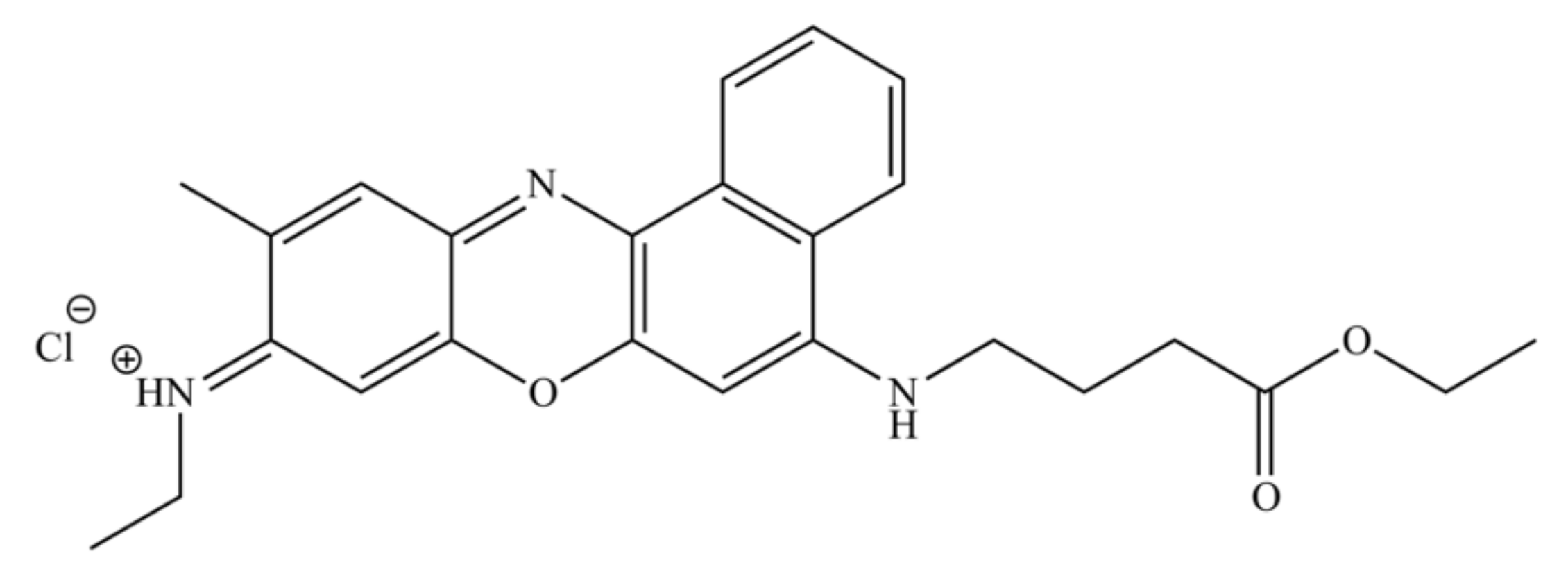
Novel Nile Blue Analogue Stains Yeast Vacuolar Membrane, Endoplasmic Reticulum, and Lipid Droplets, Inducing Cell Death through Vacuole Membrane Permeabilization
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Growth
2.3. Survival Assays
2.4. Cell Death Markers
2.5. Detection of Autophagy Induction in Yeast
2.6. Evaluation of BaP1 Intracellular Distribution and Vacuole Permeabilization by Fluorescence Microscopy
3. Results
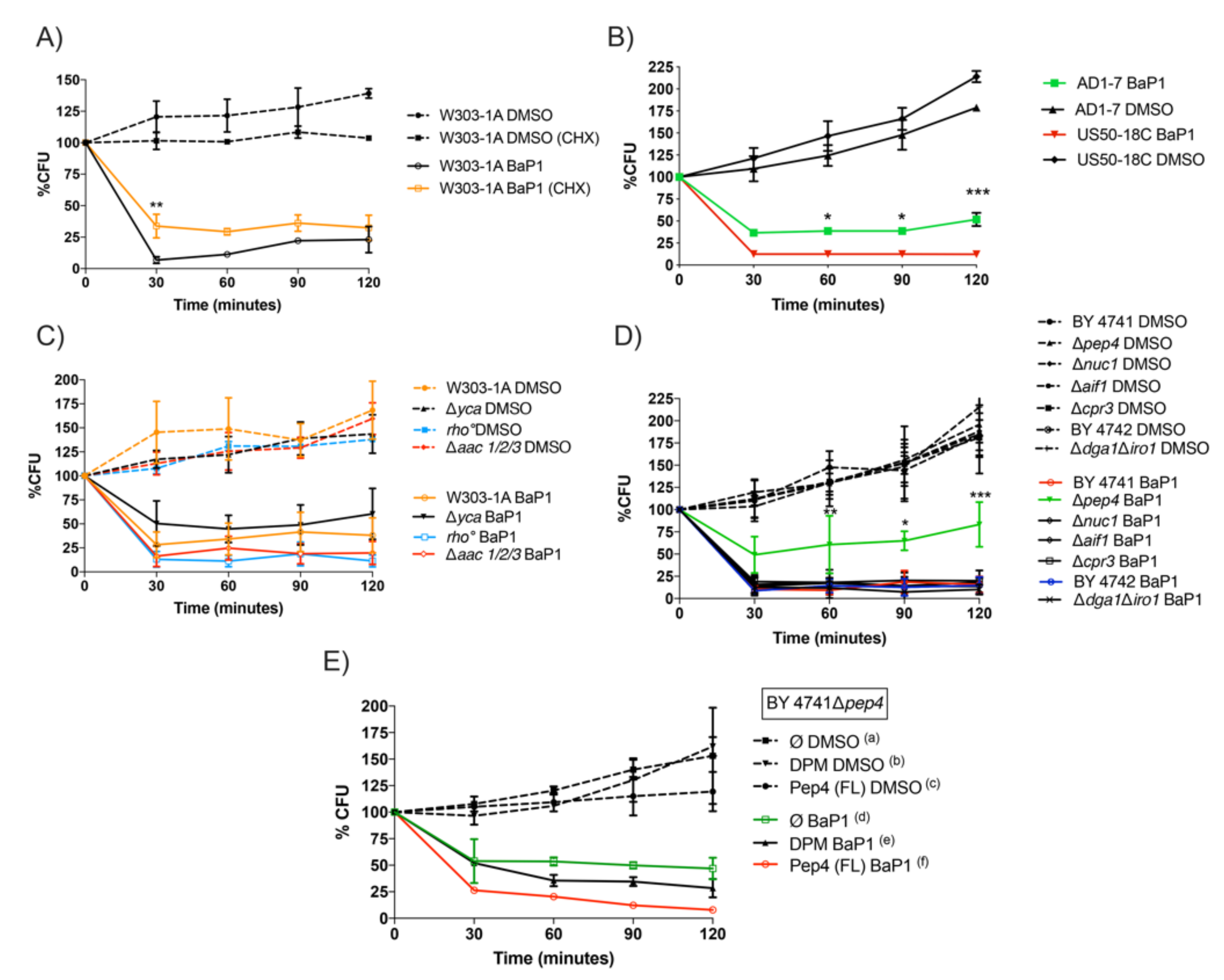
3.1. BaP1 Causes Viability Loss in Yeast Cells Partially Dependent on Protein Synthesis
3.2. Yeast PDR Transporters Do Not Confer Resistance to BaP1
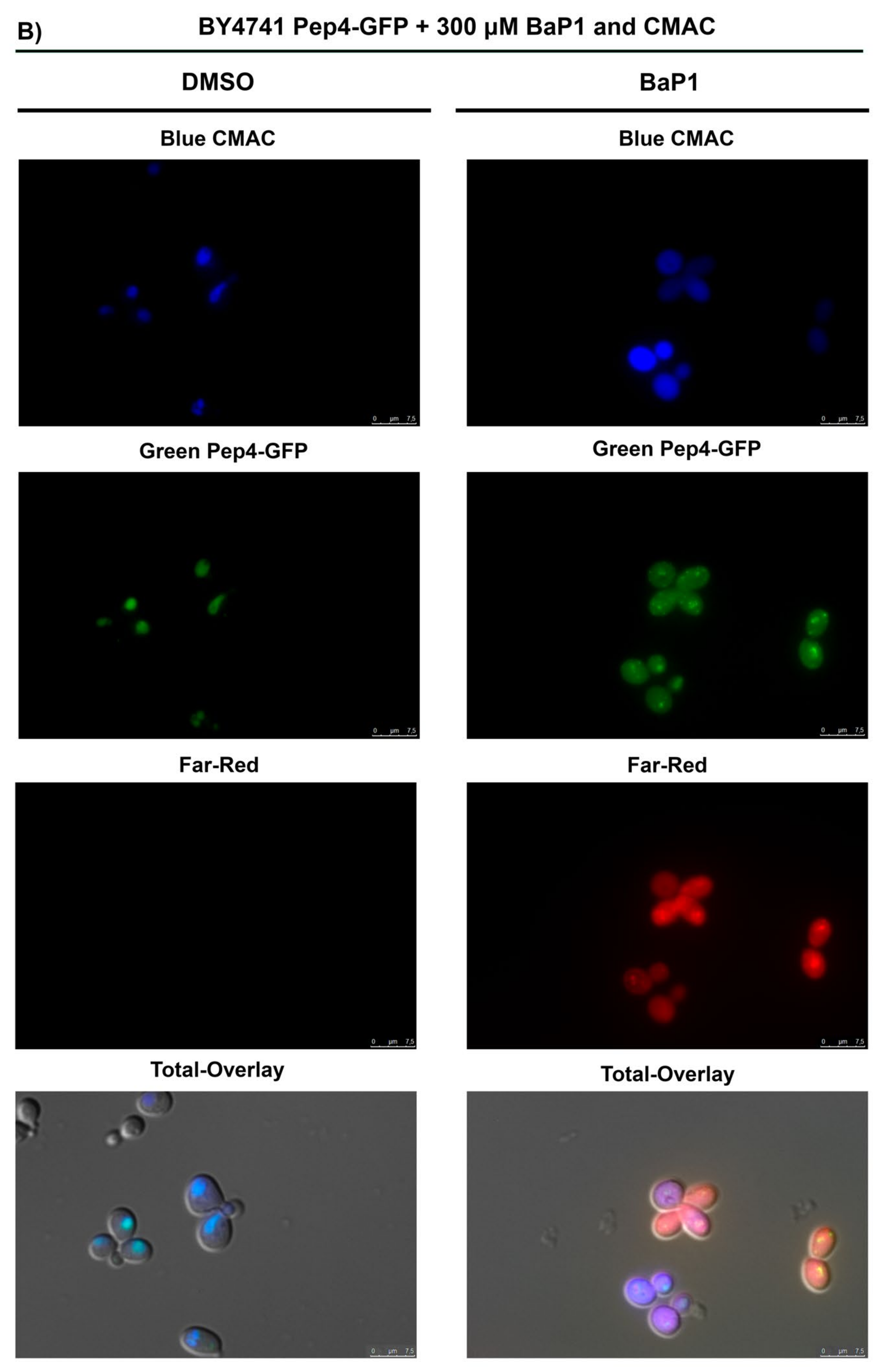
3.3. BaP1-Induced Cell Death Is Dependent on Vacuolar Protease Pep4p; However, Not on Commonly Described Mitochondrial Apoptotic Regulators
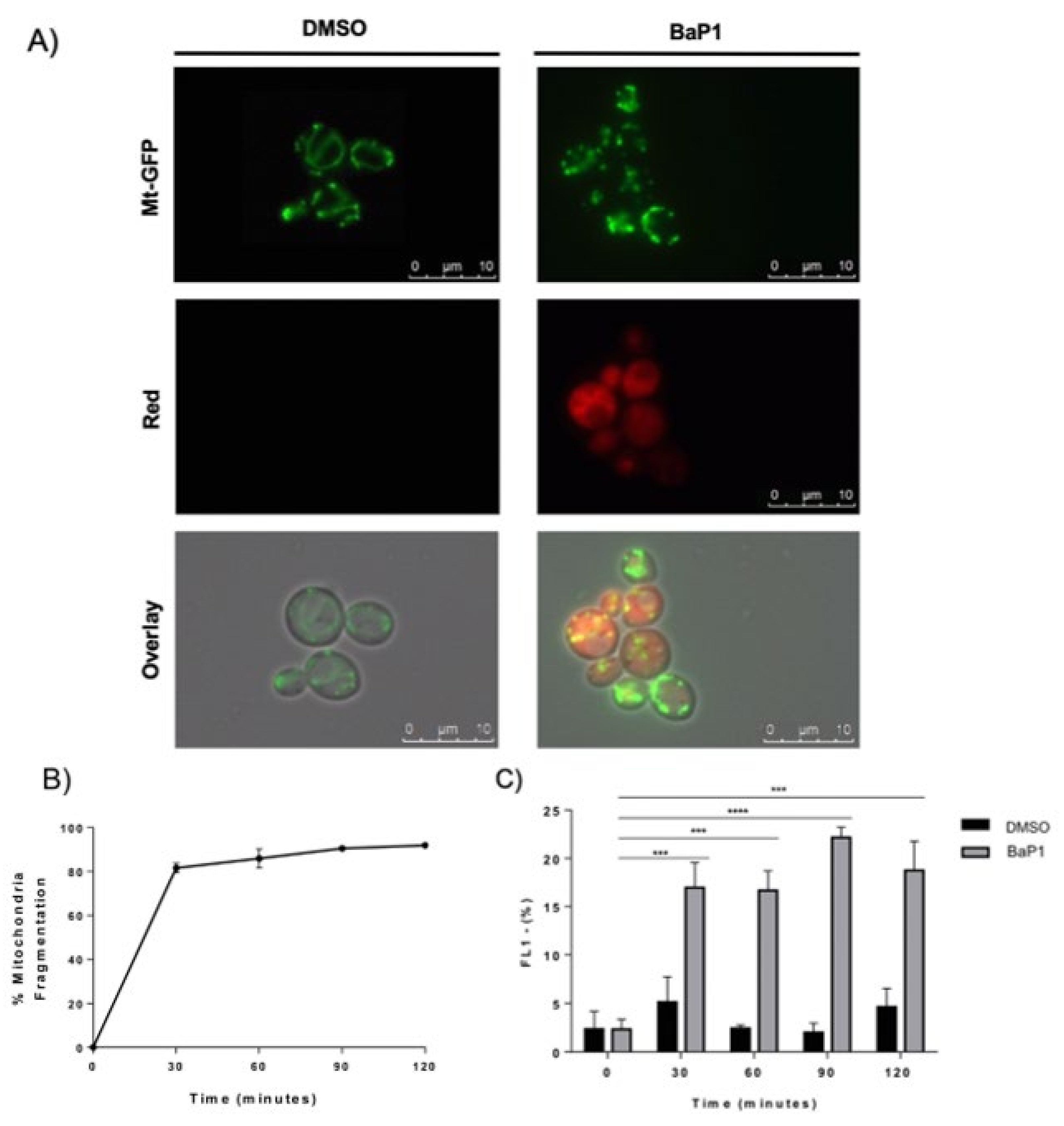
3.4. BaP1 Leads to Mitochondrial Depolarization and Fragmentation with Preserved Plasma Membrane Integrity
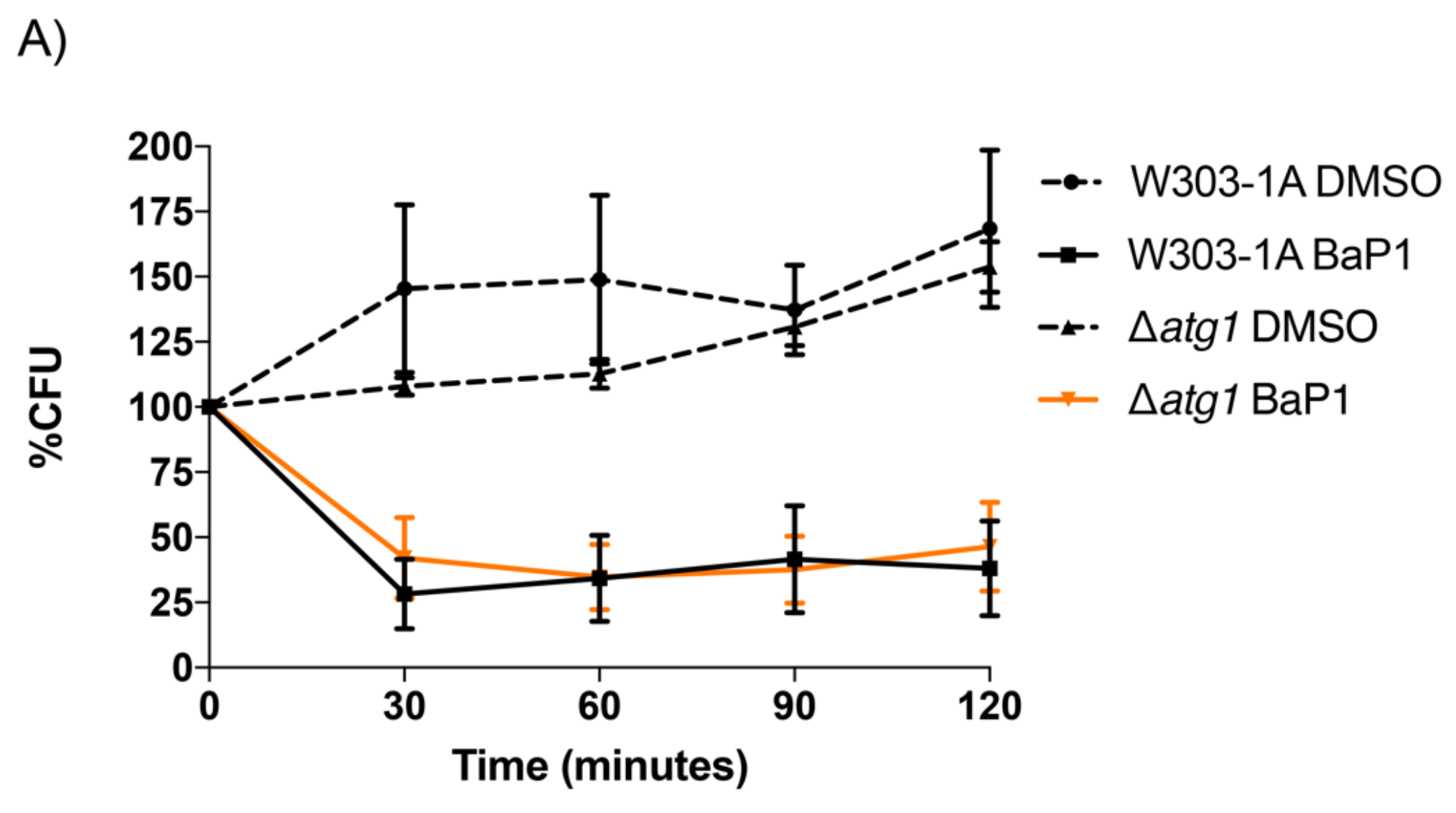
3.5. Autophagy Is Not Involved in the BaP1 Cell Death
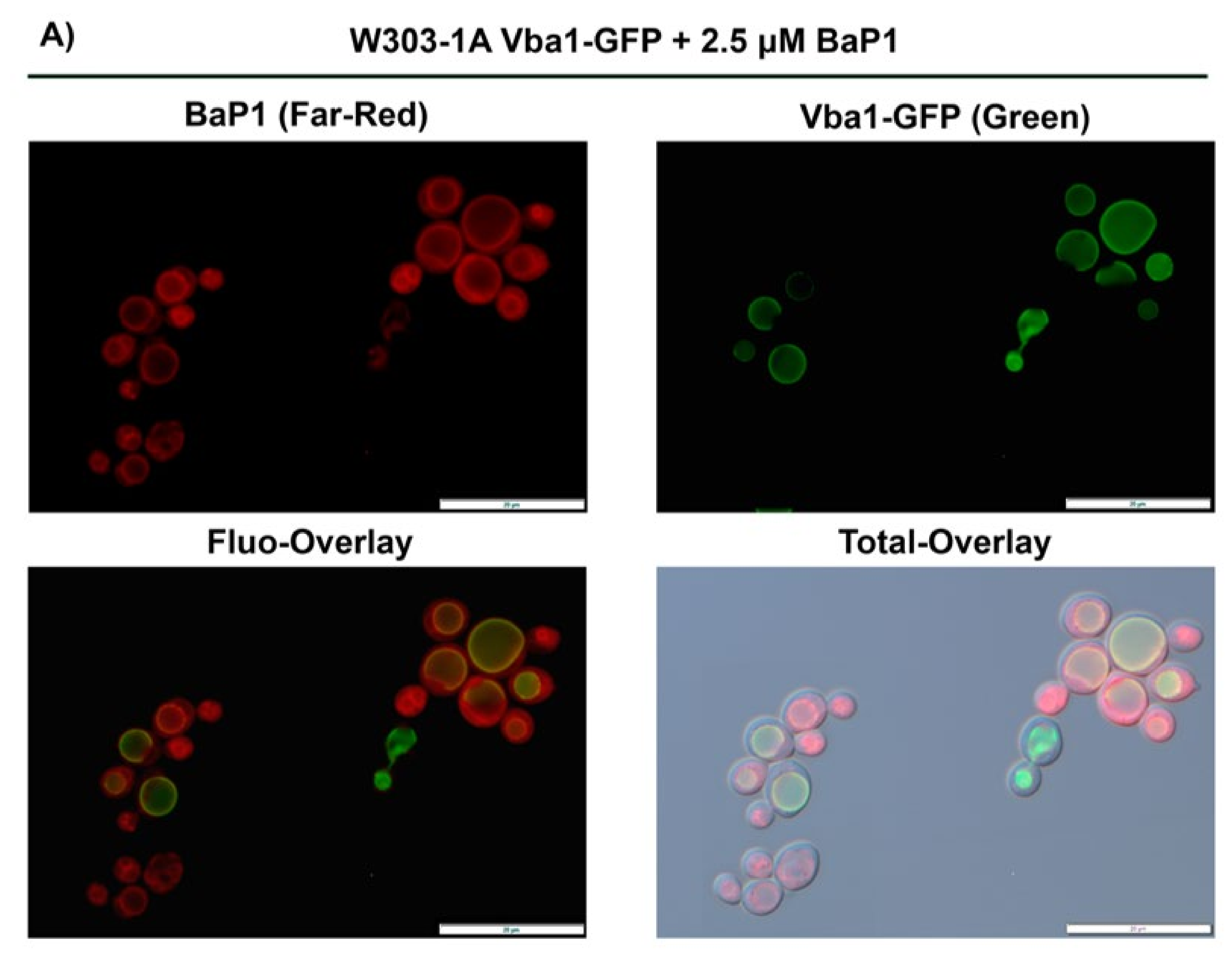
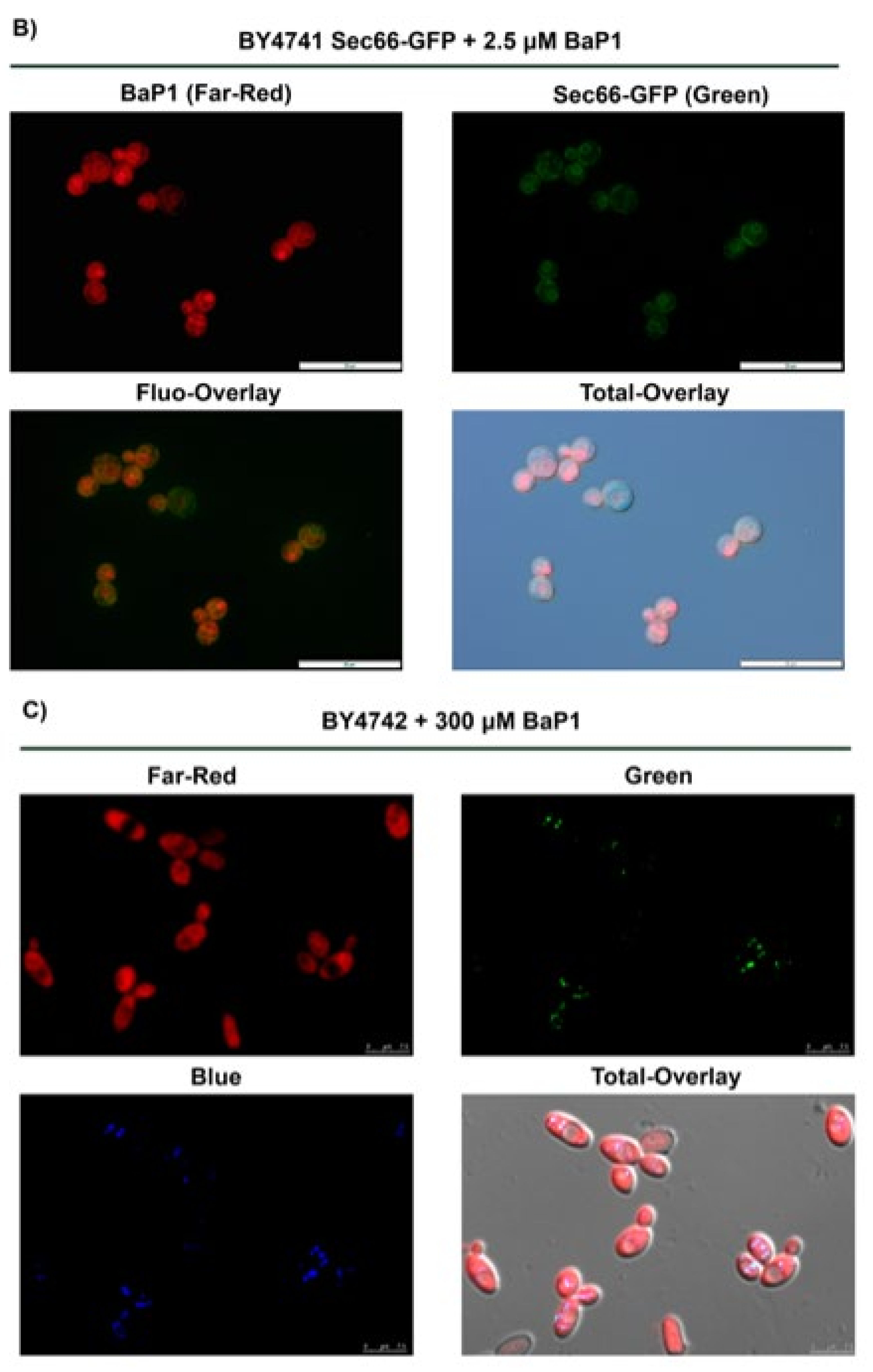
3.6. BaP1 Accumulates at the Vacuolar Membrane, Endoplasmic Reticulum, and Lipid Droplets
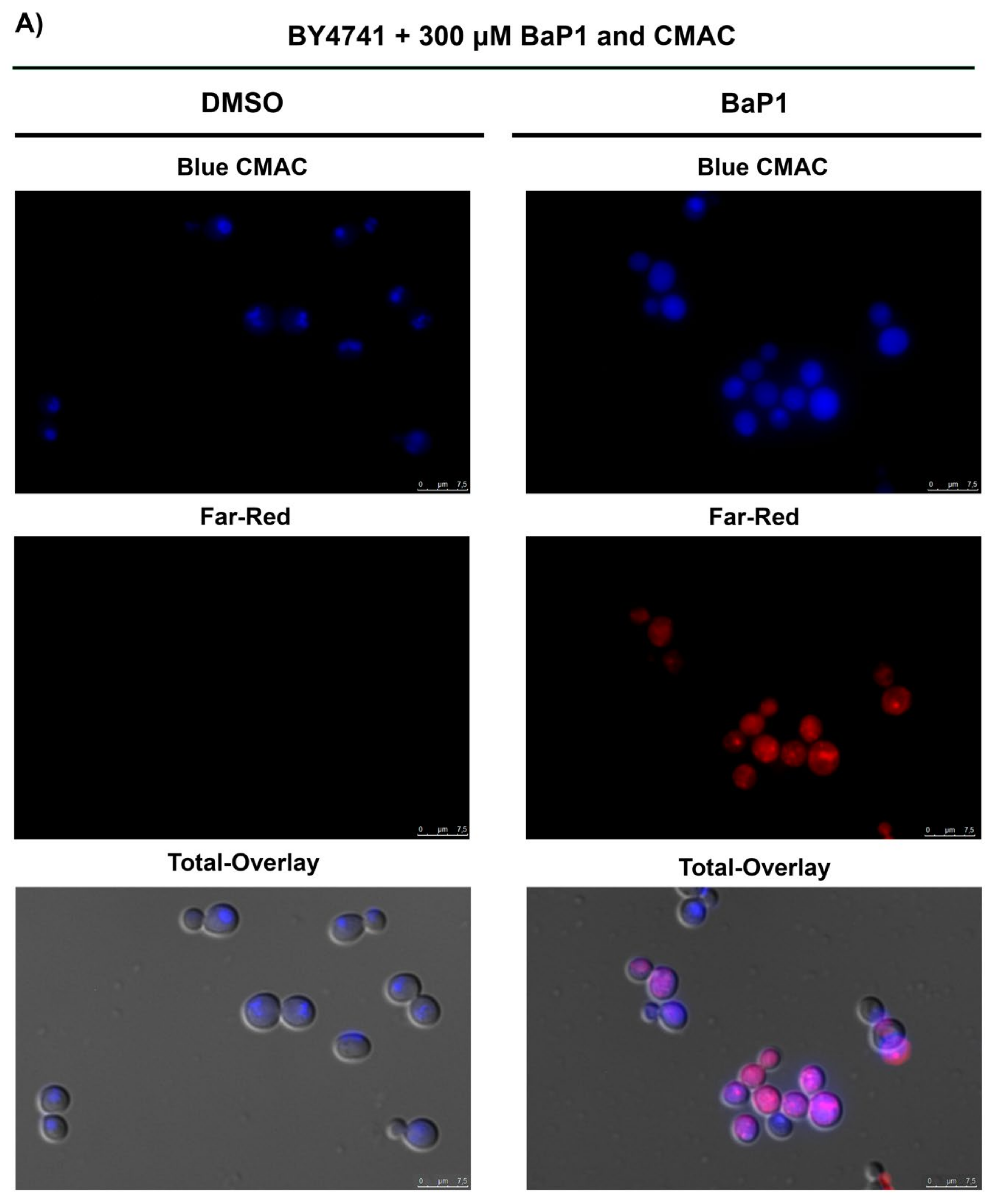
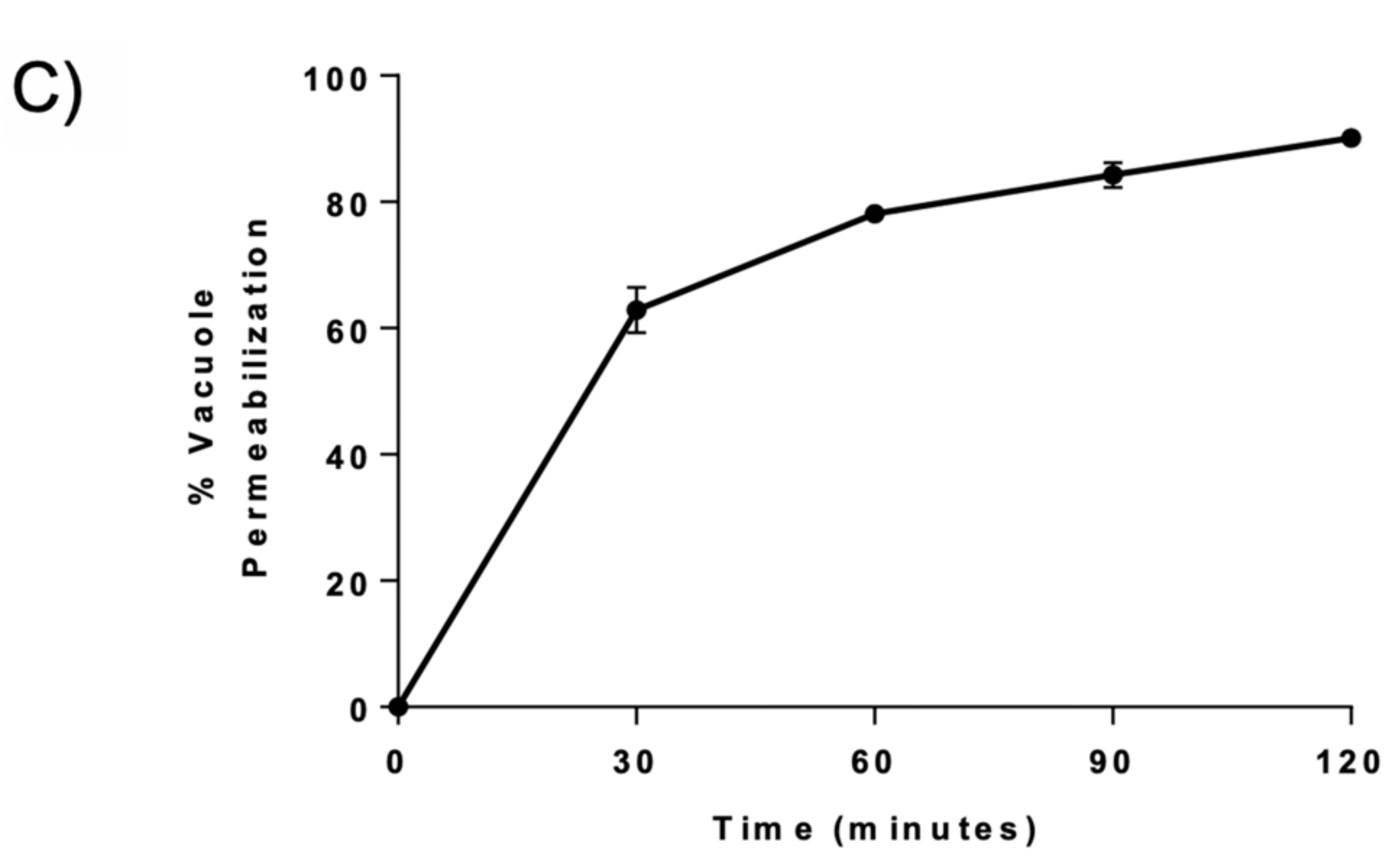
3.7. BaP1 Leads to Vacuolar Membrane Permeabilization and Cytosolic Release of Pep4p
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Jose, J.; Burgess, K. Syntheses and properties of water-soluble Nile Red derivatives. J. Org. Chem. 2006, 71, 7835–7839. [Google Scholar] [CrossRef]
- Frade, V.H.J.; Gonçalves, M.S.T.; Moura, J.C.V.P. Synthesis and fluorescence properties of side-chain carboxylated 5,9-diaminobenzo[a]phenoxazinium salts. Tetrahedron Lett. 2005, 46, 4949–4952. [Google Scholar] [CrossRef]
- Frade, V.H.J.; Gonçalves, M.S.T.; Coutinho, P.J.G.; Moura, J.C.V.P. Synthesis and spectral properties of long-wavelength fluorescent dyes. J. Photochem. Photobiol. A Chem. 2007, 185, 220–230. [Google Scholar] [CrossRef]
- Raju, B.R.; Garcia, A.M.F.; Costa, A.L.S.; Coutinho, P.J.G.; Gonçalves, M.S.T. Synthesis of new benzo[a]phenoxazinium probes possessing carboxylic ester, hydroxyl and amino functional groups: Photophysical studies in dry ethanol and conjugation with CdTe quantum dots. Dye Pigment 2014, 110, 203–213. [Google Scholar] [CrossRef]
