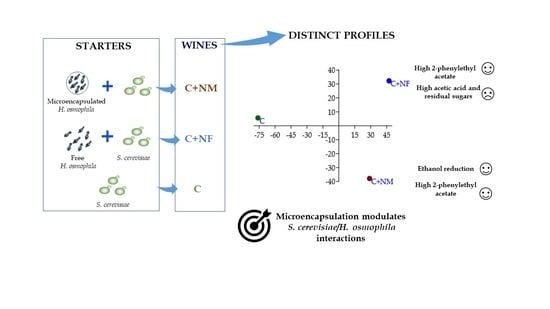
Influence of Microencapsulation on Fermentative Behavior of Hanseniaspora osmophila in Wine Mixed Starter Fermentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Biomass Production
2.3. Microencapsulation of ND1 Strain
2.4. Efficiency of Microencapsulation Process
2.5. Mixed Inoculated Fermentations at Laboratory Scale with ND1 Strain in Different Formulations
2.6. Evaluation of Yeast Viable Population during the Fermentations
2.7. Analytical Determination of Experimental Wines
2.8. Influence of Acetic Acid Addition on S. cerevisiae EC1118 Fermentative Performance
2.9. Data Analysis
3. Results
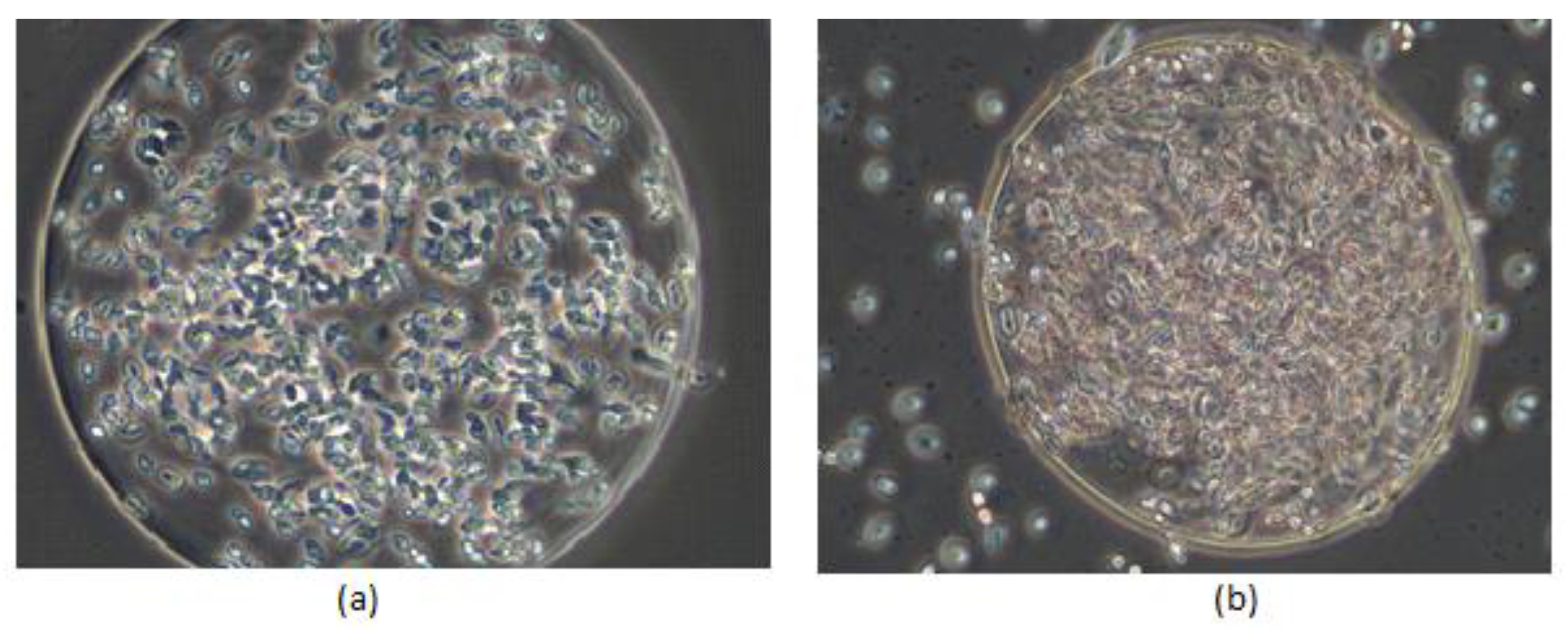
3.1. Efficiency of Microencapsulation Process
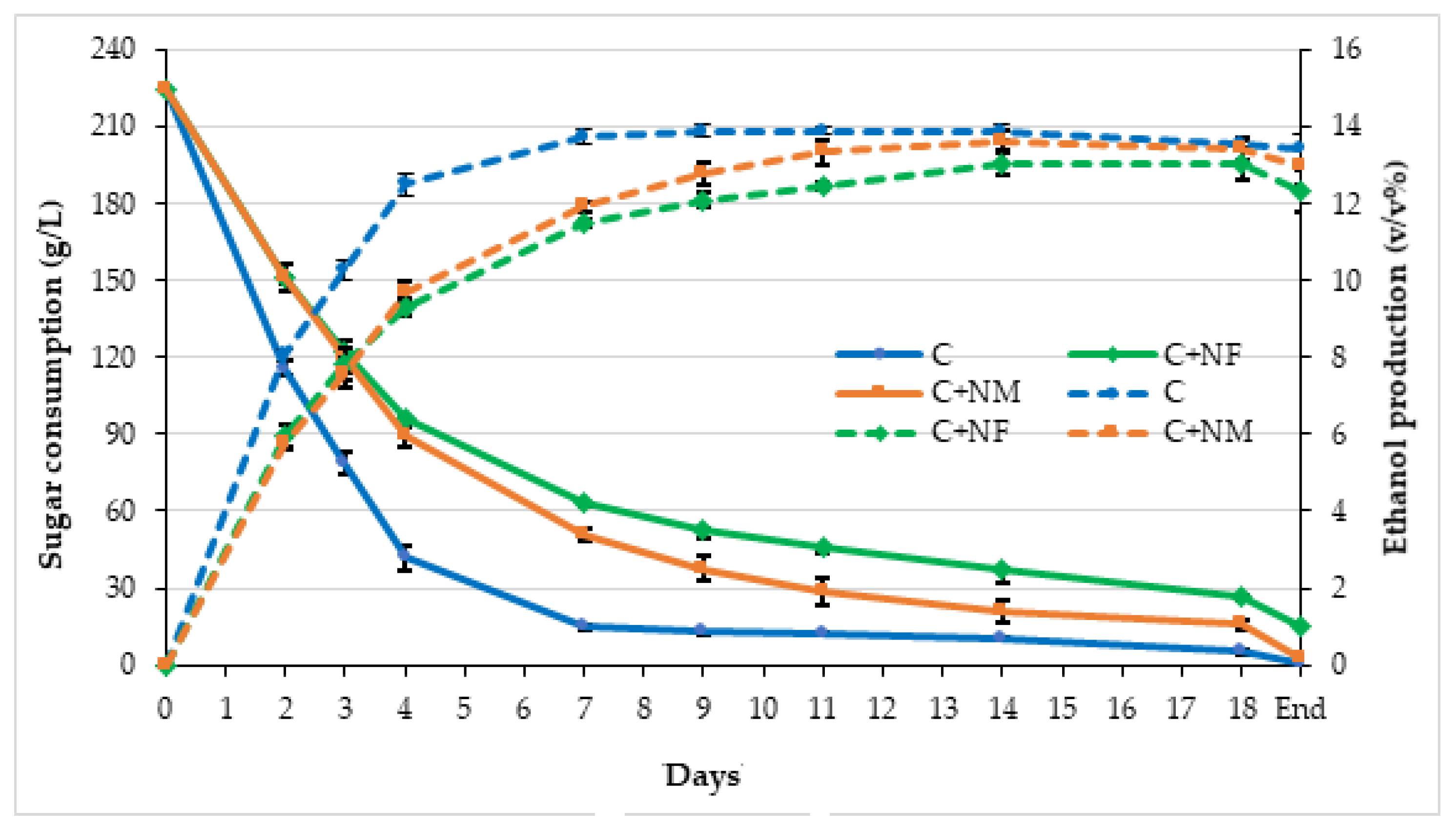
3.2. Fermentation Kinetics
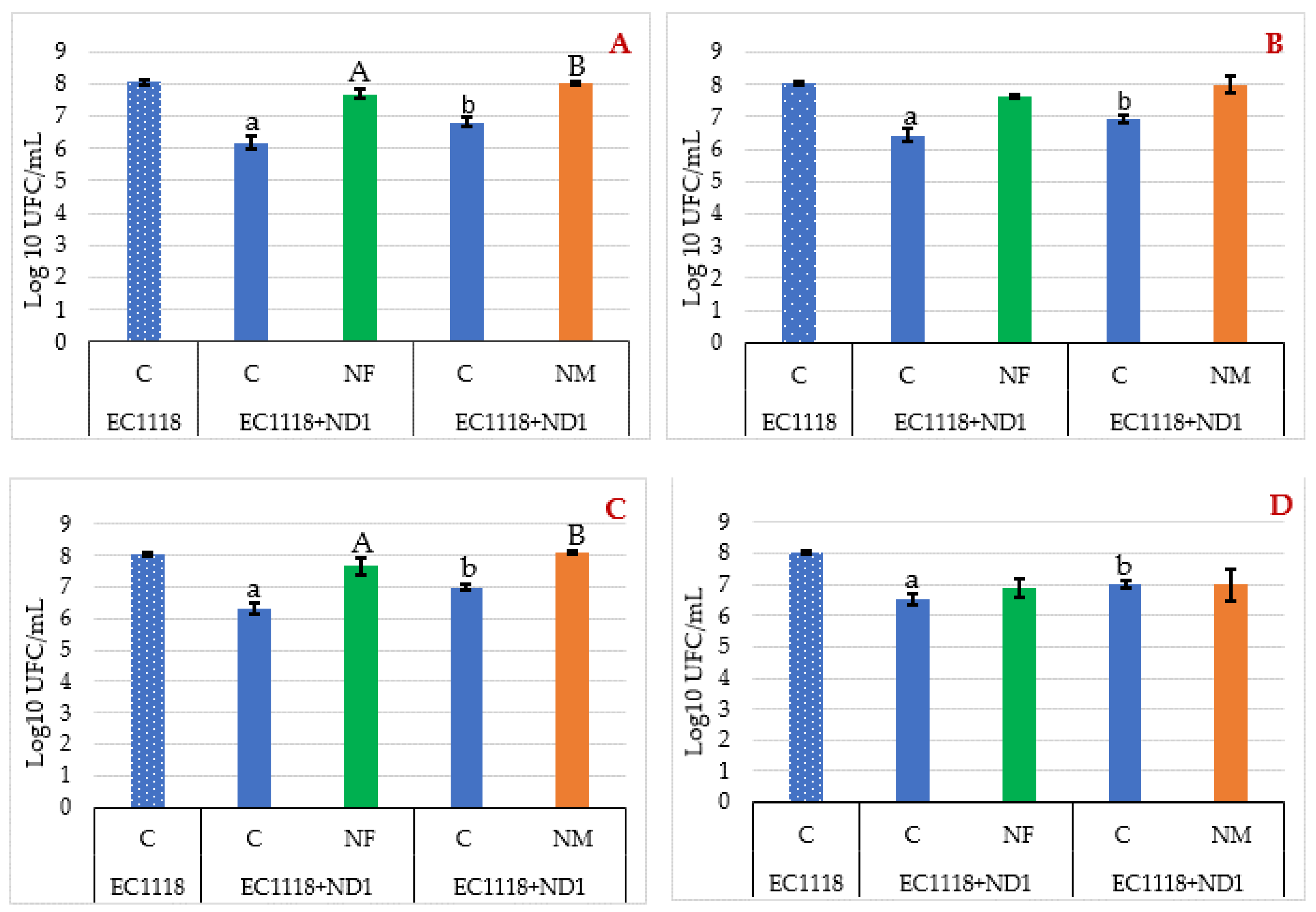
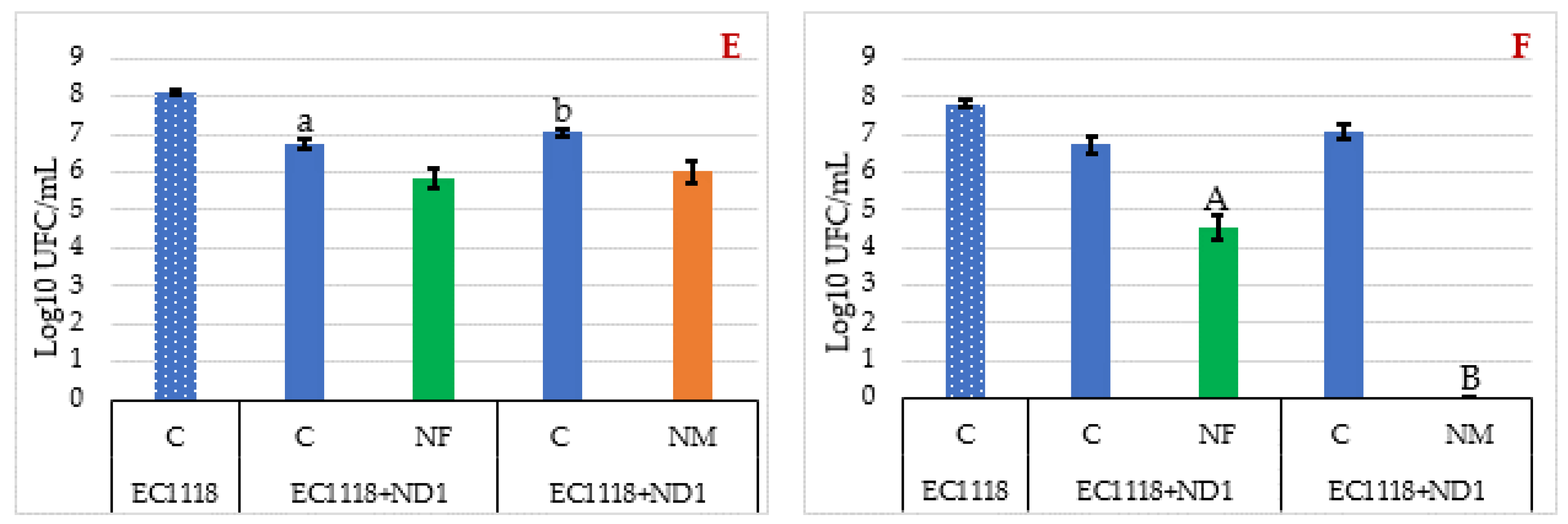
3.3. Evaluation of Yeast Viable Population during the Fermentations
3.4. Analysis of Experimental Wines for Main Oenological Parameters
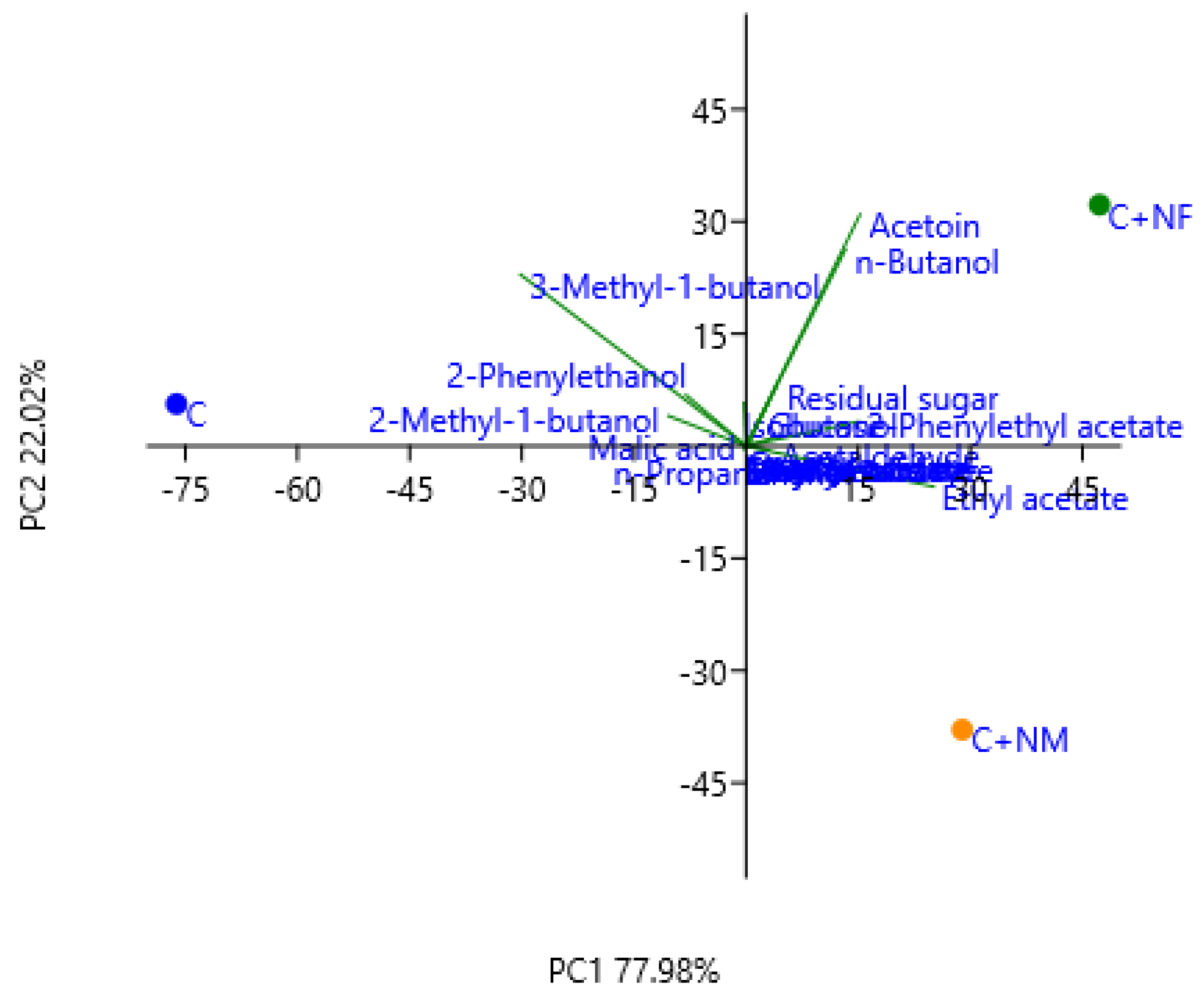
3.5. Aromatic Compounds of Experimental Wines
3.6. Influence of Acetic Acid Addition on S. cerevisiae EC1118 Fermentative Performance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Padilla, B.; Gil, J.V.; Manzanares, P. Past and Future of Non-Saccharomyces Yeasts: From Spoilage Microorganisms to Biotechnological Tools for Improving Wine Aroma Complexity. Front. Microbiol. 2016, 7, 411. [Google Scholar] [CrossRef] [Green Version]
- Ciani, M.; Comitini, F.; Mannazzu, I.; Domizio, P. Controlled mixed culture fermentation: A new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res. 2010, 10, 123–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borren, E.; Tian, B. The Important Contribution of Non-Saccharomyces Yeasts to the Aroma Complexity of Wine: A Review. Foods 2021, 10, 13. [Google Scholar] [CrossRef]
