Abstract
Three new species of Distoseptispora, viz. D. mengsongensis, D. nabanheensis, and D. sinensis, are described and illustrated from specimens collected on dead branches of unidentified plants in Yunnan Province, China. Phylogenetic analyses of LSU, ITS, and TEF1 sequence data, using maximum-likelihood (ML) and Bayesian inference (BI), reveal the taxonomic placement of D. mengsongensis, D. nabanheensis, and D. sinensis within Distoseptispora. Both morphological observations and molecular phylogenetic analyses supported D. mengsongensis, D. nabanheensis, and D. sinensis as three new taxa. To extend our knowledge of the diversity of Distoseptispora-like taxa, a list of recognized species of Distoseptispora with major morphological features, habitat, host, and locality is also provided.
1. Introduction
The genus Distoseptispora K.D. Hyde, McKenzie & Maharachch was established by Su et al. [1] with D. fluminicola McKenzie, Hong Y. Su, Z.L. Luo & K.D. Hyde as the type species, and was mainly characterized by being macronematous, septate, unbranched, smooth, olivaceous to brown conidiophores with monoblastic, integrated, terminal, determinate, cylindrical conidiogenous cells that produce acrogenous, solitary, distoseptate conidia. Subsequently, the phylogenetic analyses motivated the inclusion of taxa with euseptate conidia (e.g., D. guttulata J. Yang & K.D. Hyde and D. suoluoensis J. Yang, Maharachch. & K.D. Hyde) [2] and polyblastic conidiogenous cells (e.g., D. palmarum S.N. Zhang, K.D. Hyde & J.K. Liu) [3]. To date, 61 epithets for Distoseptispora are listed in Index Fungorum [4]. Monkai et al. [5] provided a synopsis of relevant morphological features that distinguish 29 Distoseptispora species. Zhai et al. [6] subsequently published an expanded synopsis that includes additional nine species following the same format as Monkai et al. [5], but ignored the species D. submersa Z.L. Luo, K.D. Hyde & H.Y. Su, which was regarded as the synonym of D. tectonae Doilom & K.D. Hyde by Dong et al. [7]. Thus, Distoseptispora currently contains 60 taxa, 38 of which were found in China [1,2,3,6,8,9,10,11,12,13,14,15,16,17,18,19,20,21].
Distoseptispora is one of the Sporidesmium-like genera with high morphological similarity to the Sporidesmium Link and Ellisembia Subram. Distoseptispora species with eu- or distoseptate conidia covering the criteria of Ellisembia and Sporidesmium. Accordingly, Distoseptispora species cannot be classified based on morphology alone, as phylogenetic analysis showed that these genera are not closely related [1,2,22]. Distoseptispora also appears similar in conidial ontogeny to Aquapteridospora Jiao Yang, K.D. Hyde & Maharachch., but the latter has terminal and intercalary conidiogenous cells with circular scars, and produces 3-euseptate conidia [23,24]. Additionally, Distoseptispora formed a sister clade to Aquapteridospora, and is well separated with high support in phylogenetic trees [24]. Distoseptispora and Aquapteridospora belonged to the order Distoseptisporales Z.L. Luo, K.D. Hyde & H.Y. Su, but are now, respectively, treated in the families Distoseptisporaceae K.D. Hyde & McKenzie and Aquapteridosporaceae K.D. Hyde & Hongsanan [24,25].
Distoseptispora species are primarily found as saprobes on submerged wood, dead branches, culms, or leaves in freshwater or terrestrial habitats except for D. caricis Crous and D. palmarum S.N. Zhang, K.D. Hyde & J.K. Liu occurring on the leaves of Carex sp. and rachis of Cocos nucifera [3,6,26]. Species of the genus decompose lignocellulose in wood [22,27,28], but their ecological functions, geographical distribution, alpha-taxonomy, and teleomorph relationships are poorly known. During our continuing survey (2007–2022) of saprophytic microfungi from the forest ecosystems of southwest China, several Sporidesmium-like taxa were isolated on dead branches of unidentified perennial dicotyledonous plants from terrestrial habitats in Yunnan Province, China. Using multi-gene loci of LSU, ITS, and TEF1 sequence data, the systematic placement of these isolates represented several Distoseptispora species. Based on morphological characteristics and multi-locus phylogenetic analysis, three new species of Distoseptispora, viz. D. mengsongensis, D. nabanheensis, and D. sinensis, are proposed and described in this paper.
