Classification of Cocoa Beans by Analyzing Spectral Measurements Using Machine Learning and Genetic Algorithm
Abstract
1. Introduction
- Implementation of spectral measurements of cocoa beans: we have implemented a method to measure the spectral properties of three categories of cocoa beans. These properties are measured using a spectrometer, a device that measures the amount of light reflected by an object based on its wavelength.
- Classification of spectral measurements using a set of algorithms: we used a set of algorithms to classify the spectral measurements of the three categories of cocoa beans. These algorithms are based on machine learning, a discipline of artificial intelligence that allows computers to learn from data without being explicitly programmed.
- Selection of spectra using the best algorithm coupled with the genetic algorithm: we used the genetic algorithm to select the most relevant spectra for classification. The genetic algorithm is an optimization algorithm inspired by natural selection.
- Analysis and comparison of the different classifications: We analyzed and compared them to evaluate their performance.
2. Related Work
3. Materials and Methods
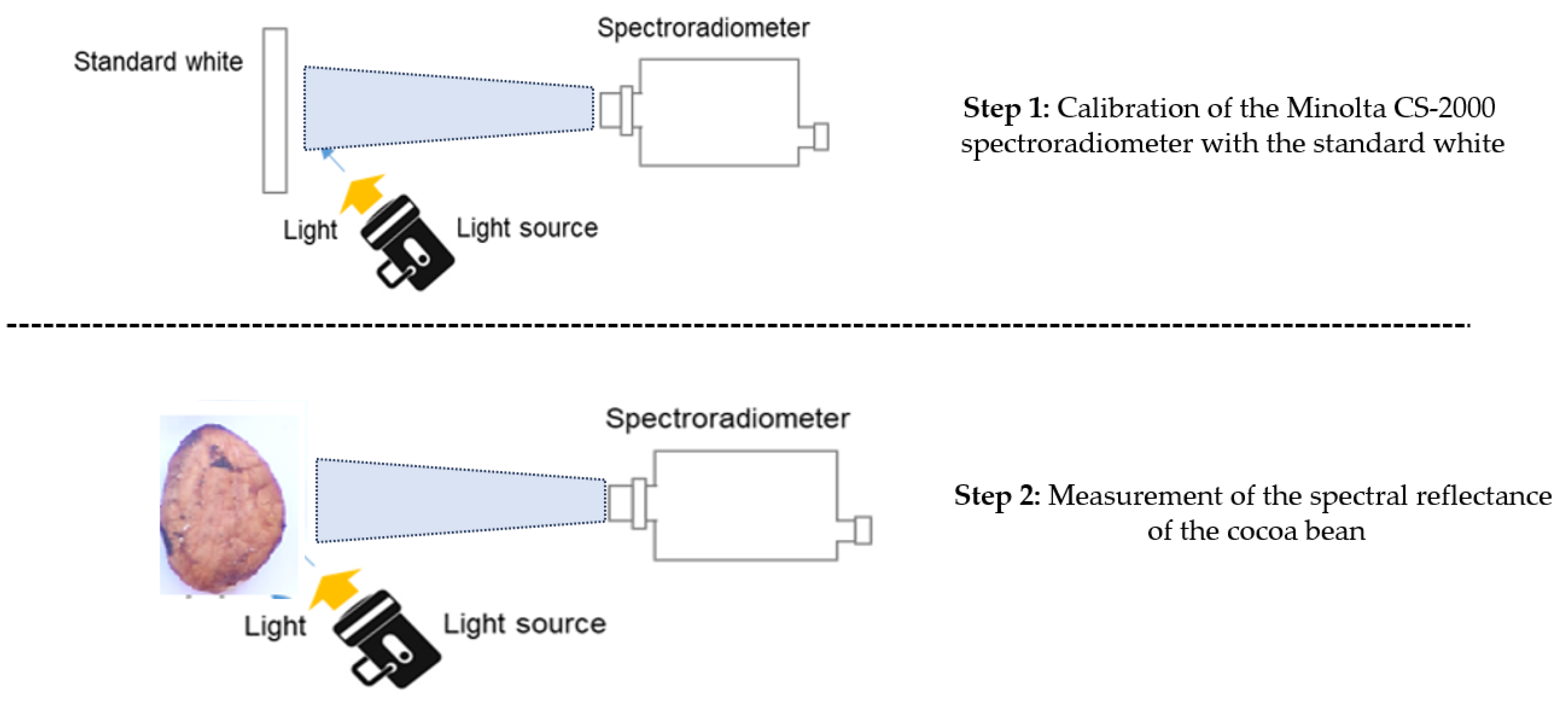
3.1. Spectral Measurement of Cocoa Beans
- Category 1: these are superior quality fermented and dried cocoa beans;
- Category 2: these are fermented and dried cocoa beans of intermediate quality;
- Category 3: these are unfermented and dried cocoa beans.
3.2. Machine Learning
- SVM (Support Vector Machine): SVMs are used for classification and regression. They work by finding a hyperplane that maximizes the margin between classes in a multidimensional data space. They are effective for binary classification but can be extended to multi-class classification [15].
- Logistic Regression: Logistic regression is mainly used for binary classification and can be extended to multi-class classification. It models the probability that an observation belongs to a particular class. It uses a logistic function to estimate the probability based on the input features [16].
- Random Forest: Random forest is an ensemble algorithm that combines multiple decision trees. Each tree is trained on a random sample of the data, and the predictions are aggregated to obtain a final prediction. It is robust, less susceptible to overfitting, and effective for classification and regression [17].