- Tufariello, M.; Fragasso, M.; Pico, J.; Panighel, A.; Castellarin, S.D.; Flamini, R.; Grieco, F. Influence of non-Saccharomyces on wine chemistry: A focus on aroma-related compounds. Molecules 2021, 26, 644. [Google Scholar] [CrossRef]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef]
- Pietrafesa, A.; Capece, A.; Pietrafesa, R.; Bely, M.; Romano, P. Saccharomyces cerevisiae and Hanseniaspora uvarum mixed starter cultures: Influence of microbial/physical interactions on wine characteristics. Yeast 2020, 37, 609–621. [Google Scholar] [CrossRef]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef] [Green Version]
- Escribano, R.; Lucía González-Arenzana, L.; Garijo, P.; Berlanas, C.; López-Alfaro, I.; López, R.; Gutiérrez, A.R.; Santamaría, P. Screening of enzymatic activities within different enological non-Saccharomyces yeasts. J. Food Sci. Technol. 2017, 54, 1555–1564. [Google Scholar] [CrossRef] [Green Version]
- López, M.C.; Mateo, J.J.; Maicas, S. Screening of β-Glucosidase and β-Xylosidase Activities in Four Non-Saccharomyces Yeast Isolates. J. Food Sci. 2015, 80, C1696–C1704. [Google Scholar] [CrossRef] [PubMed]
- Canonico, L.; Solomon, M.; Comitini, F.; Ciani, M.; Varela, C. Volatile profile of reduced alcohol wines fermented with selected non-Saccharomyces yeasts under different aeration conditions. Food Microbiol. 2019, 84, 1032472. [Google Scholar] [CrossRef]
- García, M.; Esteve-Zarzoso, B.; Cabellos, J.M.; Arroyo, T. Sequential Non-Saccharomyces and Saccharomyces cerevisiae Fermentations to Reduce the Alcohol Content in Wine. Fermentation 2020, 6, 60. [Google Scholar] [CrossRef]
- Navrátilová, M.; Beranová, M.; Severová, L.; Šrédl, K.; Svoboda, R.; Abrhám, J. The Impact of Climate Change on the Sugar Content of Grapes and the Sustainability of their Production in the Czech Republic. Sustainability 2021, 13, 222. [Google Scholar] [CrossRef]
- Varela, C.; Dry, P.R.; Kutyna, D.R.; Francis, I.L.; Henschke, P.A.; Curtin, C.D.; Chambers, P.J. Strategies for reducing alcohol concentration in wine. Aust. J. Grape Wine Res. 2015, 21, 670–679. [Google Scholar] [CrossRef]
