Transcriptional and Metabolic Response of Wine-Related Lactiplantibacillus plantarum to Different Conditions of Aeration and Nitrogen Availability
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Inoculum Preparation
2.2. MLF Conditions
2.3. Analytical Methods
2.4. RNA Sampling and Extraction
2.5. RNA Sequencing and Data Processing
2.6. Statistical Analysis
3. Results and Discussion
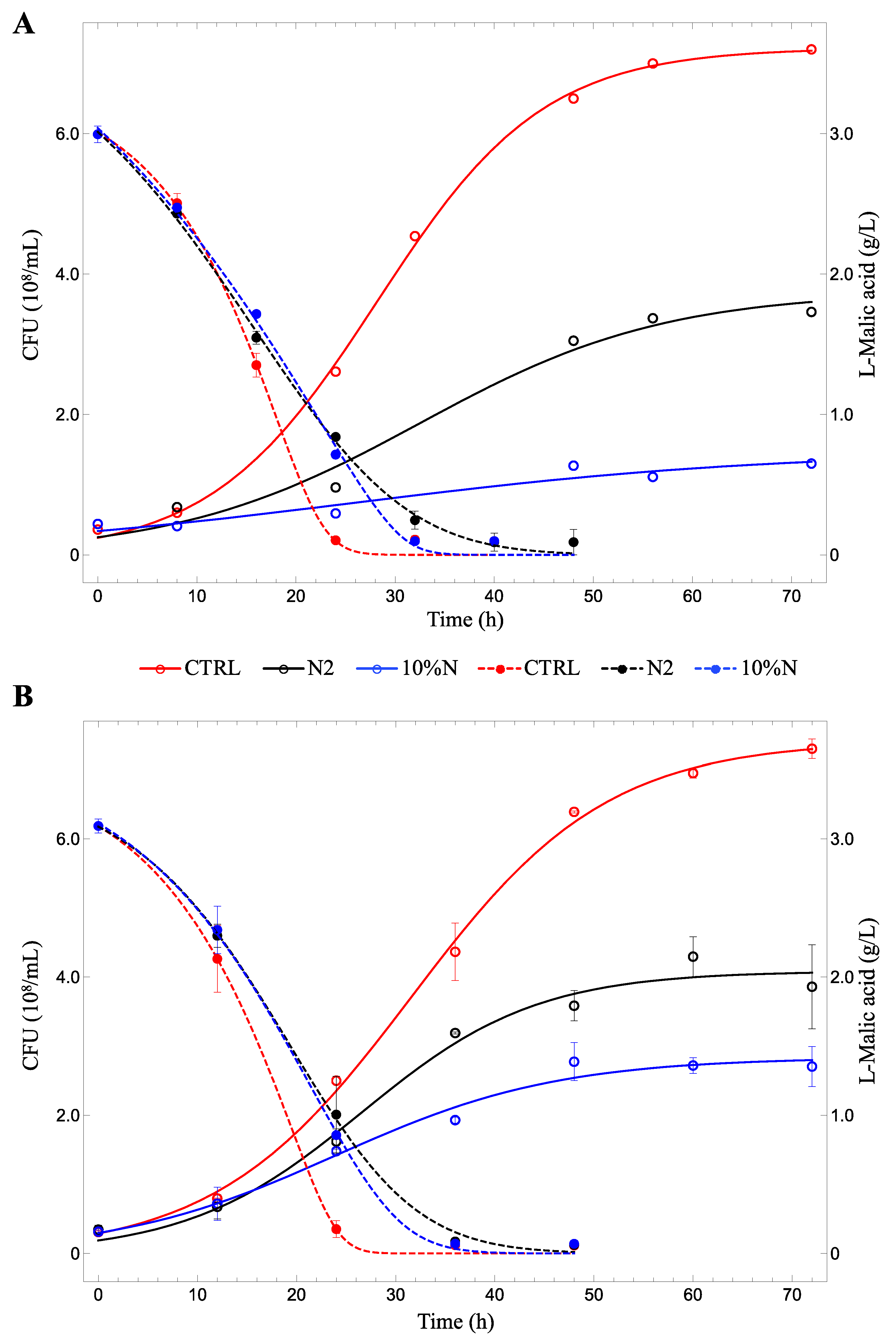
3.1. Lab-Scale Fermentations
3.2. Fermenters
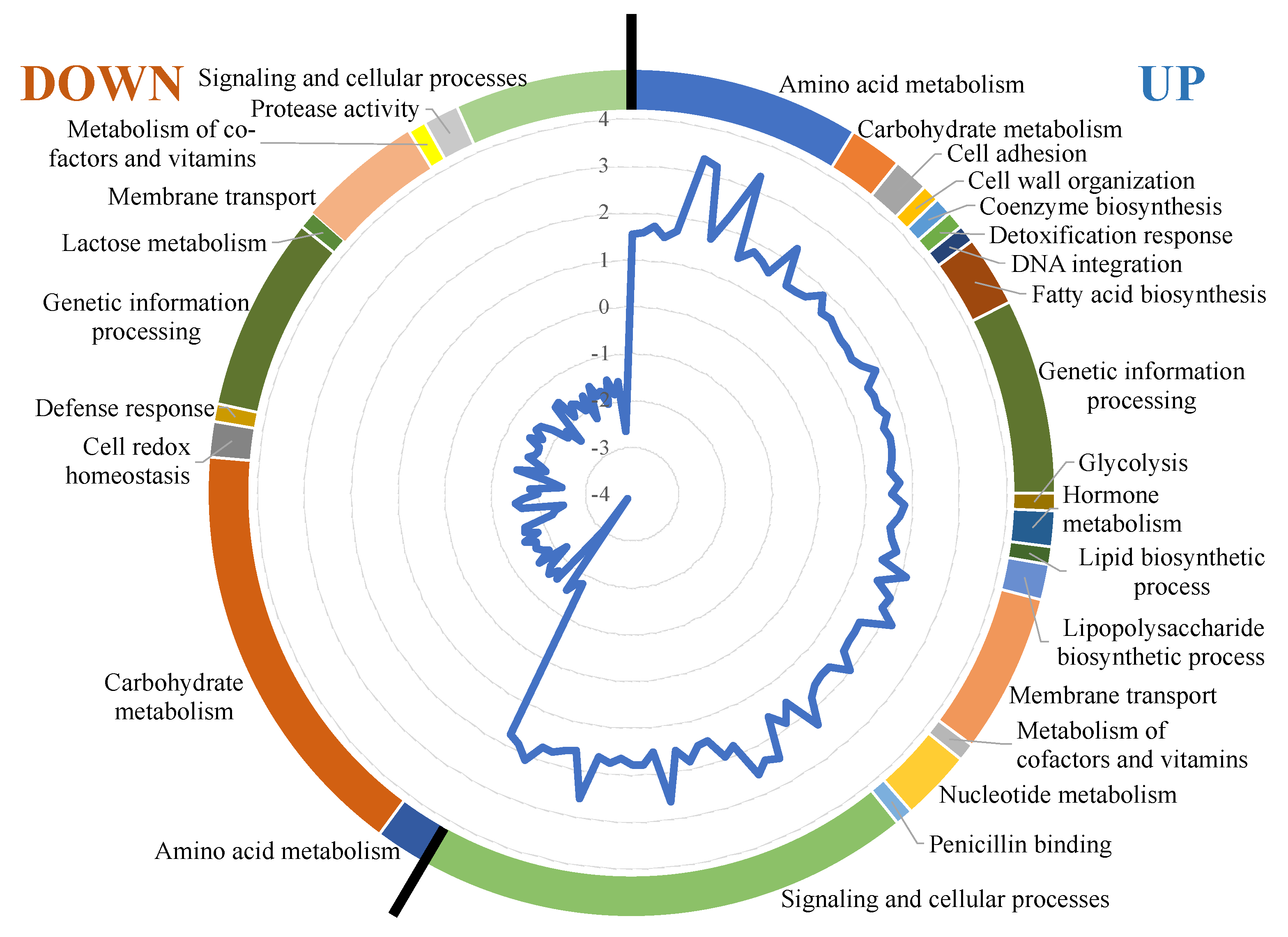
3.3. Transcriptional Response
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lerm, E.; Engelbrecht, L.; du Toit, M. Malolactic Fermentation: The ABC’s of MLF. S. Afr. J. Enol. Vitic. 2016, 31, 186–212. [Google Scholar] [CrossRef]
- Du Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: The Next Generation of Malolactic Fermentation Starter Cultures—An Overview. Food Bioprocess. Technol. 2010, 4, 876–906. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Esteban-Fernández, A.; Navascués, E.; Marquina, D.; Santos, A.; Moreno-Arribas, M.V. Microbial Contribution to Wine Aroma and Its Intended Use for Wine Quality Improvement. Molecules 2017, 22, 189. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Krieger-Weber, S.; Heras, J.M.; Suarez, C. Lactobacillus plantarum, a New Biological Tool to Control Malolactic Fermentation: A Review and an Outlook. Beverages 2020, 6, 23. [Google Scholar] [CrossRef]
- Lombardi, S.J.; Pannella, G.; Iorizzo, M.; Testa, B.; Succi, M.; Tremonte, P.; Sorrentino, E.; di Renzo, M.; Strollo, D.; Coppola, R. Inoculum Strategies and Performances of Malolactic Starter Lactobacillus plantarum M10: Impact on Chemical and Sensorial Characteristics of Fiano Wine. Microorganisms 2020, 8, 516. [Google Scholar] [CrossRef]
- Nielsen, J.C.; Richelieu, M. Control of Flavor Development in Wine during and after Malolactic Fermentation by Oenococcus oeni. Appl. Environ. Microbiol. 1999, 65, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.J.; Henschke, P.A. The ‘buttery’ attribute of wine—diacetyl—desirability, spoilage and beyond. Int. J. Food Microbiol. 2004, 96, 235–252. [Google Scholar] [CrossRef] [PubMed]
- Zotta, T.; Parente, E.; Ricciardi, A. Aerobic metabolism in the genus Lactobacillus: Impact on stress response and potential applications in the food industry. J. Appl. Microbiol. 2017, 122, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Remize, F.; Gaudin, A.; Kong, Y.; Guzzo, J.; Alexandre, H.; Krieger, S.; Guilloux-Benatier, M. Oenococcus oeni preference for peptides: Qualitative and quantitative analysis of nitrogen assimilation. Arch. Microbiol. 2006, 185, 459–469. [Google Scholar] [CrossRef]