2. Materials and Methods
2.1. Sample Collection, Isolation, and Morphological Examination
Samples of decomposing wood and bark were collected from the forest floor in Yunnan Province, China, and brought them back to the laboratory in Ziploc™ bags. Samples were treated following the methods described by Ma et al. [29]. Colonies on the surface of dead branches were examined and visually observed with a stereomicroscope (Motic SMZ-168, Xiamen, China) from low (7.5 times) to high (50 times) magnification. Fresh colonies were picked with sterile needles at a stereomicroscope magnification of 50 times, placed on a slide with a drop of lactic acid–phenol solution (lactic acid, phenol, glycerin, sterile water; 1:1:2:1, respectively), then placed under an Olympus BX 53 light microscope fitted with an Olympus DP 27 digital camera (Olympus Optical Co., Ltd., Tokyo, Japan) for microscopic morphological characterization. The tip of a sterile toothpick dipped in sterile water was used to pick conidia from the specimen; the conidia were then streaked on the surface of potato dextrose agar (PDA; 20% potato + 2% dextrose + 2% agar, w/v) and incubated at 25 °C overnight. Single-spore isolations were made on potato dextrose agar (PDA) following Goh [30]. Colony colors were assessed according to the charts of Rayner [31]. All fungal strains were stored in 10% sterilized glycerin at 4 °C for further studies. The studied specimens and cultures were deposited in the Herbarium of Jiangxi Agricultural University, Plant Pathology, Nanchang, China (HJAUP).
2.2. DNA Extraction, PCR Amplification, and Sequencing
Fungal hyphae (500 mg) were scraped from the surface of colonies growing on PDA plates, transferred to 2 mL safe-lock microtubes, and ground with liquid nitrogen. DNA was extracted using the Solarbio Fungal Genomic DNA Extraction Kit (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) according to the manufacturer’s instructions. Primer sets were used for the amplification of LSU and ITS, and TEF1: ITS5/ITS4 [32], 28S1-F/28S3-R [8], and EF1-983F/EF1-2218R [33]. The final volume of the PCR reaction was 25 μL, containing 1 μL of DNA template, 1 μL of each forward and reward primer, 12.5 μL of 2 × Power Taq PCR MasterMix, and 9.5 μL of double-distilled water (ddH2O). The PCR thermal cycling conditions of ITS and LSU were initialized at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 50 s, elongation at 72 °C for 1 min, a final extension at 72 °C for 10 min, and finally kept at 4 °C. TEF1 was initialized at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s, elongation at 72 °C for 1 min, a final extension at 72 °C for 10 min, and finally kept at 4 °C. The PCR products were checked on 1% agarose gel electrophoresis stained with ethidium bromide. Purification and DNA sequencing were carried out at Beijing Tsingke Biotechnology Co., Ltd., Beijing, China.
2.3. Sequence Alignment and Phylogenetic Analyses
Sequences, including those obtained from GenBank (Table 1), were initially aligned using MAFFTv.7 [34] on the online server (http://maffTh.cbrc.jp/alignment/server/, accessed on 1 February 2023) and optimized manually when needed. The LSU, ITS, and TEF1 sequence data were concatenated by using Phylosuite software v1.2.1 [35], and absent sequence data in the alignments were treated with the question mark and “-” as missing data. The phylogenetic tree was constructed using Phylosuite software v1.2.1 [35] based on the combined data of LSU, ITS, and TEF1 sequence (Supplementary Materials). The concatenated aligned dataset was analyzed separately using maximum likelihood (ML) and Bayesian inference (BI). Maximum-likelihood phylogenies were inferred using IQ-TREE [36] under edge-linked partition model for 10,000 ultrafast [37] bootstraps. The final tree was selected among suboptimal trees from each run by comparing the likelihood scores using the TNe+I+G4 for ITS, TNe+R3 for LSU, and TN+F+I+G4 for TEF1 substitution model. Bayesian inference phylogenies were inferred using MrBayes 3.2.6 [38] under partition model (2 parallel runs, 2,000,000 generations), in which the initial 25% of sampled data were discarded by burning. The best-fit model was GTR+F+I+G4 for ITS+TEF1, and GTR+F+I+G4 for LSU. ModelFinder was used to select the best-fit partition model (edge-linked) using BIC criterion [39]. The trees were viewed in FigTree v. 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree, accessed on 1 February 2023), and further edited in Adobe Illustrator 2021. Sequences generated in this study were deposited in GenBank (Table 1).

Table 1.
List of Distoseptispora species and GenBank accessions used in the phylogenetic analyses. New sequences are in bold.