- Neethling, E.; Petitjean, T.; Quénol, H.; Barbeau, G. Assessing local climate vulnerability and winegrowers ‘adaptive processes in the context of climate change. Mitig. Adapt. Strateg. Glob. Chang. 2017, 22, 777–803. [Google Scholar] [CrossRef]
- Varela, J.; Varela, C. Microbiological strategies to produce beer and wine with reduced ethanol concentration. Curr. Opin. Biotech. 2019, 56, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Englezos, V.; Rantsiou, K.; Cravero, F.; Torchio, F.; Ortiz-Julien, A.; Gerbi, V.; Rolle, L.; Cocolin, L. Starmerella bacillaris and Saccharomyces cerevisiae mixed fermentations to reduce ethanol content in wine. Appl. Microbiol. Biotechnol. 2016, 100, 5515–5526. [Google Scholar] [CrossRef]
- Ciani, M.; Morales, P.; Comitini, F.; Tronchoni, J.; Canonico, L.; Curiel, J.A.; Oro, L.; Rodrigues, A.J.; Gonzalez, R. Non-conventional Yeast Species for Lowering Ethanol Content of Wines. Front. Microbiol. 2016, 7, 642. [Google Scholar] [CrossRef] [Green Version]
- Canonico, L.; Comitini, F.; Oro, L.; Ciani, M. Sequential Fermentation with Selected Immobilized Non-Saccharomyces Yeast for Reduction of Ethanol Content in Wine. Front. Microbiol. 2016, 7, 278. [Google Scholar] [CrossRef] [Green Version]
- Canonico, L.; Comitini, F.; Ciani, M. Metschnikowia pulcherrima Selected Strain for Ethanol Reduction in Wine: Influence of Cell Immobilization and Aeration Condition. Foods 2019, 8, 378. [Google Scholar] [CrossRef] [Green Version]
- Kosseva, M.R. Immobilization of Microbial Cells in Food Fermentation Processes. Food Bioprocess Technol. 2011, 4, 1089–1118. [Google Scholar] [CrossRef]
- Genisheva, Z.; Teixeira, J.A.; Oliveira, J.M. Immobilized cells systems for batch and continuous winemaking. Trends Food Sci. Technol. 2014, 40, 33–47. [Google Scholar] [CrossRef] [Green Version]
- Rathore, S.; Desai, P.M.; Liew, C.V.; Chan, L.W.; Heng, P.W.S. Microencapsulation of microbial cells. J. Food Eng. 2013, 116, 369–381. [Google Scholar] [CrossRef]
