||
| Portugal [26] | 0.70% | 94.4% | 41% | ND | NA | |||||
| Germany [27,28] | PAP | DNA (31) | ST | 0.73% | 96% | 20% | 5:1 | 1/44 | ||
| US (Colorado, Texas, Wyoming) [29] | IRT | DNA (41–48) | ST | 2.10% | 96% | 20% | 10.8:1 | 1/13 | ||
| US (Wisconsin) [30] | DNA (25) | ND | 95% | 9% | 5.2:1 | 1/9.5 | ||||
| Australia (Victoria) [18] | DNA (12) | ND | 96% | 18.3% | 7.8:1 | ND | ||||
| New Zealand [19] | DNA (3) | ND | 100% | 23% | ND | ND | ||||
| Italy (Tuscany) [31] | DNA (66) | 0.85% | 89.5% | 19.4% | 2.85:1 | 1/16 | ||||
| France [32] | DNA (29) | IRT | 0.50% | 95% | 34% | 9:1 | 1/16 | |||
| Switzerland [33] | DNA (7) | IRT | 0.78% | 97% | 36% | 17:1 * | 1/11 * | |||
| Czech Republic [1,34] | DNA (50) | IRT | 0.90% | 94% | 15% | 7.5:1 * | 1/21 | |||
| Norway [35] | DNA (72) | DNA (20) | ST | 0.8% | 95% | 43% | 1:1 | 1/10 | ||
| UK [1,36] | DNA (4) | DNA (29 or 31) | IRT | IRT | 0.57% * | 96% | 76% | 10.5:1 * | 1/28 * | |
| Denmark [21] | DNA (1) | EGA | EGA | 3.70% | 92% | 85% | 7:1 | 1/20 | ||
| California [7] | DNA (40) | EGA | 1.6% | 92% | 34% | 0.65:1 | 1/25 | |||
| Netherlands [37] | PAP | DNA (35) | EGA | EGA | 0.98% | 90% | 63% | 4:1 | 1/28 | |
| Poland [1,38] | DNA(limited seq) | EGA | 0.6% * | 100% | 26% | 1.2:1 * | 1/15 * |
| Strengths | Weaknesses |
|---|---|
| High PPV | but… |
|
|
| High Sensitivity | but… |
|
|
| Technically feasible | but… requires optimal healthcare system organization |
|
|
| Increased knowledge on CFTR variants | but… raises questions on variant interpretation |
|
|
| More precise medical care | but… raises ethical questions |
|
|
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergougnoux, A.; Lopez, M.; Girodon, E. The Role of Extended CFTR Gene Sequencing in Newborn Screening for Cystic Fibrosis. Int. J. Neonatal Screen. 2020, 6, 23. https://doi.org/10.3390/ijns6010023
Bergougnoux A, Lopez M, Girodon E. The Role of Extended CFTR Gene Sequencing in Newborn Screening for Cystic Fibrosis. International Journal of Neonatal Screening. 2020; 6(1):23. https://doi.org/10.3390/ijns6010023
Chicago/Turabian StyleBergougnoux, Anne, Maureen Lopez, and Emmanuelle Girodon. 2020. "The Role of Extended CFTR Gene Sequencing in Newborn Screening for Cystic Fibrosis" International Journal of Neonatal Screening 6, no. 1: 23. https://doi.org/10.3390/ijns6010023
APA StyleBergougnoux, A., Lopez, M., & Girodon, E. (2020). The Role of Extended CFTR Gene Sequencing in Newborn Screening for Cystic Fibrosis. International Journal of Neonatal Screening, 6(1), 23. https://doi.org/10.3390/ijns6010023