- AdaBoost (Adaptive Boosting): AdaBoost is another ensemble algorithm that adjusts based on errors from previous models. It assigns different weights to observations based on their previous performance. It combines several weak models to create a robust model [18].
- Decision Tree: Decision trees are used for classification and regression. They divide the dataset into subgroups based on characteristics, using criteria such as entropy or Gini coefficient. They are easy to interpret but can suffer from overfitting [19].
- K-Nearest Neighbors (KNN): KNN is a similarity-based classification and regression algorithm. It assigns a class to an observation based on the classes of the k nearest neighbors in the feature space. It is simple to understand but can be sensitive to the distance used [20].
- XGBoost (Extreme Gradient Boosting): XGBoost is an improved implementation of gradient-boosted learning. It is efficient and effective, suitable for classification and regression. It uses regularization and advanced tree management to improve accuracy [21].
3.3. Genetic Algorithm
- Initial Population: An initial group of potential solutions is randomly generated. Each solution is represented as chromosomes or genotypes, which encode the parameters or characteristics of the solution.
- Evaluation: Each solution in the population is evaluated using an objective or fitness function. This function measures the quality of each solution to the optimization objective.
- Selection: Solutions are selected for reproduction based on their fitness. Higher-quality solutions are more likely to be chosen, thus simulating natural selection.
- Reproduction: Selected solutions are combined to create a new generation of solutions. This can include operations such as recombination (crossover) and mutation. Crossbreeding involves merging two parental solutions to produce offspring, while mutation slightly modifies one solution.
- Replacement: The new generation replaces the old generation, often using a fitness-based replacement model. This ensures that the best quality solutions are preserved.
- Stopping Criterion: The genetic algorithm runs for a certain number of iterations or until a predefined stopping criterion is reached or setting a maximum number of iterations.
- Result: The best solution found during the execution of the algorithm is returned as the optimization result.
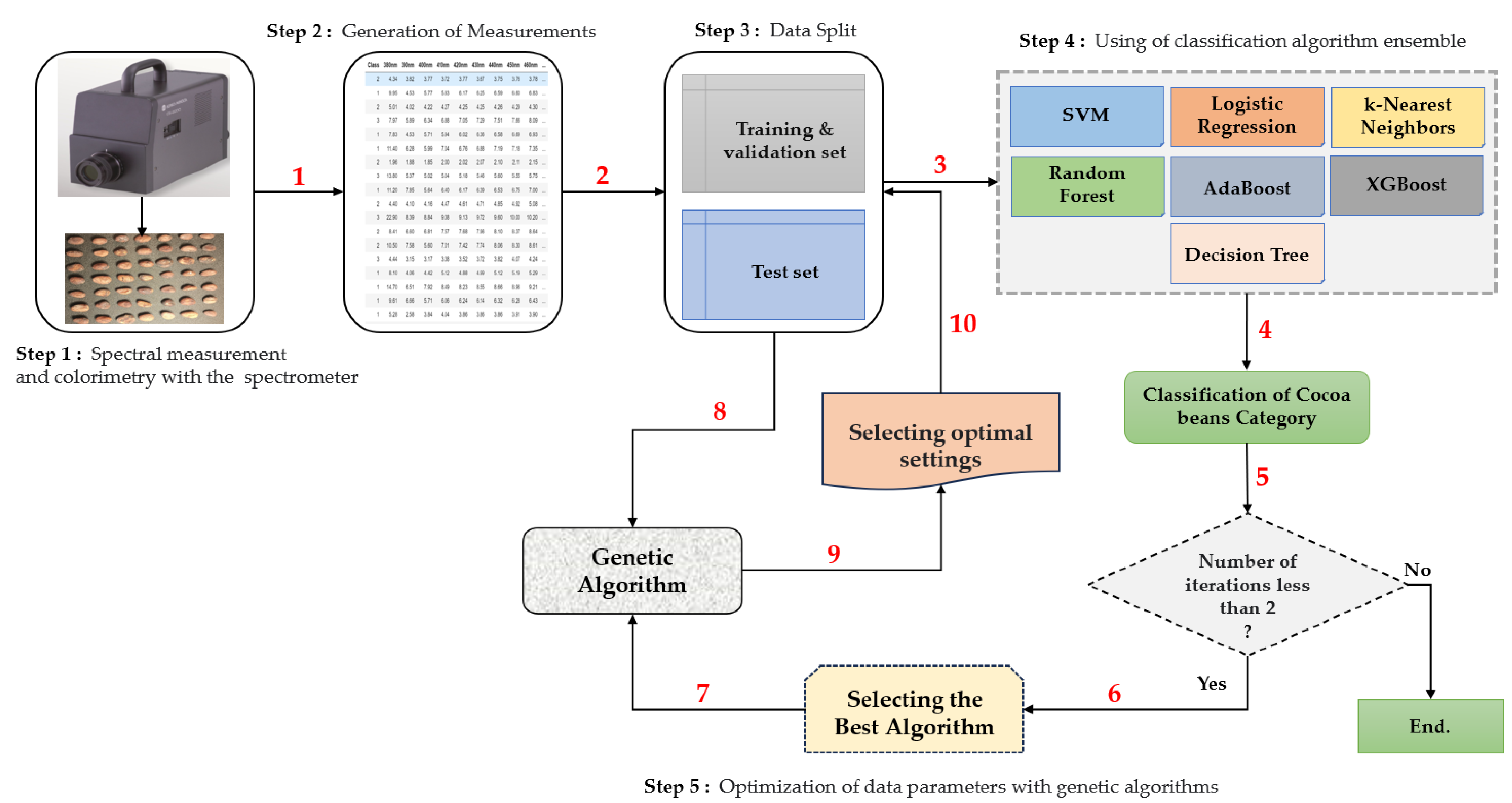
3.4. The General Architecture of the Methodology
- 1.