- Bevilacqua, A.; Campaniello, D.; Speranza, B.; Racioppo, A.; Altieri, C.; Sinigaglia, M.; Corbo, M.R. Microencapsulation of Saccharomyces cerevisiae into Alginate Beads: A Focus on Functional Properties of Released Cells. Foods 2020, 9, 1051. [Google Scholar] [CrossRef]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Evol. Microbiol. 1999, 49, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Alberico, G.; Pietrafesa, A.; Capece, A.; Pietrafesa, R.; Siesto, G.; Romano, P. Erratum to: Exploitation of technological variability among wild non-Saccharomyces yeasts to select mixed starters for the production of low alcohol wines. BIO Web Conf. 2019, 15, 02041. [Google Scholar] [CrossRef]
- De Prisco, A.; Maresca, D.; Ongeng, D.; Mauriello, G. Microencapsulation by vibrating technology of the probiotic strain Lactobacillus reuteri DSM 17938 to enhance its survival in foods and in gastrointestinal environment. LWT Food Sci. Technol. 2015, 61, 452–462. [Google Scholar] [CrossRef]
- Maresca, D.; De Prisco, A.; La Storia, A.; Cirillo, T.; Esposito, F.; Mauriello, G. Microencapsulation of nisin in alginate beads by vibrating technology: Preliminary investigation. LWT Food Sci. Technol. 2016, 66, 436–443. [Google Scholar] [CrossRef]
- Capece, A.; Siesto, G.; Romaniello, R.; Lagreca, V.M.; Pietrafesa, R.; Calabretti, A.; Romano, P. Assessment of competition in wine fermentation among wild Saccharomyces cerevisiae strains isolated from Sangiovese grapes in Tuscany region. LWT Food Sci. Technol. 2013, 54, 485–492. [Google Scholar] [CrossRef]
- Rubio-Bretón, P.; Gonzalo-Diago, A.; Iribarren, M.; Garde-Cerdán, T.; Pérez-Álvarez, E.P. Bioprotection as a tool to free additives winemaking: Effect on sensorial, anthocyanic and aromatic profile of young red wines. LWT Food Sci. Technol. 2018, 98, 458–464. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- OIV-MA-C1e01: R2011. Maximum acceptable limits of various substances. In Compendium of International Methods of Wine and Must Analysis, 2014th ed.; Organisation International de la Vigne et du Vin: Paris, France, 2011; Volume 2. [Google Scholar]