- Firmino, A.D.G.; Gonçalves, M.S.T. Bifunctionalised long-wavelength fluorescent probes for biological applications. Tetrahedron Lett. 2012, 53, 4946–4950. [Google Scholar] [CrossRef]
- Raju, B.R.; Firmino, A.D.G.; Costa, A.L.S.; Coutinho, P.J.G.; Gonçalves, M.S.T. Synthesis and photophysical properties of side-chain chlorinated benzo[a]phenoxazinium chlorides. Tetrahedron 2013, 69, 2451–2461. [Google Scholar] [CrossRef]
- Frade, V.H.J.; Sousa, M.J.; Moura, J.C.V.P.; Gonçalves, M.S.T. Synthesis, characterisation and antimicrobial activity of new benzo[a]phenoxazine based fluorophores. Tetrahedron Lett. 2007, 48, 8347–8352. [Google Scholar] [CrossRef]
- Salomi, B.S.B.; Mitra, C.K.; Gorton, L. Electrochemical and spectrophotometric studies on dyes and proteins labelled with dyes. Synth. Met. 2005, 155, 426–429. [Google Scholar] [CrossRef]
- Soto, C.Y.; Andreu, N.; Gibert, I.; Luquin, M. Simple and Rapid Differentiation of Mycobacterium tuberculosis H37Ra from M. tuberculosis Clinical Isolates through Two Cytochemical Tests Using Neutral Red and Nile Blue Stains Simple and Rapid Differentiation of Mycobacterium tuberculosis H37Ra from M. Society 2002, 40, 3021–3024. [Google Scholar]
- Rama Raju, B.; Naik, S.; Coutinho, P.J.G.; Gonçalves, M.S.T. Novel Nile Blue derivatives as fluorescent probes for DNA. Dye Pigment 2013, 99, 220–227. [Google Scholar] [CrossRef]
- Raju, B.R.; Gonçalves, M.S.T.; Coutinho, P.J.G. Fluorescent probes based on side-chain chlorinated benzo[a]phenoxazinium chlorides: Studies of interaction with DNA. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 171, 1–9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Patil, V.S.; Padalkar, V.S.; Phatangare, K.R.; Umape, P.G.; Borase, B.N.; Sekar, N. Synthesis, Characterization, and Antibacterial Activity of Novel (1H-Benzo[d]imidazole-2-yl)-6-(diethylamino)-3H-one-xanthene, Phenoxazine, and Oxazine. J. Heterocycl. Chem. 2015, 52, 124–129. [Google Scholar] [CrossRef]
- Frade, V.H.J.; Sousa, M.J.; Moura, J.C.V.P.; Gonçalves, M.S.T. Synthesis of naphtho[2,3-a]phenoxazinium chlorides: Structure-activity relationships of these heterocycles and benzo[a]phenoxazinium chlorides as new antimicrobials. Bioorganic Med. Chem. 2008, 16, 3274–3282. [Google Scholar] [CrossRef]