3. Results
3.1. Molecular Phylogeny
The combined sequence alignment comprised 72 strains representing 64 species (Table 1), 2248 total characters (ITS:1–493, LSU:494–1329, TEF1:1330–2248), including 891 distinct patterns, 537 parsimony-informative, 275 singleton sites, and 1436 constant sites), and used Aquapteridospora aquatica (MFLUCC 17-2371), A. fusiformis (MFLUCC 18-1606) and A. lignicola (MFLUCC 15-0377) as outgroup. Maximum-likelihood (ML) and Bayesian inference (BI) analyses of the combined dataset resulted in phylogenetic reconstructions with largely similar topologies, and the best-scoring ML tree (lnL = −16,099.937) is shown in Figure 1. Maximum-likelihood bootstrap support (MLBS) values above 75% and Bayesian posterior probabilities (BPP) greater than 0.90 are given above the nodes. Our three isolates in this lineage formed distinct clades with good support value, and can be recognized as three new phylogenetic species, Distoseptispora mengsongensis, D. nabanheensis, and D. sinensis. Phylogenetic analyses suggested sister group relatedness of D. nabanheensis (HJAUP C2003) and D. clematidis (MFLUCC 17-2145) (MLBS/BPP = 100/0.97); D. mengsongensis (HJAUP C2126) and D. fasciculata (KUMCC 19-0081) (MLBS/BPP = 99/0.92); and D. sinensis (HJAUP C2044), D. tectonae (MFLUCC 12-0291, MFLUCC 16-0946), and D. tectonigena (MFLUCC 12-0292) (MLBS/BPP = 85/0.99).
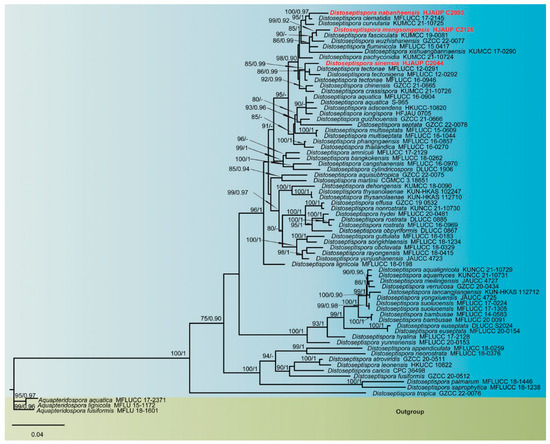
Figure 1.
Phylogenetic tree inferred from maximum-likelihood and Bayesian inference analyses based on concatenated LSU, ITS, and TEF1 sequences. Significant MLBS/BPP support values above 75% and 0.90 are indicated at the nodes. The new isolates of this study are shown in red. The tree is rooted to Aquapteridospora aquatica (MFLUCC 17-2371), A. fusiformis (MFLU 18-1601), and A. lignicola (MFLU 15-1172).
3.2. Taxonomy
Distoseptispora mengsongensis Jing W. Liu, X.G. Zhang & Jian Ma, sp. nov., Figure 2.

Figure 2.
Distoseptispora mengsongensis (HJAUP M2126, holotype). (a) Surface of colony after 2 weeks on PDA; (b) reverse of colony after 2 weeks on PDA; (c) Conidia; (d,e) Conidiophores, conidiogenous cells, and conidia; (f) Conidiophores.
Index Fungorum number: IF900033.
Etymology: In reference to the locality, Mengsong Township, where the fungus was collected.
Holotype: HJAUP M2126.
Description: Saprobic on dead branches in terrestrial habitats. Teleomorph: Undetermined. Anamorph (Figure 2): Hyphomycetes. Colonies on natural substratum effuse, brown, hairy. Mycelium is superficial and immersed, composed of branched, septate, pale brown to brown, smooth-walled hyphae. Conidiophores macronematous, mononematous, cylindrical, 1–5-septate, erect, unbranched, smooth, straight or slightly flexuous, brown to dark brown, 17–54 × 4.5–7 μm ( = 33.5 × 5.5 μm, n = 20). Conidiogenous cells monoblastic, integrated, terminal, cylindrical, flat at the conidiogenous loco, determinate, pale brown to brown, smooth, 8.5–14.2 × 3–5.7 μm ( = 11.3 × 4.1 μm, n = 20). Conidial secession schizolytic. Conidia acrogenous, solitary, obclavate, 15–31-distoseptate, sometimes constricted at the septa, especially in proximal parts, straight or slightly curved, smooth, brown to dark brown, sometimes with percurrent regeneration forming a secondary conidium from the conidial apex, 86–200 × 6–13 μm ( = 141 × 9.7 μm, n = 25), base truncate and 3–5.5 μm wide, apex rounded, 2.9–8.6 μm wide.
Culture characteristics: Colony on PDA reached 81–86 mm diam. after 2 weeks in an incubator under dark conditions at 25 °C, irregularly rounded, surface velvety, with dense, brown mycelium, margin entire, dark brown to black; the reverse is black.
Material examined: China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Menghai County, Mengsong Township, on dead branches of an unidentified broadleaf tree, 12 July 2021, J.W. Liu, HJAUP M2126 (holotype), ex-type culture permanently preserved in a metabolically inactive state, HJAUP C2126.