- Step 1: Spectral and Colorimetric Measurements
- 2.
- Step 2: Data Export
- 3.
- Step 3: Data Division
- 4.
- Step 4: Classification
- 5.
- Step 5: Selection of the Best Spectral Parameters
- a.
- First, we run a function to initialize a random population.
- b.
- The randomized population is then subjected to the fitness function, which returns the best parents (with the highest accuracy).
- c.
- The best parents will be selected according to the n-parents parameter.
- d.
- After performing the same operation, the population will be subjected to the crossover and mutation functions, respectively.
- e.
- The cross is created by combining the genes of the two most suitable parents by randomly selecting the first parent and part of the second parent.
- f.
- The mutation is obtained by randomly inverting the bits selected for the child resulting from the crossover.
- g.
- A new generation is created by selecting the most suitable parents from the previous generation and applying crossing over and mutation.
- h.
- This process is repeated for five generations.
| Algorithm 1 Train_test_evaluate |
| Input: X_train, X_test, y_train, y_test, models Output: Score, Modelchoice Begin Writing a function to train, test, and evaluate models: Creating the dataframe and initializing variables: acc_M = 0 Modelchoice = ”” Score = DataFrame({“Classifier”: classifiers}) j = 0 For each model in the models list model = i Training Phase: Train Model (X_train, y_train) Testing Phase: predictions = model.predict (X_test) Performance Evaluation: Metrics: Accuracy, Precision, Recall, F1-score, Matthews Correlation Coefficient, Mean Squared Error: Acc, Preci, Re_Sc, F1_S,Mcc,Mse = Classification_report(y_test, predictions). Saving model performance metrics: Score[j,”Accuracy”] = Acc; Score[j,”f1_score”] = F1_S; Score[j,”recall_score”] = re_sc Score[j,”matthews_corrcoef”] = Mcc; Score[j, “precision_score”] = Preci; Score[j, “Mse_score”] = Mse Identifying the best model if acc_M < acc then acc_M = acc Modelchoice = model End j = j + 1 Endfor return Score, Modelchoice End Function |
| Algorithm 2 Genetic |
| Input: X, y, X_train, X_test, y_train, y_test, new_model Output: Gene, score Begin new_model = ML Function initilization_of_population(size, n_feat) population = Empty list For i in range from 1 to size chromosome = Boolean array of size n_feat initialized to True For j in range from 0 to int(0.3 * n_feat) chromosome[j] = False Endfor Randomly shuffle the elements in chromosome Append chromosome to the population Endfor Return population Endfunction Function fitness_score (population, X_train, X_test, y_train, y_test) scores = Empty list For each chromosome in population Create a model (new_model) Train the model on X_train using the features specified by the chromosome Predict class labels on X_test with the model Calculate accuracy score by comparing predictions with y_test Append the score to scores Endfor Sort scores in descending order while maintaining the correspondence with the population Return scores, sorted population Endfunction Function selection (pop_after_fit, n_parents) population_nextgen = Empty list For i in range from 1 to n_parents Append pop_after_fit[i] to population_nextgen Endfor Return population_nextgen Endfunction Function crossover (pop_after_sel) pop_nextgen = Copy pop_after_sel For i in range from 0 to the size of pop_after_sel - 1 with a step of 2 new_par = Empty list child_1, child_2 = pop_nextgen[i], pop_nextgen[i+1] new_par = Concatenate the first half of child_1 and the second half of child_2. Append new_par to pop_nextgen Endfor Return pop_nextgen Endfunction Function mutation (pop_after_cross, mutation_rate, n_feat) mutation_range = Integer rounded down of mutation_rate times n_feat pop_next_gen = Empty list For each individual in pop_after_cross chromosome < = Copy the individual rand_posi < = Empty list For i in range from 1 to mutation_range pos < = Random integer between 0 and n_feat - 1 Append pos to rand_posi Endfor For each position in rand_posi Invert the value of the gene corresponding to that position in chromosome Endfor Append chromosome to pop_next_gen Endfor Return pop_next_gen Endfunction Function generations (df, label, size, n_feat, n_parents, mutation_rate, n_gen, X_train, X_test, y_train, y_test) best_chromo = Empty list best_score = Empty list population_nextgen = Call initilization_of_population with size and n_feat For each generation i from 1 to n_gen scores, pop_after_fit = Call fitness_score with population_nextgen, X_train, X_test, y_train, y_test Print the best score in generation i pop_after_sel = Call selection with pop_after_fit and n_parents pop_after_cross = Call crossover with pop_after_sel population_nextgen = Call mutation with pop_after_cross, mutation_rate, n_feat Append the best individual from generation i to best_chromo Append the best score from generation i to best_score Endfor Return best_chromo, best_score Endfunction Calling the function Gene,score = generations (X, y, size = 80, n_feat = X.shape[1], n_parents = 64, mutation_rate = 0.20, n_gen = 5, X_train = X_train, X_test = X_test, y_train = y_train, y_test = y_test) |
| Algorithm 3 main |
| Input: DataSpectre.xlsx Output: Score, Modelchoice Begin Reading data: data = Read Excel file: “Data.xlsx” Extracting features and labels: X = Feature selection without the target variable: “Class” y = Reading data from the target variable: “Class” Splitting data into training and testing sets: X_train, X_test, y_train, y_test = Training and testing data split (X, y) Creating classifiers and models: Classifiers= [‘SVM’, ’Logistic’, ’RandomForest’, ’AdaBoost’, ’DecisionTree’, ’KNeighbors’, ‘XGBoost’] models=[SVM(), LogisticRegression(), RandomForestClassifier(), AdaBoostClassifier(), DecisionTreeClassifier(), KNeighborsClassifier(), XGBClassifier()] Writing a function to train, test, and evaluate models: Score, Modelchoice = Train_test_evaluate(X_train, X_test, y_train, y_test, models) G, S = Genetic(X, y, X_train, X_test, y_train, y_test, Modelchoice) Choose the indices associated with the highest scores in the variable ‘ind.’ For vi in ind for i, value in enumerate(G[vi]) if value is True then append i to indices_true1 end end X_train_GA = X_train with columns selected using indices in indices_true1 X_test_GA = X_test with columns selected using indices in indices_true1 Score, Modelchoice = Train_test_evaluate(X_train_GA, X_test_GA, y_train, y_test, models) end |
3.5. Performance Metric
- Accuracy: This assesses the proportion of correctly predicted observations.