- Lopes, M.; Alves, C.T.; Rama Raju, B.; Gonçalves, M.S.T.; Coutinho, P.J.G.; Henriques, M.; Belo, I. Application of benzo[a]phenoxazinium chlorides in antimicrobial photodynamic therapy of Candida albicans biofilms. J. Photochem. Photobiol. B Biol. 2014, 141, 93–99. [Google Scholar] [CrossRef]
- Shimamoto, T.; Tomoda, A.; Ishida, R.; Ohyashiki, K. Antitumor effects of a novel phenoxazine derivative on human leukemia cell lines in vitro and in vivo. Clin. Cancer Res. 2001, 7, 704–708. [Google Scholar]
- Bolognese, A.; Correale, G.; Manfra, M.; Lavecchia, A.; Novellino, E.; Pepe, S. Antitumor agents. 5. Synthesis, structure - activity relationships, and biological evaluation of dimethyl-5H-pyridophenoxazin-5-ones, tetrahydro-5H-benzopyridophenoxazin-5-ones, and 5H-benzopyridophenoxazin-5-ones with potent antiproliferative activity. J. Med. Chem. 2006, 49, 5110–5118. [Google Scholar] [CrossRef] [PubMed]
- Alberti, A.; Bolognese, A.; Guerra, M.; Lavecchia, A.; Macciantelli, D.; Marcaccio, M.; Novellino, E.; Paolucci, F. Antitumor agents 4. Characterization of free radicals produced during reduction of the antitumor drug 5H-pyridophenoxazin-5-one: An EPR study. Biochemistry 2003, 42, 11924–11931. [Google Scholar] [CrossRef] [PubMed]
- Küçükkılınç, T.; Özer, İ. Multi-site inhibition of human plasma cholinesterase by cationic phenoxazine and phenothiazine dyes. Arch. Biochem. Biophys. 2007, 461, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Konkimalla, V.B.; Kathawate, L.; Rao, S.S.; Gejji, S.P.; Puranik, V.G.; Weyhermüller, T.; Salunke-Gawali, S. Targeting a chemorefractory COLO205 (BRAF V600E) cell line using substituted benzo[α]phenoxazines. RSC Adv. 2015, 5, 82549–82563. [Google Scholar] [CrossRef]
- Suzuki, F.; Hashimoto, K.; Ishihara, M.; Westman, G.; Samuelsson, K.; Kawase, M.; Motohashi, N.; Sakagami, H. Tumor-specificity and type of cell death induced by phenoxazines. Anticancer Res. 2007, 27, 4233–4238. [Google Scholar] [PubMed]