Notes: Phylogenetic analyses showed that D. mengsongensis (HJAUP C2126) clusters with D. fasciculata (KUMCC 19-0081). BLASTn analysis of D. mengsongensis (HJAUP C2126) and D. fasciculata (KUMCC 19-0081) showed 99% identity (544/551, no gaps) using ITS, 99% identity (565/567, two gaps) using LSU, and 100% identity (927/927, no gaps) using TEF1. Moreover, D. mengsongensis morphologically differs from D. fasciculata W. Dong, H. Zhang & K.D. Hyde, which occurs in freshwater habitats and has smaller conidiophores (12–16 × 5–6 μm) and wider conidia (10–16.5 μm wide) [7]. Distoseptispora mengsongensis is morphologically similar to D. xishuangbannaensis Tibpromma & K.D. Hyde, but the latter differs by occurring on dead leaf sheath and not on wood, and further differs by its shorter and narrower conidiophores (12–17 × 2–5 μm) and bigger conidia (160–305 × 8–15 μm) with up to 40 distosepta [10].
Distoseptispora nabanheensis Jing W. Liu, X.G. Zhang & Jian Ma, sp. nov., Figure 3.

Figure 3.
Distoseptispora nabanheensis (HJAUP M2003, holotype). (a) Surface of colony after 2 weeks on PDA; (b) reverse of colony after 2 weeks on PDA; (c) Conidia; (d,e) Conidiophores, conidiogenous cells, and conidia.
Index Fungorum number: IF900054.
Etymology: In reference to the locality, Nabanhe Nature Reserve, in which the fungus was collected.
Holotype: HJAUP M2003.
Description: Saprobic on dead branches in terrestrial habitats. Teleomorph: Undetermined. Anamorph (Figure 3): Hyphomycetes. Colonies on natural substratum effuse, brown, hairy. Mycelium is superficial and immersed, composed of branched, septate, pale brown to brown, smooth-walled hyphae. Conidiophores macronematous, mononematous, cylindrical, 3–8-septate, erect, unbranched, solitary, smooth, straight or slightly flexuous, brown to dark brown, 29–42 × 8–10 μm ( = 37.5 × 9.2 μm, n = 9). Conidiogenous cells monoblastic, integrated, terminal, cylindrical, flat at the conidiogenous loco, determinate, brown to pale brown, smooth, 4–6 × 4–5 μm ( = 5.2 × 4.6 μm, n = 9). Conidial secession schizolytic. Conidia acrogenous, solitary, obclavate, 18–31-distoseptate, slightly constricted at the septa, smooth, brown to dark brown, 102–214.5 × (7–)11–14.5 μm ( = 171 × 11.3 μm, n = 20), base truncate and 3.5–7.5 μm wide, apex rounded, 3.2–8 μm wide.
Culture characteristics: Colony on PDA reached 83–88 mm diam. after 2 weeks in an incubator under dark conditions at 25 °C, circular, surface velvety, with dense, gray mycelium on the surface along the entire margin; the reverse is dark brown to black.
Material examined: China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, the Nabanhe National Nature Reserve, on dead branches of an unidentified broadleaf tree, 12 July 2021, J.W. Liu, HJAUP M2003 (holotype), ex-type culture permanently preserved in a metabolically inactive state, HJAUP C2003.
Notes: Phylogenetic analyses showed that D. nabanheensis (HJAUP C2003) clusters with D. clematidis (MFLUCC 17-2145). BLASTn analysis of D. nabanheensis (HJAUP C2003) and D. clematidis (MFLUCC 17-2145) shows 99% identity (531/533, no gaps) using ITS, 99% identity (471/477, two gaps) using LSU. Moreover, D. nabanheensis morphologically differs from D. clematidis Phukhams., M.V. de Bult & K.D. Hyde, which has smaller conidiophores (22–40 × 4–10 μm) and wider conidia (12–20 μm wide) with 28–35 distosepta [41]. Distoseptispora nabanheensis is morphologically similar to D. chinensis X. Tang, Jayaward., J.C. Kang & K.D. Hyde, and D. tectonigena Doilom & K.D. Hyde, but D. chinensis lives in freshwater habitats and differs by its narrower conidiophores (5.5–9 μm wide) and bigger conidia (81–283 × 10–19 μm), with up to 40 distosepta [12]; D. tectonigena differs by its longer conidiophores (up to 110 μm long) and longer conidia (83–360 μm long), with 20–46 distosepta [42].
Distoseptispora sinensis Jing W. Liu, X.G. Zhang & Jian Ma, sp. nov., Figure 4.

Figure 4.
Distoseptispora sinensis (HJAUP M2044, holotype). (a) Surface of colony after 2 weeks on PDA; (b) reverse of colony after 2 weeks on PDA; (c,d) Conidia; (e) Conidiophores, conidiogenous cells, and conidia.
Index Fungorum number: IF900055.