- Precision: This measures the proportion of accurate positive predictions.
- Mean Squared Error (MSE) calculates the average difference between predicted and actual values.
- Recall: This evaluates the proportion of actual positive observations that are correctly predicted.
- F1 Score: This is a weighted average of precision and recall.
- Matthews Correlation Coefficient (MCC): This correlation coefficient assesses the quality of binary and multiclass classifications.
4. Results and Discussion
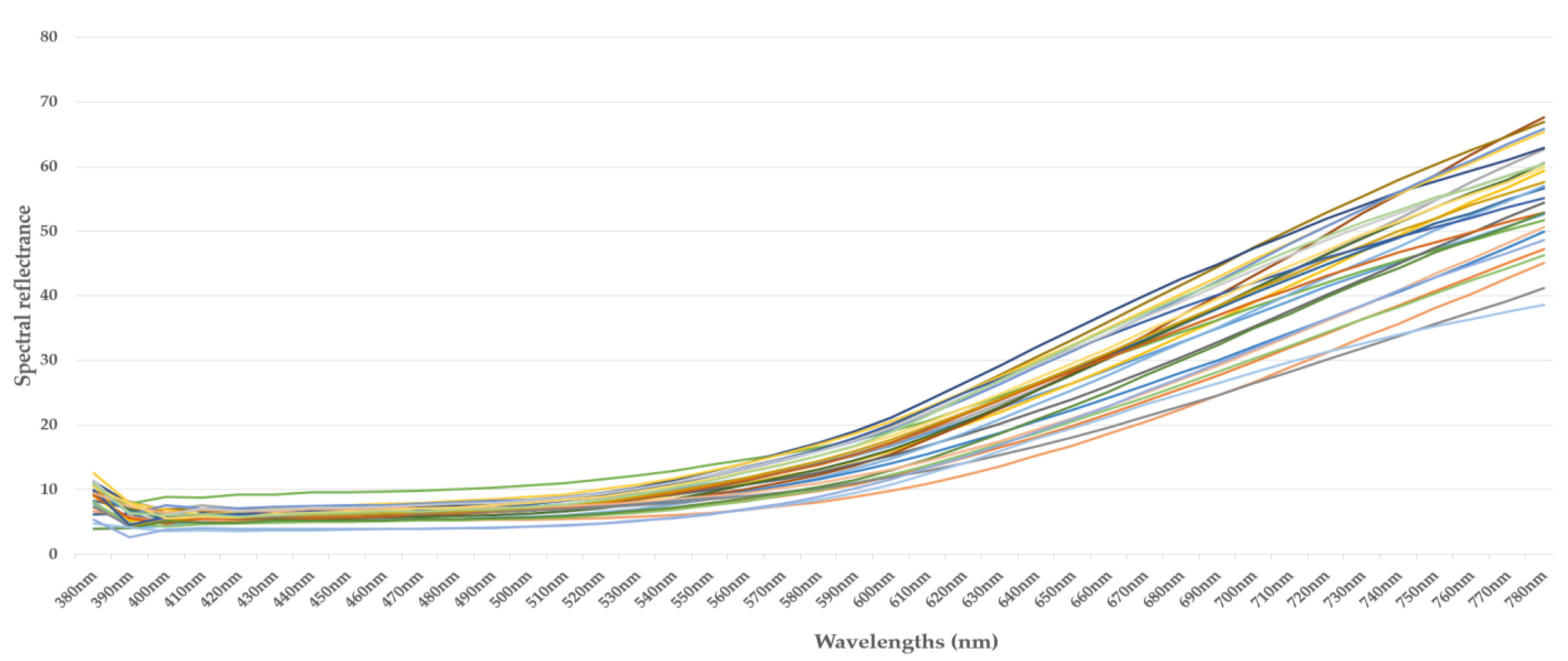
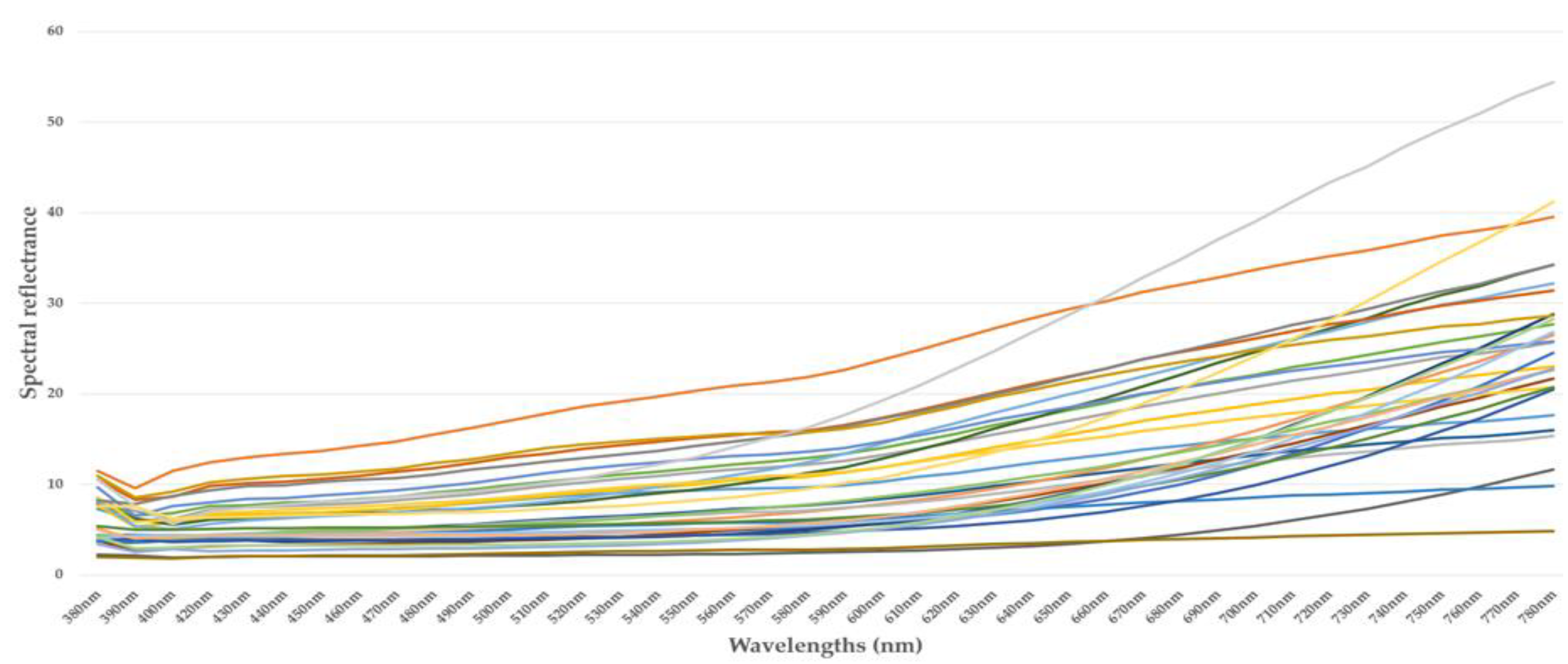
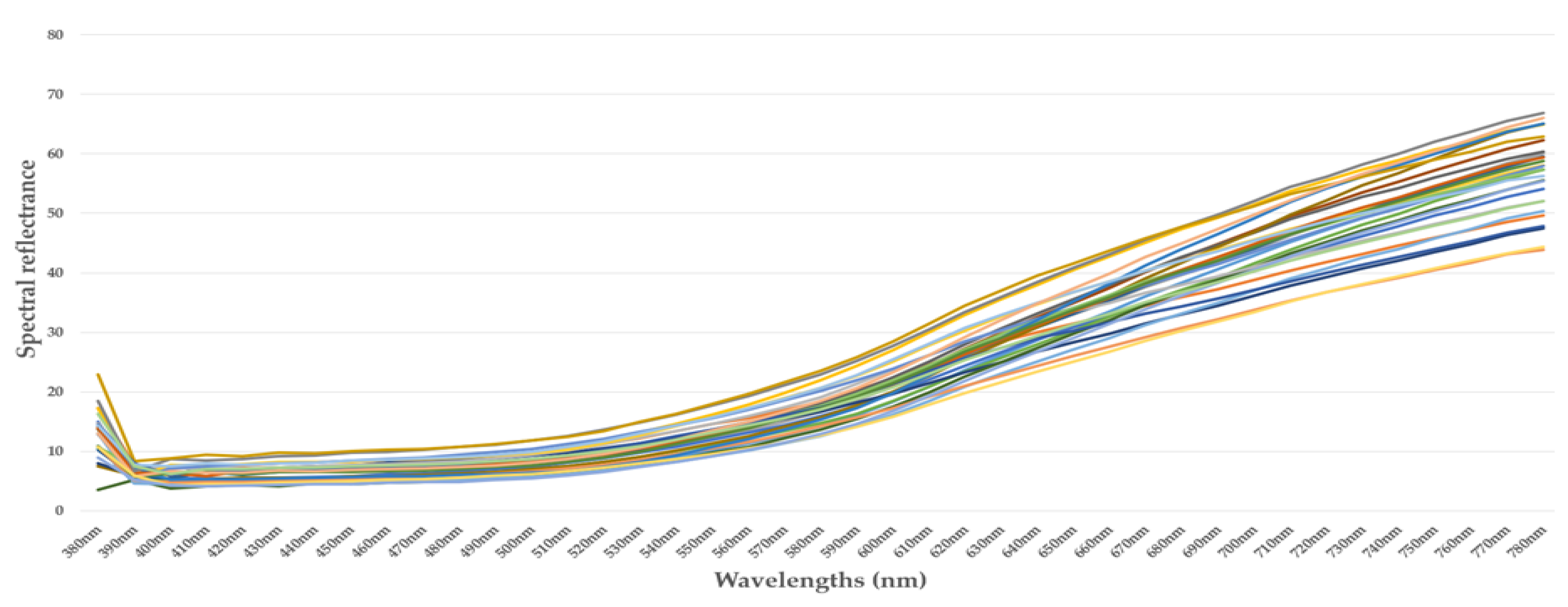
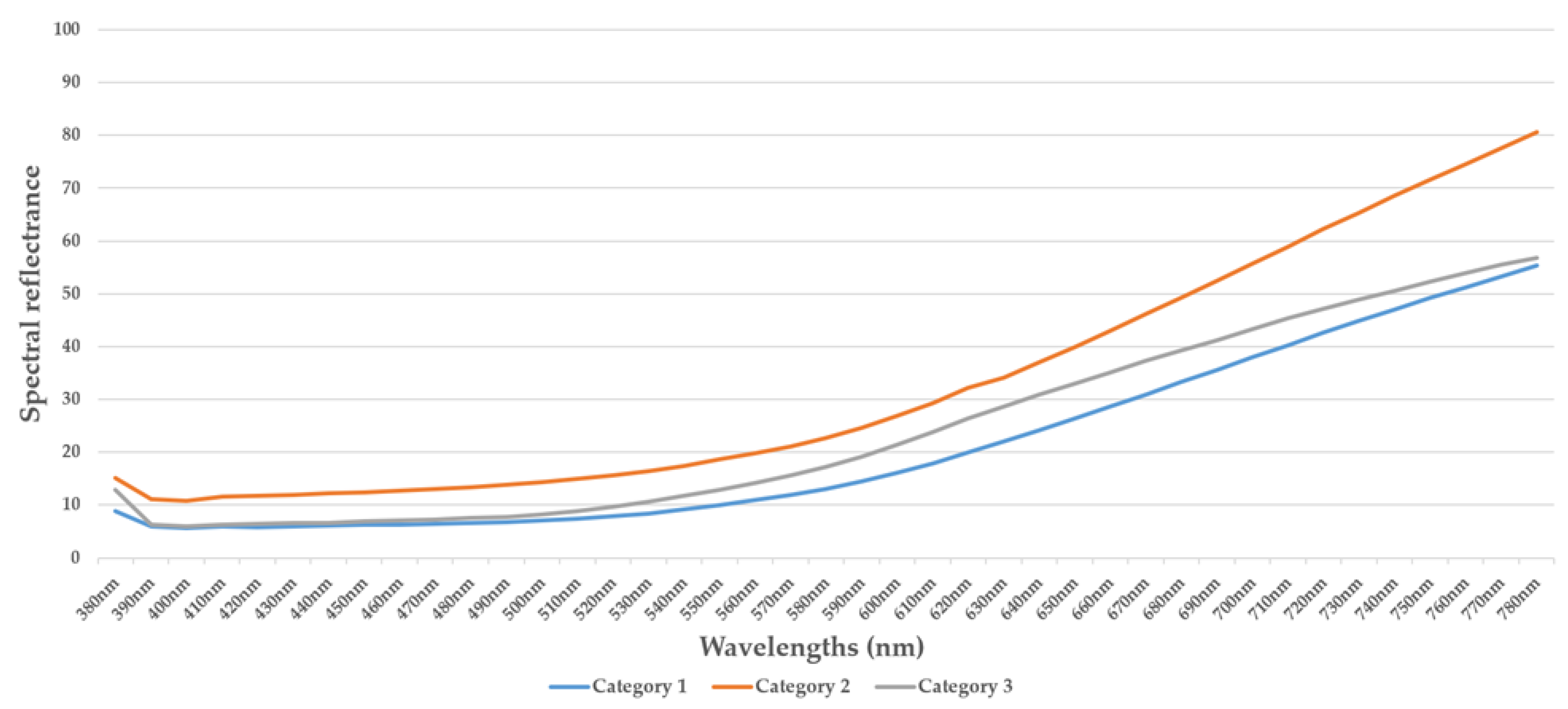
4.1. Spectral Measurements
4.2. Summary of Spectral Measurements