- Shirato, K.; Imaizumi, K.; Abe, A.; Tomoda, A. Phenoxazine derivatives 2-amino-4,4alpha-dihydro-4alpha-phenoxazine-3-one and 2-aminophenoxazine-3-one-induced apoptosis through a caspase-independent mechanism in human neuroblastoma cell line NB-1 cells. Biol. Pharm. Bull. 2007, 30, 331–336. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abe, A.; Yamane, M.; Tomoda, A. Prevention of growth of human lung carcinoma cells and induction of apoptosis by a novel phenoxazinone, 2-amino-4,4alpha-dihydro-4alpha,7-dimethyl-3H-phenoxazine-3-one. Anticancer. Drugs 2001, 12, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Salunke-Gawali, S.; Konkimalla, V.B. Induction of Autophagic Cell Death in Apoptosis-resistant Pancreatic Cancer Cells using Benzo[α]phenoxazines Derivatives, 10-methyl-benzo[α]phenoxazine-5-one and benzo[α]phenoxazine-5-one. Anticancer Agents Med. Chem. 2017, 17, 115–125. [Google Scholar] [PubMed]
- Lewis, M.R.; Goland, P.P.; Sloviter, H.A. The action of oxazine dyes on tumors in mice. Cancer Res. 1949, 9, 736–740. [Google Scholar] [PubMed]
- Lin, C.W.; Shulok, J.R.; Wong, Y.K.; Schanbacher, C.F.; Cincotta, L.; Foley, J.W. Photosensitization, Uptake, and Retention of Phenoxazine Nile Blue Derivatives in Human Bladder Carcinoma Cells. Cancer Res. 1991, 51, 1109–1116. [Google Scholar] [PubMed]
- Frade, V.H.J.; Gonçalves, M.S.T.; Moura, J.C.V.P. Synthesis of fluorescent water-soluble functionalised benzo[a]phenoxazinium salts. Tetrahedron Lett. 2006, 47, 8567–8570. [Google Scholar] [CrossRef]
- Frade, V.H.J.; Coutinho, P.J.G.; Moura, J.C.V.P.; Gonçalves, M.S.T. Functionalised benzo[a]phenoxazine dyes as long-wavelength fluorescent probes for amino acids. Tetrahedron 2007, 63, 1654–1663. [Google Scholar] [CrossRef]
- Popa, C.-V.; Dumitru, I.; Ruta, L.L.; Danet, A.F.; Farcasanu, I.C. Exogenous oxidative stress induces Ca2+ release in the yeast Saccharomyces cerevisiae. FEBS J. 2010, 277, 4027–4038. [Google Scholar] [CrossRef]
- Ludovico, P.; Sousa, M.J.; Silva, M.T.; Leão, C.; Côrte-Real, M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology 2001, 147, 2409–2415. [Google Scholar] [CrossRef]
- Fortuna, M.; Sousa, M.J.; Côrte-Real, M.; Leão, C.; Salvador, A.; Sansonetty, F. Cell cycle analysis of yeasts. Curr. Protoc. Cytom. 2001, 13, 11–13. [Google Scholar] [CrossRef]
- Camougrand, N.; Kiššová, I.; Salin, B.; Devenish, R.J. Monitoring Mitophagy in Yeast. Autophagy 2008, 451, 89–107. [Google Scholar]