- Melkonian, C.; Gottstein, W.; Blasche, S.; Kim, Y.; Abel-Kistrup, M.; Swiegers, H.; Saerens, S.; Edwards, N.; Patil, K.R.; Teusink, B.; et al. Finding Functional Differences Between Species in a Microbial Community: Case Studies in Wine Fermentation and Kefir Culture. Front. Microbiol. 2019, 10, 1347. [Google Scholar] [CrossRef]
- Pretorius, N.; Engelbrecht, L.; Du Toit, M. Influence of sugars and pH on the citrate metabolism of different lactic acid bacteria strains in a synthetic wine matrix. J. Appl. Microbiol. 2019, 127, 1490–1500. [Google Scholar] [CrossRef]
- Reverón, I.; Plaza-Vinuesa, L.; Santamaría, L.; Oliveros, J.C.; Rivas, B.D.L.; Muñoz, R.; de Felipe, F.L. Transcriptomic Evidence of Molecular Mechanisms Underlying the Response of Lactobacillus plantarum WCFS1 to Hydroxytyrosol. Antioxidants 2020, 9, 442. [Google Scholar] [CrossRef]
- Barbosa, C.; Mendes-Faia, A.; Lage, P.; Mira, N.P.; Mendes-Ferreira, A. Genomic expression program of Saccharomyces cerevisiae along a mixed-culture wine fermentation with Hanseniaspora guilliermondii. Microb. Cell Factories 2015, 14, 1–17. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, H.; Peng, S.; Wang, T.; Su, J.; Liang, Y.; Li, H.; Wang, H. Transcriptomic Analysis of Oenococcus oeni SD-2a Response to Acid Shock by RNA-Seq. Front. Microbiol. 2017, 8, 1586. [Google Scholar] [CrossRef] [PubMed]
- Güell, M.; van Noort, V.; Yus, E.; Chen, W.-H.; Leigh-Bell, J.; Michalodimitrakis, K.; Yamada, T.; Arumugam, M.; Doerks, T.; Kühner, S.; et al. Transcriptome Complexity in a Genome-Reduced Bacterium. Science 2009, 326, 1268–1271. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.M.; Hoffmann, S.; Darfeuille, F.; Reignier, J.; Findeiss, S.; Sittka, A.; Chabas, S.; Reiche, K.; Hackermüller, J.; Reinhardt, R.; et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nat. Cell Biol. 2010, 464, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Leimena, M.M.; Wels, M.; Bongers, R.S.; Smid, E.J.; Zoetendal, E.G.; Kleerebezem, M. Comparative Analysis of Lactobacillus plantarum WCFS1 Transcriptomes by Using DNA Microarray and Next-Generation Sequencing Technologies. Appl. Environ. Microbiol. 2012, 78, 4141–4148. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, L.; Lu, Y.; Yu, H.; Tian, H. Comparative Transcriptional Analysis of Lactobacillus plantarum and Its ccpA-Knockout Mutant Under Galactooligosaccharides and Glucose Conditions. Front. Microbiol. 