Etymology: In reference to the country “China” in which the fungus was collected.
Holotype: HJAUP M2044.
Description: Saprobic on dead branches in terrestrial habitats. Teleomorph: Undetermined. Anamorph: Hyphomycetes. Colonies on natural substratum effuse, brown, hairy. Mycelium is superficial and immersed, composed of branched, septate, pale brown to brown, smooth-walled hyphae. Conidiophores macronematous, mononematous, cylindrical, 2–5-septate, erect, unbranched, solitary, smooth, straight or slightly flexuous, brown to dark brown, 23.5–56.5 × 3.5–7 μm ( = 39.6 × 5 μm, n = 20). Conidiogenous cells monoblastic, integrated, terminal, cylindrical, flat at the conidiogenous loco, determinate, pale brown, smooth, 6.5–10 × 3.3–3.6 μm ( = 8.3 × 3.5 μm, n = 20). Conidial secession schizolytic. Conidia acrogenous, solitary, obclavate, straight or slightly curved, smooth, 10–25-distoseptate, brown to dark brown, apical cell paler, 40–107(–137) × 10–12 μm ( = 78.8 × 10.5 μm, n = 30), base truncate and 3–3.5 μm wide, apex rounded, 3.5–10 μm wide.
Culture characteristics: Colony on PDA reaching 64–69 mm diam. after 2 weeks in an incubator under dark conditions at 25 °C, irregularly rounded, surface velvety, with dense, gray mycelium, black at the entire margin; the reverse is dark brown to black.
Material examined: China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Jinghong City, Gasa Township, on dead branches of an unidentified broadleaf tree, 12 July 2021, J.W. Liu, HJAUP M2044 (holotype), ex-type culture permanently preserved in a metabolically inactive state, HJAUP C2044.
Notes: Phylogenetic analyses showed that D. sinensis (HJAUP C2044) clusters with D. tectonae (MFLUCC 12–0291T, MFLUCC 16–0946) and D. tectonigena (MFLUCC 12–0292). BLASTn analysis of D. sinensis (HJAUP C2044) and D. tectonae (MFLUCC 12-0291T) shows 98% identity (559/569, five gaps) using ITS, 98% identity (574/583, five gaps) using LSU, and 99% identity (926/930, no gaps) using TEF1; BLASTn analysis of D. sinensis (HJAUP C2044) and D. tectonigena (MFLUCC 12–0292) shows 96% identity (549/570, seven gaps) using ITS and 99% identity (575/583, five gaps) using LSU. Moreover, D. sinensis morphologically differs from D. tectonae, which has smaller conidiophores (up to 40 × 4–6 μm) and larger conidia (90–170 × 11–16 μm) with 20–28 distosepta [42], as well as from D. tectonigena, which has larger conidiophores (up to 110 × 5–11 μm) and larger conidia (83–360 × 10–13 μm), with 20–46 distosepta [42].
4. Discussion
Sporidesmium-like taxa have undergone convergent evolution, and the morphological characteristics used to delimit Sporidesmium-like genera are shown to be insignificant in a phylogenetic context. Responding to the heterogeneity of Sporidesmium, the genus Distoseptispora was introduced by Su et al. [1] based on multi-locus phylogenies together with morphology. In recent years, the number of Distoseptispora species steadily increased and currently reached 63 species, including D. mengsongensis, D. nabanheensis, and D. sinensis. In the phylogenetic tree (Figure 1), some Distoseptispora species form sister clades, but they show different morphological characteristics, such as how D. mengsongensis and D. fasciculata are clustered, but the conidia of D. mengsongensis are obclavate, constricted at the septa, especially in proximal parts, sometimes with percurrent regeneration forming a secondary conidium from the conidial apex, with an average conidial length/width ratio of 14.54, while the conidia of D. fasciculata are subcylindrical to obclavate, with an average conidial length/width ratio of 8.44. Distoseptispora nabanheensis and D. clematidis have a close phylogenetic relationship, but D. nabanheensis has obclavate, slightly constricted at the septa, brown to dark brown conidia, with an average conidial length/width ratio of 15.13, while D. clematidis has oblong, obclavate, cylindrical or rostrate, brown with green tinge conidia, with an average conidial length/width ratio of 10.29. Distoseptispora sinensis, D. tectonigena, and D. tectonae have different morphology of conidiophores and conidia, and comparisons of nucleotides between D. tectonae (MFLUCC 12-0291T) and our isolate (HJAUP C2044) showed 10 and 9 (2%, including five gaps) nucleotide differences in ITS and LSU regions, respectively; D. tectonigena (MFLUCC 12–0292T) and our isolate (HJAUP C2044) showed 21 and 8 (4%, including seven gaps; 1.2%, including five gaps) nucleotide differences in ITS and LSU regions, respectively. Considering this scenario, additional molecular data and morphological characteristics are required for verification and expansion.