- Cannon, R.D.; Lamping, E.; Holmes, A.R.; Niimi, K.; Baret, P.V.; Keniya, M.V.; Tanabe, K.; Niimi, M.; Goffeau, A.; Monk, B.C. Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 2009, 22, 291–321. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Goffeau, A. Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu. Rev. Microbiol. 2012, 66, 39–63. [Google Scholar] [CrossRef] [PubMed]
- Decottignies, A.; Grant, A.M.; Nichols, J.W.; de Wet, H.; McIntosh, D.B.; Goffeau, A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 1998, 273, 12612–12622. [Google Scholar] [CrossRef] [PubMed]
- Hallstrom, T.C.; Lambert, L.; Schorling, S.; Balzi, E.; Goffeau, A.; Moye-Rowley, W.S. Coordinate Control of Sphingolipid Biosynthesis and Multidrug Resistance in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 23674–23680. [Google Scholar] [CrossRef]
- Kamei, Y.; Koushi, M.; Aoyama, Y.; Asakai, R. The yeast mitochondrial permeability transition is regulated by reactive oxygen species, endogenous Ca(2+) and Cpr3, mediating cell death. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 1313–1326. [Google Scholar] [CrossRef]
- Pereira, C.; Camougrand, N.; Manon, S.; Sousa, M.J.; Côrte-Real, M. ADP/ATP carrier is required for mitochondrial outer membrane permeabilization and cytochrome c release in yeast apoptosis. Mol. Microbiol. 2007, 66, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Buttner, S.; Eisenberg, T.; Carmona-Gutierrez, D.; Ruli, D.; Knauer, H.; Ruckenstuhl, C.; Sigrist, C.; Wissing, S.; Kollroser, M.; Frohlich, K.-U.; et al. Endonuclease G regulates budding yeast life and death. Mol. Cell 2007, 25, 233–246. [Google Scholar] [CrossRef]
- Mason, D.A.; Shulga, N.; Undavai, S.; Ferrando-May, E.; Rexach, M.F.; Goldfarb, D.S. Increased nuclear envelope permeability and Pep4p-dependent degradation of nucleoporins during hydrogen peroxide-induced cell death. FEMS Yeast Res. 2005, 5, 1237–1251. [Google Scholar] [CrossRef]
- Pereira, C.; Chaves, S.; Alves, S.; Salin, B.; Camougrand, N.; Manon, S.; Sousa, M.J.; Corte-Real, M. Mitochondrial degradation in acetic acid-induced yeast apoptosis: The role of Pep4 and the ADP/ATP carrier. Mol. Microbiol. 2010, 76, 1398–1410. [Google Scholar] [CrossRef]