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cocoa Bean Production Ivory Coast 2022/2023. Available online: https://www.statista.com/statistics/497838/production-of-cocoa-beans-in-ivory-coast/ (accessed on 20 October 2023).
- UNSDG|Sustainable Cocoa Farming in Côte d’Ivoire: UN Deputy Chief Notes Significant Progress and Calls for Greater International Support. Available online: https://unsdg.un.org/latest/stories/sustainable-cocoa-farming-cote-divoire-un-deputy-chief-notes-significant-progress (accessed on 20 October 2023).
- Teye, E.; Anyidoho, E.; Agbemafle, R.; Sam-Amoah, L.K.; Elliott, C. Cocoa Bean and Cocoa Bean Products Quality Evaluation by NIR Spectroscopy and Chemometrics: A Review. Infrared Phys. Technol. 2020, 104, 103127. [Google Scholar] [CrossRef]
- Santika, G.D.; Wulandari, D.A.R.; Dewi, F. Quality Assessment Level of Quality of Cocoa Beans Export Quality Using Hybrid Adaptive Neuro-Fuzzy Inference System (ANFIS) and Genetic Algorithm. In Proceedings of the 2018 International Conference on Electrical Engineering and Computer Science (ICECOS), Pangkal Pinang, Indonesia, 2–4 October 2018; pp. 195–200. [Google Scholar]
- Essah, R.; Anand, D.; Singh, S. An Intelligent Cocoa Quality Testing Framework Based on Deep Learning Techniques. Meas. Sens. 2022, 24, 100466. [Google Scholar] [CrossRef]
- Agronomy|Free Full-Text|Feature Extraction for Cocoa Bean Digital Image Classification Prediction for Smart Farming Application. Available online: https://www.mdpi.com/2073-4395/10/11/1642 (accessed on 21 October 2023).