2019, 10, 1584. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Li, H.; Song, D.; Zhang, L.; Zhao, X.; Xu, X. Genome, transcriptome and fermentation analyses of Lactobacillus plantarum LY-78 provide new insights into the mechanism of phenyllactate biosynthesis in lactic acid bacteria. Biochem. Biophys. Res. Commun. 2019, 519, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Rossouw, D.; du Toit, M.; Bauer, F.F. The impact of co-inoculation with Oenococcus oeni on the trancriptome of Saccharomyces cerevisiae and on the flavour-active metabolite profiles during fermentation in synthetic must. Food Microbiol. 2012, 29, 121–131. [Google Scholar] [CrossRef]
- Shekhawat, K.; Patterton, H.; Bauer, F.F.; Setati, M.E. RNA-seq based transcriptional analysis of Saccharomyces cerevisiae and Lachancea thermotolerans in mixed-culture fermentations under anaerobic conditions. BMC Genom. 2019, 20, 1–15. [Google Scholar] [CrossRef]
- Li, J.; Zhu, K.; Zhao, H. Transcriptome analysis reveals the protection mechanism of proanthocyanidins for Saccharomyces cerevisiae during wine fermentation. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Tronchoni, J.; Curiel, J.A.; Morales, P.; Torres-Pérez, R.; Gonzalez, R. Early transcriptional response to biotic stress in mixed starter fermentations involving Saccharomyces cerevisiae and Torulaspora delbrueckii. Int. J. Food Microbiol. 2017, 241, 60–68. [Google Scholar] [CrossRef]
- Giorello, F.; Valera, M.J.; Martin, V.; Parada, A.; Salzman, V.; Camesasca, L.; Fariña, L.; Boido, E.; Medina, K.; Dellacassa, E.; et al. Genomic and Transcriptomic Basis of Hanseniaspora vineae’s Impact on Flavor Diversity and Wine Quality. Appl. Environ. Microbiol. 2018, 85, 01959-18. [Google Scholar] [CrossRef] [PubMed]
- Valdetara, F.; Škalič, M.; Fracassetti, D.; Louw, M.; Compagno, C.; du Toit, M.; Foschino, R.; Petrovič, U.; Divol, B.; Vigentini, I. Transcriptomics unravels the adaptive molecular mechanisms of Brettanomyces bruxellensis under SO2 stress in wine condition. Food Microbiol. 2020, 90, 103483. [Google Scholar] [CrossRef]
- Sternes, P.R.; Costello, P.J.; Chambers, P.J.; Bartowsky, E.J.; Borneman, A.R. Whole transcriptome RNAseq analysis of Oenococcus oeni reveals distinct intra-specific expression patterns during malolactic fermentation, including genes involved in diacetyl metabolism. Int. J. Food Microbiol. 2017, 257, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Henschke, P.A.; Jiranek, V. Yeasts-metabolism of nitrogen compounds. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic: Chur, Switzerland, 1993; pp. 77–164. [Google Scholar]