- Westermann, B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010, 11, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Suen, D.; Norris, K.L.; Youle, R.J. Mitochondrial dynamics and apoptosis. Genes Dev. 2008, 22, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- Herrero, E.; Ros, J.; Bellí, G.; Cabiscol, E. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta—Gen. Subj. 2008, 1780, 1217–1235. [Google Scholar] [CrossRef]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003, 552, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Orrenius, S.; Zhivotovsky, B.; Nicotera, P. Regulation of cell death: The calcium-apoptosis link. Nat. Rev. Mol. Cell Biol. 2003, 4, 552–565. [Google Scholar] [CrossRef]
- Azzopardi, M.; Farrugia, G.; Balzan, R. Cell-cycle involvement in autophagy and apoptosis in yeast. Mech. Ageing Dev. 2017, 161, 211–224. [Google Scholar] [CrossRef]
- Zhang, J.H.; Xu, M. DNA fragmentation in apoptosis. Cell Res. 2000, 10, 205–211. [Google Scholar] [CrossRef]
- Papinski, D.; Schuschnig, M.; Reiter, W.; Wilhelm, L.; Barnes, C.A.; Maiolica, A.; Hansmann, I.; Pfaffenwimmer, T.; Kijanska, M.; Stoffel, I.; et al. Early Steps in Autophagy Depend on Direct Phosphorylation of Atg9 by the Atg1 Kinase. Mol. Cell 2014, 53, 471–483. [Google Scholar] [CrossRef]
- Shintani, T.; Klionsky, D.J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004, 279, 29889–29894. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Ohbayashi, S.; Sakoh-Nakatogawa, M.; Kakuta, S.; Suzuki, S.W.; Kirisako, H.; Kondo-Kakuta, C.; Noda, N.N.; Yamamoto, H.; Ohsumi, Y. The autophagy-related protein kinase Atg1 interacts with the ubiquitin-like protein Atg8 via the Atg8 family interacting motif to facilitate autophagosome formation. J. Biol. Chem. 2012, 287, 28503–28507. [Google Scholar] [CrossRef]
- Shpilka, T.; Weidberg, H.; Pietrokovski, S.; Elazar, Z. Atg8: An autophagy-related ubiquitin-like protein family. Genome Biol. 2011, 12, 226. [Google Scholar] [CrossRef]
- Martinez, V.; Henary, M. Nile Red and Nile Blue: Applications and Syntheses of Structural Analogues. Chem. Eur. J. 2016, 22, 13764–13782. [Google Scholar] [CrossRef]
- Guicciardi, M.E.; Leist, M.; Gores, G.J. Lysosomes in cell death. Oncogene 2004, 23, 2881–2890. [Google Scholar] [CrossRef]