- Swiegers, J.H.; Bartowsky, P.A.; Henschke, P.A.; Pretorius, I.S. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Moreira, N.; Pina, C.; Mendes, F.; Couto, J.A.; Hogg, T.; Vasconcelos, I. Volatile compounds contribution of Hanseniaspora guilliermondii and Hanseniaspora uvarum during red wine vinifications. Food Control 2011, 22, 662–667. [Google Scholar] [CrossRef]
- Martin, V.; Valera, M.J.; Medina, K.; Boido, E.; Carrau, F. Oenological impact of the Hanseniaspora/Kloeckera yeast genus on wines—A review. Fermentation 2018, 4, 76. [Google Scholar] [CrossRef] [Green Version]
- Valera, M.J.; Boido, E.; Dellacassa, E.; Carrau, F. Comparison of the Glycolytic and Alcoholic Fermentation Pathways of Hanseniaspora vineae with Saccharomyces cerevisiae Wine Yeasts. Fermentation 2020, 6, 78. [Google Scholar] [CrossRef]
- Benucci, I.; Cecchi, T.; Lombardelli, C.; Maresca, D.; Mauriello, G.; Esti, M. Novel microencapsulated yeast for the primary fermentation of green beer: Kinetic behavior, volatiles and sensory profile. Food Chem. 2021, 340, 127900. [Google Scholar] [CrossRef]
- Moreno-García, J.; García-Martínez, T.; Mauricio, J.C.; Moreno, J. Yeast Immobilization Systems for Alcoholic Wine Fermentations: Actual Trends and Future Perspectives. Front. Microbiol. 2018, 9, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, K.; Boido, E.; Fariña, L.; Gioia, O.; Gomez, M.E.; Barquet, M.; Gaggero, C.; Dellacassa, E.; Carrau, F. Increased flavour diversity of Chardonnay wines by spontaneous fermentation and co-fermentation with Hanseniaspora vineae. Food Chem. 2013, 141, 2513–2521. [Google Scholar] [CrossRef]
- Ciani, M.; Comitini, F. Yeast interactions in multi-starter wine fermentation. Curr. Opin. Food Sci. 2015, 1, 1–6. [Google Scholar] [CrossRef]