- Louw, L.; Tredoux, A.; van Rensburg, P.; Kidd, M.; Naes, T.; Nieuwoudt, H. Fermentation-derived Aroma Compounds in Varietal Young Wines from South Africa. S. Afr. J. Enol. Vitic. 2016, 31, 213–225. [Google Scholar] [CrossRef][Green Version]
- Miller, B.J.; Franz, C.M.A.P.; Cho, G.-S.; du Toit, M. Expression of the Malolactic Enzyme Gene (mle) from Lactobacillus plantarum Under Winemaking Conditions. Curr. Microbiol. 2011, 62, 1682–1688. [Google Scholar] [CrossRef]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; deJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Kal, A.J.; van Zonneveld, A.J.; Benes, V.; Berg, M.V.D.; Koerkamp, M.G.; Albermann, K.; Strack, N.; Ruijter, J.M.; Richter, A.; Dujon, B.; et al. Dynamics of Gene Expression Revealed by Comparison of Serial Analysis of Gene Expression Transcript Profiles from Yeast Grown on Two Different Carbon Sources. Mol. Biol. Cell 1999, 10, 1859–1872. [Google Scholar] [CrossRef] [PubMed]
- Betteridge, A.; Grbin, P.; Jiranek, V. Improving Oenococcus oeni to overcome challenges of wine malolactic fermentation. Trends Biotechnol. 2015, 33, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.-J.; Henschke, P.A. Implications of nitrogen nutrition for grapes, fermentation and wine. Aust. J. Grape Wine Res. 2005, 11, 242–295. [Google Scholar] [CrossRef]
- Lytra, G.; Miot-Sertier, C.; Moine, V.; Coulon, J.; Barbe, J.-C. Influence of must yeast-assimilable nitrogen content on fruity aroma variation during malolactic fermentation in red wine. Food Res. Int. 2020, 135, 109294. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, S.C.; Rossouw, D.; du Toit, M.; Bauer, F.F. Enforced Mutualism Leads to Improved Cooperative Behavior between Saccharomyces cerevisiae and Lactobacillus plantarum. Microorganisms 2020, 8, 1109. [Google Scholar] [CrossRef]
- Gänzle, M.G. Lactic metabolism revisited: Metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr. Opin. Food Sci. 2015, 2, 106–117. [Google Scholar] [CrossRef]
- Bron, P.A.; Wels, M.; Bongers, R.S.; Veen, H.V.B.-V.D.; Wiersma, A.; Overmars, L.; Marco, M.L.; Kleerebezem, M. Transcriptomes Reveal Genetic Signatures Underlying Physiological Variations Imposed by Different Fermentation Conditions in Lactobacillus plantarum. PLoS ONE 2012, 7, e38720. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Moslehi-Jenabian, S.; Werner, B.B.; Jensen, M.L.; Garrigues, C.; Vogensen, F.K.; Jespersen, L. Transcriptome analysis of Lactococcus lactis subsp. lactis during milk acidification as affected by dissolved oxygen and the redox potential. Int. J. Food Microbiol. 