- Bursch, W. The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ. 2001, 8, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Sousa, R.P.C.L.; Ferreira, J.C.C.; Sousa, M.J.; Gonçalves, M.S.T. N-(5-Amino-9H-benzo[a]phenoxazin-9-ylidene)propan-1-aminium chlorides as antifungal agents and NIR fluorescent probes. New J. Chem. 2021, 45, 7808–7815. [Google Scholar] [CrossRef]
- Kolaczkowska, A.; Goffeau, A. Regulation of pleiotropic drug resistance in yeast. Drug Resist. Updates 1999, 2, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Terman, A.; Kurz, T.; Gustafsson, B.; Brunk, U.T. Lysosomal labilization. IUBMB Life 2006, 58, 531–539. [Google Scholar] [CrossRef]
- Serrano-Puebla, A.; Boya, P. Lysosomal membrane permeabilization in cell death: New evidence and implications for health and disease. Ann. N. Y. Acad. Sci. 2016, 1371, 30–44. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C.; Wattiaux, R. Functions of lysosomes. Annu. Rev. Physiol. 1966, 28, 435–492. [Google Scholar] [CrossRef]
- Kim, H.; Kim, A.; Cunningham, K.W. Vacuolar H+-ATPase (V-ATPase) promotes vacuolar membrane permeabilization and nonapoptotic death in stressed yeast. J. Biol. Chem. 2012, 287, 19029–19039. [Google Scholar] [CrossRef]
- Leitão, M.I.P.S.; Rama Raju, B.; Cerqueira, N.M.F.S.A.; Sousa, M.J.; Gonçalves, M.S.T. Benzo[a]phenoxazinium chlorides: Synthesis, antifungal activity, in silico studies and evaluation as fluorescent probes. Bioorg. Chem. 2020, 98, 103730. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, E.; Prelle, A. Application of nile blue and nile red, two fluorescent probes, for detection of lipid droplets in human skeletal muscle. J. Histochem. Cytochem. Off. J. Histochem. Soc. 1987, 35, 619–621. [Google Scholar] [CrossRef]
- Greenspan, P.; Mayer, E.P.; Fowler, S.D. Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985, 100, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Tatenaka, Y.; Kato, H.; Ishiyama, M.; Sasamoto, K.; Shiga, M.; Nishitoh, H.; Ueno, Y. Monitoring Lipid Droplet Dynamics in Living Cells by Using Fluorescent Probes. Biochemistry 2019, 58, 499–503. [Google Scholar] [CrossRef]
- Zhang, J.; De Sousa, W.T.; Da Silva, V.C.M.; Rodrigues, M.C.; Morais, J.A.V.; Song, J.L.; Cheng, Z.Q.; Longo, J.P.F.; Azevedo, R.B.; Jiang, C.S.; et al. Synthesis and evaluation of new potential benzo[a]phenoxazinium photosensitizers for anticancer photodynamic therapy. Molecules 2018, 23, 1436. [Google Scholar] [CrossRef]