- Ludovico, P.; Sousa, M.J.; Silva, M.T.; Leao, C.; Corte-Real, M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology 2001, 147, 2409–2415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleet, G.H. Yeast interactions and wine flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Vasserot, Y.; Mornet, F.; Jeandet, P. Acetic acid removal by Saccharomyces cerevisiae during fermentation in oenological conditions. Metabolic consequences. Food Chem. 2010, 119, 1220–1223. [Google Scholar] [CrossRef]
- Shang, Y.; Zeng, Y.; Zhu, P.; Zhong, Q. Acetate metabolism of Saccharomyces cerevisiae at different temperatures during lychee wine fermentation. Biotechnol. Biotechnol. Equip. 2016, 30, 512–520. [Google Scholar] [CrossRef] [Green Version]
- Pampulha, M.E.; Loureiro-Dias, M.C. Energetics of the effect of acetic acid on growth of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2000, 184, 69–72. [Google Scholar] [CrossRef]
- Vilera-Moura, A.; Schuller, D.; Mendes-Faia, A.; Real, M. Reduction of volatile acidity of wines by selected yeasts strains. Appl. Microbiol. Biotechnol. 2008, 80, 881–890. [Google Scholar] [CrossRef] [Green Version]
- Viana, F.; Gil, J.V.; Vallés, S.; Manzanares, P. Increasing the levels of 2-phenylethyl acetate in wine through the use of a mixed culture of Hanseniaspora osmophila and Saccharomyces cerevisiae. Int. J. Food Microbiol. 2009, 135, 68–74. [Google Scholar] [CrossRef]
- Zhang, B.-Q.; Shen, J.-Y.; Duan, C.-Q.; Yan, G.-L. Use of Indigenous Hanseniaspora vineae and Metschnikowia pulcherrima Co-fermentation With Saccharomyces cerevisiae to Improve the Aroma Diversity of Vidal Blanc Icewine. Front. Microbiol. 2018, 9, 2303. [Google Scholar] [CrossRef]
- Jackson, R.S. Sensory perception and wine assessment. In Wine Science; Jackson, R.S., Jackson, R.S., Eds.; Academic Press: San Diego, CA, USA, 1994; p. 447. [Google Scholar]
- Viana, F.; Gil, J.V.; Genovés, S.; Vallés, S.; Manzanares, P. Rational selection of non-Saccharomyces wine yeast for mixed starters based on ester formation and enological traits. Food Microbiol. 2008, 25, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Cordente, A.G.; Solomon, M.; Schulkin, A.; Francis, I.L.; Barker, A.; Borneman, A.R.; Curtin, C.D. Novel wine yeast with ARO4 and TYR1 mutations that overproduce ‘floral’ aroma compounds 2-phenylethanol and 2-phenylethyl acetate. Appl. Microbiol. Biotechnol. 2018, 102, 5977–5988. [Google Scholar] [CrossRef] [PubMed]