2016, 226, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Mazzeo, M.F.; Cacace, G.; Peluso, A.; Zotta, T.; Muscariello, L.; Vastano, V.; Parente, E.; Siciliano, R.A. Effect of inactivation of ccpA and aerobic growth in Lactobacillus plantarum: A proteomic perspective. J. Proteom. 2012, 75, 4050–4061. [Google Scholar] [CrossRef] [PubMed]
- Echave, P.; Tamarit, J.; Cabiscol, E.; Ros, J. Novel Antioxidant Role of Alcohol Dehydrogenase E from Escherichia coli. J. Biol. Chem. 2003, 278, 30193–30198. [Google Scholar] [CrossRef] [PubMed]
- Andreevskaya, M.; Johansson, P.; Jääskeläinen, E.; Rämö, T.; Ritari, J.; Paulin, L.; Björkroth, J.; Auvinen, P. Lactobacillus oligofermentans glucose, ribose and xylose transcriptomes show higher similarity between glucose and xylose catabolism-induced responses in the early exponential growth phase. BMC Genom. 2016, 17, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.J.A.; Molenaar, D.; de Jong, A.; de Vos, W.M.; Kleerebezem, M. Involvement of the Mannose Phosphotransferase System of Lactobacillus plantarum WCFS1 in Peroxide Stress Tolerance. Appl. Environ. Microbiol. 2010, 76, 3748–3752. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yin, J.; Liu, J.; Xu, Q.; Lan, T.; Ren, F.; Hao, Y. The Copper Homeostasis Transcription Factor CopR Is Involved in H2O2 Stress in Lactobacillus plantarum CAUH2. Front. Microbiol. 2017, 8, 2015. [Google Scholar] [CrossRef]
- Huang, T.; Xiong, T.; Peng, Z.; Xiao, Y.-S.; Liu, Z.-G.; Hu, M.; Xie, M.-Y. Genomic analysis revealed adaptive mechanism to plant-related fermentation of Lactobacillus plantarum NCU116 and Lactobacillus spp. Genomics 2020, 112, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Filannino, P.; de Angelis, M.; di Cagno, R.; Gozzi, G.; Riciputi, Y.; Gobbetti, M. How Lactobacillus plantarum shapes its transcriptome in response to contrasting habitats. Environ. Microbiol. 2018, 20, 3700–3716. [Google Scholar] [CrossRef]
- Wang, X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 147. [Google Scholar] [CrossRef]
- Delgado, S.; Flórez, A.B.; Guadamuro, L.; Mayo, B. Genetic and biochemical characterization of an oligo-α-1,6-glucosidase from Lactobacillus plantarum. Int. J. Food Microbiol. 2017, 246, 32–39. [Google Scholar] [CrossRef] [PubMed]
| Group | Component | Amount per Litre |
|---|---|---|
| Carbon Sources | Glucose | 115 g |
| Fructose | 115 g | |