To date, all Distoseptispora species were identified by morphological and phylogenetic analyses, which led to a better evaluation of their phylogenetic relationships and taxonomic placements. However, studies conducted on Distoseptispora have no universally accepted standards with respect to barcode selection for phylogenetic analyses. For instance, Su et al. [1] established the genus Distoseptispora, but the initial species had only LSU sequences. Tibpromma et al. [10] introduced D. thailandica Tibpromma & K.D. Hyde and D. xishuangbannaensis using LSU and ITS. Monkai et al. [5] introduced D. hydei Monkai & Phookamsak using LSU, ITS, and RPB2. Hyde et al. [12] introduced D. chinensis X. Tang, Jayaward., J.C. Kang & K.D. Hyde and D. guizhouensis X. Tang, Jayaward., J.C. Kang & K.D. Hyde using ITS, LSU, SSU, TEF1, and RPB2. Zhai et al. [6] introduced D. meilingensis Z.J. Zhai & D.M. Hu, D. yongxiuensis Z.J. Zhai & D.M. Hu, and D. yunjushanensis Z.J. Zhai & D.M. Hu using ITS, LSU, SSU, and TEF1. Ma et al. [20] and Zhang et al. [21] introduced 10 Distoseptispora species using LSU, ITS, TEF1, and RPB2. The recent studies also indicated that the use of only LSU and ITS sequences might be problematic in resolving the phylogeny of Distoseptisporaceae, as RPB2 and/or TEF1 usually increased phylogenetic resolution significantly. In our study, we conducted phylogenetic analyses based on combined LSU, ITS, and TEF1 sequences, and obtained good phylogenetic support. Our three species, viz. D. mengsongensis, D. nabanheensis, and D. sinensis, are considerably distinct from all other described Distoseptispora species by morphological characteristics and multi-locus phylogenetic analysis, and thus we are convinced that the newly introduced species are new to science.
Studies conducted to date on Distoseptispora are mainly focused on their alpha-taxonomy, and most species of this genus are known from dead parts of plants as saprobic fungi in aquatic and terrestrial habitats [5,11,12,17,19,20,21,43], except D. caricis and D. palmarum which are reported, respectively, on leaves of Carex sp. and rachis of Cocos nucifera on [3,6,26], whereas we have little attention on their roles in ecosystem function. They total 63 valid species (Table 2 and Table 3), 44 of which are from freshwater, and 19 are from terrestrial habitats. Most Distoseptispora species are described based on their anamorph alone, and only two species, D. hyalina and D. licualae (Table 3), are reported as sexual morphs based on molecular DNA data, but the connection of teleomorph and anamorph has not been proved by pure culture or sequence data. The genus Distoseptispora has mainly been reported in China (41 species) and Thailand (22 species) [4], and only a small amount of published information is recorded in other regions (e.g., Hungary, Malaysia, Puerto Rico, Sierra Leone) [4,6,44]. Thus, it is unclear whether it has a close relationship with geographic regions, but we expect that large-scale surveys of Distoseptispora in aquatic and terrestrial habitats within different geographic regions, ecological environments, and climatic conditions are needed. This will contribute to a comprehensive knowledge of the species diversity of this genus, and further evaluate their phylogenetic relationships and taxonomic placements by molecular methods.

Table 2.
Synopsis of morphological characteristics, habitat, host, and locality compared across Distoseptispora anamorph species.

Table 3.
Synopsis of morphological characteristics, habitat, host, and locality compared across Distoseptispora teleomorph species.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof9040470/s1.
Author Contributions
Conceptualization, J.L. and Y.H.; methodology, J.M.; software, X.L. and Z.X.; validation, J.X.; formal analysis, Y.H.; investigation, J.L.; resources, J.L. and X.L.; data curation, J.L. and Z.X.; writing—original draft preparation, J.L. and Y.H.; writing—review and editing, R.F.C.-R., X.Z. and R.C.; visualization, L.Z.; supervision, J.M.; project administration, J.M.; funding acquisition, J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31970018, 32160006).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences generated in this study were submitted to GenBank.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Su, H.Y.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Ariyawansa, H.A.; Luo, Z.L.; Promputtha, I.; Tian, Q.; Lin, C.G.; Shang, Q.J.; Zhao, Y.C.; et al. The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers. 2016, 80, 375–409. [Google Scholar] [CrossRef]
- Yang, J.; Maharachchikumbura, S.S.N.; Liu, J.K.; Hyde, K.D.; Jones, E.B.G.; Al-Sadi, A.M.; Liu, Z.Y. Pseudostanjehughesia aquitropica gen. et sp. nov. and Sporidesmium sensu lato species from freshwater habitats. Mycol. Prog. 2018, 17, 591–616. [Google Scholar] [CrossRef]
- Hyde, K.D.; Tennakoon, D.S.; Jeewon, R.; Bhat, D.J.; Maharachchikumbura, S.S.N.; Rossi, W.; Leonardi, M.; Lee, H.B.; Mun, H.Y.; Houbraken, J.; et al. Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019, 96, 1–242. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: http://www.indexfungorum.org/Names/Names.asp (accessed on 1 March 2023).