- Tanimowo, A.O.; Eludiora, S.; Ajayi, A. A Decision Tree Classification Model for Cocoa Beans Quality Evaluation Based on Digital Imaging. Ife J. Technol. 2020, 27, 6–13. [Google Scholar]
- Wei, J.; Liu, S.; Qileng, A.; Qin, W.; Liu, W.; Wang, K.; Liu, Y. A Photoelectrochemical/Colorimetric Immunosensor for Broad-Spectrum Detection of Ochratoxins Using Bifunctional Copper Oxide Nanoflowers. Sens. Actuators B Chem. 2021, 330, 129380. [Google Scholar] [CrossRef]
- Lin, H.; Kang, W.; Han, E.; Chen, Q. Quantitative Analysis of Colony Number in Mouldy Wheat Based on near Infrared Spectroscopy Combined with Colorimetric Sensor. Food Chem. 2021, 354, 129545. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Hernández, C.; Fernández-Cabanás, V.M.; Rodríguez-Gutiérrez, G.; Fernández-Prior, Á.; Morales-Sillero, A. Rapid Screening of Unground Cocoa Beans Based on Their Content of Bioactive Compounds by NIR Spectroscopy. Food Control 2022, 131, 108347. [Google Scholar] [CrossRef]
- Karadağ, K.; Tenekeci, M.E.; Taşaltın, R.; Bilgili, A. Detection of Pepper Fusarium Disease Using Machine Learning Algorithms Based on Spectral Reflectance. Sustain. Comput. Inform. Syst. 2020, 28, 100299. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, J.; Chen, Y.; Wan, S.; Zhang, L. Detection of Peanut Leaf Spots Disease Using Canopy Hyperspectral Reflectance. Comput. Electron. Agric. 2019, 156, 677–683. [Google Scholar] [CrossRef]
- CS-2000 Spectroradiometer. Konica Minolta Sensing. Available online: https://www.konicaminolta.fr/fr-fr/hardware/instruments-de-mesure/lumiere-ecrans/spectroradiometres/cs-2000a-cs-2000 (accessed on 22 October 2023).
- Ayikpa, K.J.; Mamadou, D.; Gouton, P.; Adou, K.J. Classification of Cocoa Pod Maturity Using Similarity Tools on an Image Database: Comparison of Feature Extractors and Color Spaces. Data 2023, 8, 99. [Google Scholar] [CrossRef]
- Mamadou, D.; Ayikpa, K.J.; Ballo, A.B.; Kouassi, B.M. Cocoa Pods Diseases Detection by MobileNet Confluence and Classification Algorithms. IJACSA 2023, 14, 9. [Google Scholar] [CrossRef]
- Bailly, A.; Blanc, C.; Francis, É.; Guillotin, T.; Jamal, F.; Wakim, B.; Roy, P. Effects of Dataset Size and Interactions on the Prediction Performance of Logistic Regression and Deep Learning Models. Comput. Methods Programs Biomed. 2022, 213, 106504. [Google Scholar] [CrossRef] [PubMed]
- Sensors|Free Full-Text|A Hybrid Intrusion Detection Model Using EGA-PSO and Improved Random Forest Method. Available online: https://www.mdpi.com/1424-8220/22/16/5986 (accessed on 22 October 2023).
- Ding, Y.; Zhu, H.; Chen, R.; Li, R. An Efficient AdaBoost Algorithm with the Multiple Thresholds Classification. Appl. Sci. 2022, 12, 5872. [Google Scholar] [CrossRef]
- Costa, V.G.; Pedreira, C.E. Recent Advances in Decision Trees: An Updated Survey. Artif. Intell. Rev. 2023, 56, 4765–4800. [Google Scholar] [CrossRef]
- Skin Lesion Classification System Using a K-Nearest Neighbor Algorithm|Visual Computing for Industry, Biomedicine, and Art|Full Text. Available online: https://vciba.springeropen.com/articles/10.1186/s42492-022-00103-6 (accessed on 22 October 2023).
- Applied Sciences|Free Full-Text|Prediction of Pile Bearing Capacity Using XGBoost Algorithm: Modeling and Performance Evaluation. Available online: https://www.mdpi.com/2076-3417/12/4/2126 (accessed on 22 October 2023).