| Acids | Potassium L-Tartrate Monobasic | 2.5 g |
| L-Malic acid | 3 g | |
| Citric acid | 0.2 g | |
| Salts | Potassium phosphate dibasic (K2HPO4) | 1.14 g |
| Magnesium sulphate heptahydrate (MgSO4.7H2O) | 1.23 g | |
| Calcium chloride dihydrate (CaCl2.2H2O) | 0.44 g | |
| Nitrogen Sources | Ammonium chloride (NH4Cl) | 0.46 g |
| Amino acids (prepared as 100X stock solution in 20 g/L NaHCO3 buffer solution) | ||
| -Tyrosine | 0.014 g | |
| -Tryptophane | 0.137 g | |
| -Isoleucine | 0.025 g | |
| -Aspartic acid | 0.034 g | |
| -Glutamic acid | 0.092 g | |
| -Arginine | 0.286 g | |
| -Leucine | 0.037 g | |
| -Threonine | 0.058 g | |
| -Glycine | 0.014 g | |
| -Glutamine | 0.386 g | |
| -Alanine | 0.111 g | |
| -Valine | 0.034 g | |
| -Methionine | 0.024 g | |
| -Phenylalanine | 0.029 g | |
| -Serine | 0.060 g | |
| -Histidine | 0.025 g | |
| -Lysine | 0.013 g | |
| -Cysteine | 1 g | |
| -Proline | 0.468 g | |
| Trace Elements (prepared as 100X stock solution) | Manganese(II) chloride tetrahydrate (MnCl2.4H2O) | 200 µg |
| Zinc(II) chloride (ZnCl2) | 135 µg | |
| Iron(II) chloride (FeCl2) | 30 µg | |
| Copper(II) chloride (CuCl2) | 15 µg | |
| Boric acid (H3BO3) | 5 µg | |
| Cobalt(II) nitrate hexahydrate (Co(NO3)2.6H2O) | 30 µg | |
| Sodium molybdate dihydrate (NaMoO4.2H2O) | 25 µg | |
| Potassium iodate (KIO3) | 10 µg | |
| Vitamins (prepared as 100X stock solution) | Myo-inositol | 100 mg |
| Pyridoxine hydrochloride | 2 mg | |
| Nicotinic acid | 2 mg | |
| Calcium pantothenate | 1 mg | |
| Thiamin hydrochloride | 0.5 mg | |
| PABA.K | 0.2 mg | |
| Riboflavin | 0.2 mg | |
| Biotin | 0.125 mg | |
| Folic acid | 0.2 mg | |
| Anaerobic Factors (prepared as 10X stock solution in hot 96% EtOH) | Ergosterol | 10 mg |
| Tween 80 | 0.5 mL |
| Parameter | Lab-Scale | Fermenter | ||||
|---|---|---|---|---|---|---|
| CTRL | N2 | 10%N | CTRL | N2 | 10%N | |
| µmax (h−1) | 0.12 ± 0.01 | 0.08 ± 0.02 | 0.05 ± 0.02 | 0.10 ± 0.01 | 0.11 ± 0.02 | 0.09 ± 0.02 |
| K (108 CFU) | 7.22 ± 0.10 | 3.75 ± 0.39 | 1.47 ± 0.37 | 7.41 ± 0.14 | 4.08 ± 0.18 | 2.84 ± 0.14 |
| Compound | Condition | Significance | ||
|---|---|---|---|---|
| CTRL | N2 | 10%N | ||
| L-lactic acid (g/L) | 2.273 ± 0.001a | 1.951 ± 0.077b | 1.892 ± 0.055b | p = 4.21 × 10−6 |
| Acetic acid (g/L) | 0.137 ± 0.006a | 0.073 ± 0.007c | 0.087 ± 0.001b | p = 0.01124 |
| Acetoin (mg/L) | 15.492 ± 0.985 | n.d. | 15.499 ± 0.114 | |
| Gene | Function | Category | Fold-Change |
|---|---|---|---|
| adeC | Adenine deaminase | Nucleotide metabolism | 2.00 |