- Monkai, J.; Boonmee, S.; Ren, G.C.; Wei, D.P.; Phookamsak, R.; Mortimer, P.E. Distoseptispora hydei sp. nov. (Distoseptisporaceae), a novel lignicolous fungus on decaying bamboo in Thailand. Phytotaxa 2020, 459, 93–107. [Google Scholar] [CrossRef]
- Zhai, Z.-J.; Yan, J.-Q.; Li, W.-W.; Gao, Y.; Hu, H.-J.; Zhou, J.-P.; Song, H.-Y.; Hu, D.-M. Three novel species of Distoseptispora (Distoseptisporaceae) isolated from bamboo in Jiangxi Province, China. Mycokeys 2022, 88, 35–54. [Google Scholar] [CrossRef]
- Dong, W.; Hyde, K.D.; Jeewon, R.; Doilom, M.; Yu, X.-D.; Wang, G.-N.; Liu, N.G.; Hu, D.M.; Nalumpang, S.; Zhang, H. Towards a natural classification of annulatascaceae-like taxa II: Introducing five new genera and eighteen new species from freshwater. Mycosphere 2021, 12, 1–88. [Google Scholar] [CrossRef]
- Xia, J.W.; Ma, Y.R.; Li, Z.; Zhang, X.G. Acrodictys-like wood decay fungi from southern China, with two new families Acrodictyaceae and Junewangiaceae. Sci. Rep. 2017, 7, 7888. [Google Scholar] [CrossRef]
- Luo, Z.; Hyde, K.; Liu, J.; Bhat, D.; Bao, D.; Li, W.; Su, H. Lignicolous freshwater fungi from China II: Novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 2018, 9, 444–461. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; McKenzie, E.H.C.; Bhat, D.J.; Phillips, A.J.L.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018, 93, 1–160. [Google Scholar] [CrossRef]
- Yang, J.; Liu, L.-L.; Jones, E.B.G.; Li, W.-L.; Hyde, K.D.; Liu, Z.-Y. Morphological variety in Distoseptispora and introduction of six novel species. J. Fungi 2021, 7, 945. [Google Scholar] [CrossRef]
- Hyde, K.D.; Suwannarach, N.; Jayawardena, R.S.; Manawasinghe, I.S.; Liao, C.F.; Doilom, M.; Cai, L.; Zhao, P.; Buyck, B.; Phukhamsakda, C.; et al. Mycosphere notes 325–344—Novel species and records of fungal taxa from around the world. Mycosphere 2021, 12, 1101–1156. [Google Scholar] [CrossRef]
- Phookamsak, R.; Hyde, K.D.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Raspé, O.; Karunarathna, S.C.; Wanasinghe, D.N.; Hongsanan, S.; et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019, 95, 1–273. [Google Scholar] [CrossRef]
- Song, H.Y.; El Sheikha, A.F.; Zhai, Z.J.; Zhou, J.P.; Chen, M.H.; Huo, G.H.; Huang, X.G.; Hu, D.M. Distoseptispora longispora sp. nov. from freshwater habitats in China. Mycotaxon 2020, 135, 513–523. [Google Scholar] [CrossRef]
- Sun, Y.; Goonasekara, I.D.; Thambugala, K.M.; Jayawardena, R.S.; Wang, Y.; Hyde, K.D. Distoseptispora bambusae sp. nov. (Distoseptisporaceae) on bamboo from China and Thailand. Biodivers. Data J. 2020, 8, e53678. [Google Scholar] [CrossRef]
- Li, W.-L.; Liu, Z.-P.; Zhang, T.; Dissanayake, A.J.; Luo, Z.-L.; Su, H.-Y.; Liu, J.-K. Additions to Distoseptispora (Distoseptisporaceae) associated with submerged decaying wood in China. Phytotaxa 2021, 520, 75–86. [Google Scholar] [CrossRef]
- Shen, H.W.; Bao, D.F.; Hyde, K.D.; Su, H.Y.; Bhat, D.J.; Luo, Z.L. Two novel species and two new records of Distoseptispora from freshwater habitats in China and Thailand. MycoKeys 2021, 84, 79–101. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; Wang, S.; Sun, Y.R.; Suwannarach, N.; Sysouphanthong, P.; Abdel-Wahab, M.A.; Abdel-Aziz, F.A.; Abeywickrama, P.D.; Abreu, V.P.; et al. Fungal diversity notes 1512–1610: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2022, 117, 1–272. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; Nilsson, R.H.; Bhunjun, C.S.; de Farias, A.R.G.; Sun, Y.-R.; Wijesinghe, S.N.; Raza, M.; Bao, D.-F.; Lu, L.; Tibpromma, S.; et al. The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Divers. 2022, 114, 327–386. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, J.-Y.; Xiao, X.-J.; Xiao, Y.-P.; Tang, X.; Boonmee, S.; Kang, J.-C.; Lu, Y.-Z. Multi-gene phylogenetic analyses revealed five new species and two new records of Distoseptisporales from China. J. Fungi 2022, 8, 1202. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, R.; Qing, Y.; Yang, H.; Li, C.; Wang, G.; Zhang, D.; Ning, P. Polyphasic identification of Distoseptispora with six new species from fresh water. J. Fungi 2022, 8, 1063. [Google Scholar] [CrossRef]