- Chhabria, M.T.; Jani, M.H. Design, synthesis and antimycobacterial activity of some novel imidazo[1,2-c]pyrimidines. Eur. J. Med. Chem. 2009, 44, 3837–3844. [Google Scholar] [CrossRef]
- Mickevičienė, K.; Baranauskaitė, R.; Kantminienė, K.; Stasevych, M.; Komarovska-Porokhnyavets, O.; Novikov, V. Synthesis and antimicrobial activity of N-substituted-β-amino acid derivatives containing 2-hydroxyphenyl, benzo[b]phenoxazine and quinoxaline moieties. Molecules 2015, 20, 3170–3189. [Google Scholar] [CrossRef] [PubMed]
- Kozlovskaya, L.I.; Andrei, G.; Orlov, A.A.; Khvatov, E.V.; Koruchekov, A.A.; Belyaev, E.S.; Nikolaev, E.N.; Korshun, V.A.; Snoeck, R.; Osolodkin, D.I.; et al. Antiviral activity spectrum of phenoxazine nucleoside derivatives. Antiviral Res. 2019, 163, 117–124. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira, J.C.C.; Lopes, C.; Preto, A.; Gonçalves, M.S.T.; Sousa, M.J. Novel Nile Blue Analogue Stains Yeast Vacuolar Membrane, Endoplasmic Reticulum, and Lipid Droplets, Inducing Cell Death through Vacuole Membrane Permeabilization. J. Fungi 2021, 7, 971. https://doi.org/10.3390/jof7110971
Ferreira JCC, Lopes C, Preto A, Gonçalves MST, Sousa MJ. Novel Nile Blue Analogue Stains Yeast Vacuolar Membrane, Endoplasmic Reticulum, and Lipid Droplets, Inducing Cell Death through Vacuole Membrane Permeabilization. Journal of Fungi. 2021; 7(11):971. https://doi.org/10.3390/jof7110971
Chicago/Turabian StyleFerreira, João Carlos Canossa, Carla Lopes, Ana Preto, Maria Sameiro Torres Gonçalves, and Maria João Sousa. 2021. "Novel Nile Blue Analogue Stains Yeast Vacuolar Membrane, Endoplasmic Reticulum, and Lipid Droplets, Inducing Cell Death through Vacuole Membrane Permeabilization" Journal of Fungi 7, no. 11: 971. https://doi.org/10.3390/jof7110971
APA StyleFerreira, J. C. C., Lopes, C., Preto, A., Gonçalves, M. S. T., & Sousa, M. J. (2021). Novel Nile Blue Analogue Stains Yeast Vacuolar Membrane, Endoplasmic Reticulum, and Lipid Droplets, Inducing Cell Death through Vacuole Membrane Permeabilization. Journal of Fungi, 7(11), 971. https://doi.org/10.3390/jof7110971