| argO | arginine transmembrane transporter activity | Signaling and cellular processes | 2.31 |
| csbC_3 | Major facilitator superfamily protein | Membrane transport | 2.11 |
| cycA_2 | Cyclin-A2-4 | Signaling and cellular processes | 2.46 |
| cysE | Serine acetyltransferase | Amino acid metabolism | 3.33 |
| cysM | Cysteine synthase B | Amino acid metabolism | 3.23 |
| dtpT_1 | Amino acid/peptide transporter (Peptide:H+ symporter) | Signaling and cellular processes | 2.35 |
| dtpT_2 | Di-/tripeptide transporter | Signaling and cellular processes | 2.56 |
| licB_1 | Protein LicB | Carbohydrate metabolism | 2.31 |
| lysN | LysN | Amino acid metabolism | 2.28 |
| metB | metB | Amino acid metabolism | 3.31 |
| oppA_4 | Oligopeptide-binding protein | Signaling and cellular processes | 2.61 |
| sdcS | Sodium-dependent dicarboxylate transporter SdcS | Signaling and cellular processes | 2.57 |
| yjeM | - | Membrane transport | 2.20 |
| ywqD_2 | Cytokinin riboside 5’-monophosphate phosphoribohydrolase | Signaling and cellular processes | 2.11 |
| S_00258 | Putative protein of unknown function | NrdR-regulated deoxyribonucleotide transporter, PnuC-like | 2.33 |
| S_00271 | Putative protein of unknown function | 3.81 | |
| S_00272 | Extracellular protein | 3.83 | |
| S_00273 | Putative protein of unknown function | 2.80 | |
| S_00282 | Putative protein of unknown function | 2.11 | |
| S_01162 | Sulfotransferase/hypothetical protein | 2.33 | |
| S_01534 | Putative protein of unknown function | 2.09 | |
| S_01773 | Putative protein of unknown function | 2.44 | |
| S_01864 | Putative protein of unknown function | 2.50 | |
| S_02063 | Prophage protein | Mobilome: prophages and transposons | 2.11 |
| S_02789 | Hydrolases of the alpha/beta superfamily | 2.24 | |
| S_02790 | HTH_MerR-SF transcriptional regulator | Transcription | 2.33 |
| S_02820 | Glycopeptide antibiotic-resistance protein | 2.06 | |
| S_03325 | Putative protein of unknown function | 3.06 |
| Gene | Function | Category | Fold-Change |
|---|---|---|---|
| adhE_1 | Aldehyde-alcohol dehydrogenase | Carbohydrate metabolism | −3.87 |
| adhE_2 | Aldehyde-alcohol dehydrogenase | Carbohydrate metabolism | −3.01 |
| asnS_1 | Asparagine–tRNA ligase | Genetic information processing | −2.10 |
| bceA_2 | Bacitracin export ATP-binding protein BceA | Signaling and cellular processes | −2.23 |
| copA_1 | Metal transporting atpase Mta72 | Membrane transport | −2.19 |
| copB | CopB | Membrane transport | −2.06 |
| davT | 5-aminovalerate aminotransferase DavT | Carbohydrate metabolism | −2.22 |