- Luo, Z.-L.; Hyde, K.D.; Liu, J.-K.J.; Maharachchikumbura, S.S.N.; Jeewon, R.; Bao, D.-F.; Bhat, D.J.; Lin, C.-G.; Li, W.-L.; Yang, J.; et al. Freshwater Sordariomycetes. Fungal Divers. 2019, 99, 451–660. [Google Scholar] [CrossRef]
- Yang, J.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, D.J.; McKenzie, E.H.C.; Bahkali, A.H.; Gareth Jones, E.B.G.; Liu, Z.Y. Aquapteridospora lignicola gen. et sp. nov., a new hyphomycetous taxon (Sordariomycetes) from wood submerged in a freshwater stream. Cryptogam. Mycol. 2015, 36, 469–478. [Google Scholar] [CrossRef]
- Hyde, K.D.; Bao, D.F.; Hongsanan, S.; Chethana, K.; Yang, J.; Suwannarach, N. Evolution of freshwater Diaporthomycetidae (Sordariomycetes) provides evidence for five new orders and six new families. Fungal Divers. 2021, 107, 71–105. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Crous, P.; Wingfield, M.; Lombard, L.; Roets, F.; Swart, W.; Alvarado, P.; Carnegie, A.; Moreno, G.; Luangsaard, J.; Thangavel, R.; et al. Fungal Planet description sheets: 951–1041. Persoonia 2019, 43, 223–425. [Google Scholar] [CrossRef]
- Wong, K.M.K.; Goh, T.K.; Hodgkiss, I.J.; Hyde, K.D.; Ranghoo, V.M.; Tsui, C.K.M.; Ho, W.H.; Wong, S.W.; Yuen, T.K. Role of fungi in freshwater ecosystems. Biodivers. Conserv. 1998, 7, 1187–1206. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Ba, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Y.; Ma, L.G.; Zhang, Y.D.; Castañeda-Ruíz, R.F.; Zhang, X.G. Three new species of Neosporidesmium from Hainan, China. Mycol. Prog. 2011, 10, 157–162. [Google Scholar] [CrossRef]
- Goh, T.K. Single-spore isolation using a hand-made glass needle. Fungal Divers. 1999, 2, 47–63. [Google Scholar]
- Rayner, R.W. A Mycological Colour Chart; Commonwealth Mycological Institute, Kew, Surrey & British Mycological Society: Kew, UK, 1970. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovli’c, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Shenoy, B.D.; Jeewon, R.; Wu, W.P.; Bhat, D.J.; Hyde, K.D. Ribosomal and RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycol. Res. 2006, 110, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Phukhamsakda, C.; McKenzie, E.H.C.; Phillips, A.J.L.; Jones, E.B.G.; Bhat, D.J.; Marc, S.; Bhunjun, C.S.; Wanasinghe, D.N.; Thongbai, B.; Camporesi, E.; et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020, 102, 1–203. [Google Scholar] [CrossRef]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Konta, S.; Tibpromma, S.; Karunarathna, S.C.; Samarakoon, M.C.; Steven, L.S.; Mapook, A.; Boonmee, S.; Senwanna, C.; Balasuriya, A.; Eungwanichayapant, P.D.; et al. Morphology and multigene phylogeny reveal ten novel taxa in Ascomycota from terrestrial palm substrates (Arecaceae) in Thailand. Mycosphere 2023, 14, 107–152. [Google Scholar]
- Farr, D.F.; Rossman, A.Y. Fungal Databases. Systematic Mycology and Microbiology Laboratory, ARS, USDA. Available online: http://nt.ars-grin.gov/fungaldatabases/ (accessed on 1 March 2023).
- Wu, W.; Zhuang, W. Sporidesmium, Endophragmiella and Related Genera from China; Fungal Diversity Press: Hong Kong, China, 2005. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).