- Sohail, A. Genetic Algorithms in the Fields of Artificial Intelligence and Data Sciences. Ann. Data. Sci. 2023, 10, 1007–1018. [Google Scholar] [CrossRef]
- Neumann, A.; Hajji, A.; Rekik, M.; Pellerin, R. Genetic Algorithms for Planning and Scheduling Engineer-to-Order Production: A Systematic Review. Int. J. Prod. Res. 2023, 1–30. [Google Scholar] [CrossRef]
- Gen, M.; Lin, L. Genetic Algorithms and Their Applications. In Springer Handbook of Engineering Statistics; Pham, H., Ed.; Springer Handbooks; Springer: London, UK, 2023; pp. 635–674. ISBN 978-1-4471-7503-2. [Google Scholar]
- Cheuque, C.; Querales, M.; León, R.; Salas, R.; Torres, R. An Efficient Multi-Level Convolutional Neural Network Approach for White Blood Cells Classification. Diagnostics 2022, 12, 248. [Google Scholar] [CrossRef] [PubMed]
- Bechelli, S.; Delhommelle, J. Machine Learning and Deep Learning Algorithms for Skin Cancer Classification from Dermoscopic Images. Bioengineering 2022, 9, 97. [Google Scholar] [CrossRef] [PubMed]


| Algorithms | Accuracy (%) | Precision (%) | MSE | F-Score (%) | Recall (%) | MCC (%) |
|---|---|---|---|---|---|---|
| SVM | 78.26 | 79.42 | 0.6086 | 77.98 | 78.26 | 68.10 |
| Decision Tree | 60.86 | 60.86 | 1.3043 | 60.31 | 60.86 | 41.66 |
| Random Forest | 73.91 | 74.12 | 0.7826 | 73.82 | 73.91 | 60.96 |
| XGBoost | 65.21 | 64.34 | 1.13 | 64.18 | 65.21 | 48.27 |
| KNeighbors | 65.21 | 66.66 | 1.13 | 65.28 | 65.21 | 48.27 |
| Logistic | 82.60 | 83.76 | 0.69 | 82.33 | 82.60 | 74.71 |
| AdaBoost | 69.56 | 72.46 | 0.95 | 67.82 | 69.56 | 56.90 |
| Generation | Score (%) |
|---|---|
| 1 | 82.60 |
| 2 | 86.95 |
| 3 | 82.60 |
| 4 | 82.60 |
| 5 | 86.95 |
| Generation | 2 | 5 |
|---|---|---|
| Wavelength (nm) | 410; 430; 490; 500; 510; 520; 530; 540; 550; 560; 580; 600; 610; 630; 690; 700; 710; 720; 740; 760. | 380; 390; 400; 420; 430; 440; 450; 500; 510; 520; 550; 580; 590; 620; 630; 640; 660; 670; 680; 730. |
| Algorithms | Accuracy (%) | Precision (%) | MSE | F-Score (%) | Recall (%) | MCC (%) |
|---|---|---|---|---|---|---|
| SVM | 78.26 | 79.42 | 0. 60 | 77.98 | 78.26 | 68.10 |
| Decision Tree | 65.21 | 65.21 | 1.13 | 65.21 | 65.21 | 47.72 |
| Random Forest | 69.56 | 69.15 | 0.95 | 69.19 | 69.56 | 54.41 |
| XGBoost | 78.26 | 79.08 | 0.60 | 78.46 | 78.26 | 67.52 |
| KNeighbors | 65.21 | 65.21 | 1.13 | 65.21 | 65.21 | 47.72 |
| Logistic | 82.60 | 84.71 | 0.56 | 82.22 | 82.60 | 75.31 |
| AdaBoost | 60.86 | 43.47 | 1.30 | 49.27 | 60.86 | 51.63 |
| Algorithms | Accuracy (%) | Precision (%) | MSE | F-Score (%) | Recall (%) | MCC (%) |
|---|---|---|---|---|---|---|
| SVM | 73.91 | 75.36 | 0.91 | 74.38 | 73.91 | 60.85 |
| Decision Tree | 65.21 | 65.28 | 1.13 | 65.09 | 65.21 | 47.86 |
| Random Forest | 73.91 | 74.12 | 0.78 | 73.82 | 73.91 | 60.96 |
| XGBoost | 69.56 | 70.14 | 0.95 | 69.15 | 69.56 | 54.88 |
| KNeighbors | 65.21 | 67.14 | 1.26 | 65.94 | 65.21 | 47.71 |
| Logistic | 69.56 | 71.49 | 1.08 | 70.29 | 69.56 | 54.28 |
| AdaBoost | 60.86 | 43.47 | 1.30 | 49.27 | 60.86 | 51.63 |
| Algorithms | Precision (%)–All | Precision (%)–Generation 2 | Precision (%)–Generation 5 |
|---|---|---|---|
| SVM | 79.42 | 79.42 | 75.36 |
| Decision Tree | 60.86 | 65.21 | 65.28 |
| Random Forest | 74.12 | 69.15 | 74.12 |
| XGBoost | 64.34 | 79.08 | 70.14 |
| KNeighbors | 66.66 | 65.21 | 67.14 |
| Logistic | 83.76 | 84.71 | 71.49 |
| AdaBoost | 72.46 | 43.47 | 43.47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayikpa, K.J.; Gouton, P.; Mamadou, D.; Ballo, A.B. Classification of Cocoa Beans by Analyzing Spectral Measurements Using Machine Learning and Genetic Algorithm. J. Imaging 2024, 10, 19. https://doi.org/10.3390/jimaging10010019
Ayikpa KJ, Gouton P, Mamadou D, Ballo AB. Classification of Cocoa Beans by Analyzing Spectral Measurements Using Machine Learning and Genetic Algorithm. Journal of Imaging. 2024; 10(1):19. https://doi.org/10.3390/jimaging10010019
Chicago/Turabian StyleAyikpa, Kacoutchy Jean, Pierre Gouton, Diarra Mamadou, and Abou Bakary Ballo. 2024. "Classification of Cocoa Beans by Analyzing Spectral Measurements Using Machine Learning and Genetic Algorithm" Journal of Imaging 10, no. 1: 19. https://doi.org/10.3390/jimaging10010019
APA StyleAyikpa, K. J., Gouton, P., Mamadou, D., & Ballo, A. B. (2024). Classification of Cocoa Beans by Analyzing Spectral Measurements Using Machine Learning and Genetic Algorithm. Journal of Imaging, 10(1), 19. https://doi.org/10.3390/jimaging10010019