- Moreira, N.; Mendes, F.; Guedes de Pinho, P.; Hogg, T.; Vasconcelos, I. Heavy sulphur compounds, higher alcohols and esters production profile of Hansenispora uvarum and Hanseniaspora guilliermondii grown as pure and mixed cultures in grape must. Int. J. Food Microbiol. 2008, 124, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Capece, A.; Comitini, F.; Canonico, L.; Siesto, G.; Romano, P. Yeast Interactions in Inoculated Wine Fermentation. Front. Microbiol. 2016, 7, 555. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Parameters. | C | C + NF | C + NM | Sign. |
|---|---|---|---|---|
| Ethanol | 13.45 ± 0.35 | 12.32 ± 0.55 | 12.98 ± 0.50 | n.s. |
| Residual sugar | 0.95 ± 0.03 a | 15.16 ± 1.00 b | 2.51 ± 0.46 a | ** |
| Glucose | 0.33 ± 0.15 a | 11.30 ± 1.97 b | 1.60 ± 0.10 a | ** |
| Fructose | 0.51 ± 0.01 a | 2.77 ± 0.90 b | 1.43 ± 0.15 a | * |
| Volatile acidity | 0.40 ± 0.09 a | 1.56 ± 0.20 b | 1.05 ± 0.09 c | ** |
| Malic acid | 1.60 ± 0.09 a | 1.29 ± 0.07 b | 1.30 ± 0.04 b | * |
| Volatile Compounds (mg/L) | C | C + NF | C + NM |
|---|---|---|---|
| Acetaldehyde | 32.59 ± 2.82 a | 45.85 ± 3.88 b | 41.80 ± 2.56 b |
| Acetoin | 3.16 ± 0.52 a | 58.22 ± 8.67 b | 8.32 ± 1.28 a |
| n-Propanol | 11.91 ± 1.00 a | 19.44 ± 1.59 b | 19.06 ± 1.23 b |
| Isobutanol | 33.68 ± 1.43 | 36.07 ± 2.44 | 27.97 ± 2.28 |
| n-Butanol | 8.75 ± 0.78 a | 56.43 ± 2.52 c | 13.93 ± 1.97 b |
| 2-Methyl-1-butanol | 71.13 ± 9.68 a | 47.31 ± 3.01 b | 45.31 ± 3.74 b |
| 3-Methyl-1-butanol | 235.26 ± 10.26 a | 171.76 ± 9.66 b | 150.32 ± 12.25 b |
| 2-Phenylethanol | 62.54 ± 4.53 a | 46.05 ± 1.81 b | 39.32 ± 1.84 b |
| 1-Hexanol * | 0.22 ± 0.02 a | 0.26 ± 0.01 ab | 0.28 ± 0.03 b |
| Methionol * | 0.17 ± 0.01 a | 0.04 ± 0.01 b | 0.05 ± 0.01 b |
| Ethyl acetate | 29.44 ± 2.84 a | 89.58 ± 7.88 b | 87.94 ± 7.79 b |