| dppE_2 | Dipeptide-binding protein DppE | Membrane transport | −2.41 |
| lrgB | Antiholin-like protein LrgB | Signaling and cellular processes | −2.03 |
| manX_1 | EIIAB-Man | Carbohydrate metabolism | −2.23 |
| manY | - | Carbohydrate metabolism | −2.32 |
| manZ | ManZ protein | Carbohydrate metabolism | −2.51 |
| nox_6 | - | Cell redox homeostasis | −2.50 |
| npr_2 | Regulatory protein NPR2 | Defense response | −2.11 |
| yhdG_3 | Putative amino acid permease YhdG | Signaling and cellular processes | −2.64 |
| yodC_3 | - | Cell redox homeostasis | −2.34 |
| S_00347 | L-lactate dehydrogenase | Carbohydrate metabolism | −2.24 |
| S_02279 | MSF transporter | −2.27 | |
| S_02428 | Putative protein of unknown function | −2.04 | |
| S_02979 | MSF sugar transporters | −2.20 | |
| S_03213 | Putative protein of unknown function | −2.03 | |
| S_03235 | LtrC-like protein, unknown function | −2.01 | |
| S_03252 | Putative protein of unknown function | −2.04 | |
| S_03253 | ATP-binding protein | −2.13 | |
| S_03254 | Putative protein of unknown function | −2.11 | |
| S_03255 | Putative protein of unknown function | −2.14 | |
| S_03256 | Putative protein of unknown function | −2.43 | |
| S_03399 | DNA topoisomerase III | Replication, recombination, and repair | −2.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Binati, R.L.; Du Toit, M.; Snoep, J.L.; Salvetti, E.; Torriani, S. Transcriptional and Metabolic Response of Wine-Related Lactiplantibacillus plantarum to Different Conditions of Aeration and Nitrogen Availability. Fermentation 2021, 7, 68. https://doi.org/10.3390/fermentation7020068
Binati RL, Du Toit M, Snoep JL, Salvetti E, Torriani S. Transcriptional and Metabolic Response of Wine-Related Lactiplantibacillus plantarum to Different Conditions of Aeration and Nitrogen Availability. Fermentation. 2021; 7(2):68. https://doi.org/10.3390/fermentation7020068
Chicago/Turabian StyleBinati, Renato L., Maret Du Toit, Jacky L. Snoep, Elisa Salvetti, and Sandra Torriani. 2021. "Transcriptional and Metabolic Response of Wine-Related Lactiplantibacillus plantarum to Different Conditions of Aeration and Nitrogen Availability" Fermentation 7, no. 2: 68. https://doi.org/10.3390/fermentation7020068
APA StyleBinati, R. L., Du Toit, M., Snoep, J. L., Salvetti, E., & Torriani, S. (2021). Transcriptional and Metabolic Response of Wine-Related Lactiplantibacillus plantarum to Different Conditions of Aeration and Nitrogen Availability. Fermentation, 7(2), 68. https://doi.org/10.3390/fermentation7020068