| Isoamyl acetate * | 1.45 ± 0.20 a | 0.63 ± 0.06 b | 0.51 ± 0.06 b |
| 2-Phenylethyl acetate * | 1.20 ± 0.14 a | 40.90 ± 0.87 b | 30.57 ± 2.42 c |
| Ethyl hexanoate * | 0.16 ± 0.03 a | 0.05 ± 0.01 b | 0.06 ± 0.01 b |
| Ethyl lactate * | 0.18 ± 0.02 a | 0.08 ± 0.01 b | 0.17 ± 0.01 a |
| Ethyl octanoate * | 0.09 ± 0.01 a | 0.01 ± 0.01 b | 0.01 ± 0.01 b |
| Diethyl succinate * | 0.20 ± 0.03 a | 0.05 ± 0.01 b | 0.11 ± 0.01 c |
| C | C-0 h | C-48 h | C-72 h | |
|---|---|---|---|---|
| Residual sugar | 1.45 ± 0.02 a | 1.81 ± 0.01 a | 3.1 ± 0.14 b | 3.7 ± 0.15 c |
| Acetic acid | 0.36 ± 0.04 a | 0.94 ± 0.08 b | 1.44 ± 0.03 c | 1.68 ± 0.09 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alberico, G.; Capece, A.; Mauriello, G.; Pietrafesa, R.; Siesto, G.; Garde-Cerdán, T.; Maresca, D.; Romano, R.; Romano, P. Influence of Microencapsulation on Fermentative Behavior of Hanseniaspora osmophila in Wine Mixed Starter Fermentation. Fermentation 2021, 7, 112. https://doi.org/10.3390/fermentation7030112
Alberico G, Capece A, Mauriello G, Pietrafesa R, Siesto G, Garde-Cerdán T, Maresca D, Romano R, Romano P. Influence of Microencapsulation on Fermentative Behavior of Hanseniaspora osmophila in Wine Mixed Starter Fermentation. Fermentation. 2021; 7(3):112. https://doi.org/10.3390/fermentation7030112
Chicago/Turabian StyleAlberico, Grazia, Angela Capece, Gianluigi Mauriello, Rocchina Pietrafesa, Gabriella Siesto, Teresa Garde-Cerdán, Diamante Maresca, Raffaele Romano, and Patrizia Romano. 2021. "Influence of Microencapsulation on Fermentative Behavior of Hanseniaspora osmophila in Wine Mixed Starter Fermentation" Fermentation 7, no. 3: 112. https://doi.org/10.3390/fermentation7030112
APA StyleAlberico, G., Capece, A., Mauriello, G., Pietrafesa, R., Siesto, G., Garde-Cerdán, T., Maresca, D., Romano, R., & Romano, P. (2021). Influence of Microencapsulation on Fermentative Behavior of Hanseniaspora osmophila in Wine Mixed Starter Fermentation. Fermentation, 7(3), 112. https://doi.org/10.3390/fermentation7030112