Abstract
Circular RNAs (circRNAs) are covalently closed RNA molecules, and have been identified in many crops. However, there are few datasets for circRNA junctions from common wheat during Fusarium head blight disease. In the present study, we used RNA-seq to determine the changes in circRNAs among the control (CK) and 1, 3, and 5 days post-Fusarium graminearum inoculation (dpi) samples. More than one billion reads were produced from 12 libraries, and 99.99% of the reads were successfully mapped to a wheat reference genome. In total, 2091 high-confidence circRNAs—which had two or more junction reads and were supported by at least two circRNA identification algorithms—were detected. The completed expression profiling revealed a distinct expression pattern of circRNAs among the CK, 1dpi, 3dpi and 5dpi samples. This study provides a valuable resource for identifying F. graminearum infection-responsive circRNAs in wheat and for further functional characterization of circRNAs that participated in the Fusarium head blight disease response of wheat.
Dataset: The FastQ CASAVA (v1.8.2, Illumina) format sequencing reads have been deposited in the US National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) with the Bioproject ID: PRJNA826345.
Dataset License: CC-BY
1. Summary
The long noncoding RNAs (lncRNA) [1], microRNAs (miRNA) [2], small interference RNAs (siRNA), piwi-interacting RNAs (piRNA) [3], and transfer RNA-derived small RNAs (tsRNA) [4] have been demonstrated to play an indispensable role in regulating plant growth and stress response [5]. In addition to these linear RNAs, the identification and function characterization of circular RNAs, a new biotype on the noncoding RNA list, have gained wide attention. With the development of next-generation sequencing techniques and the supporting bioinformatic analysis strategies, genome-wide identification for circRNAs has been conducted in a wide range of species [6].
Circular RNAs (circRNAs), which are covalently closed with a 2′,5′-phosphodiester bond or a 3′,5′-phosphodiester bond at the back-splicing junction sites, are single-stranded noncoding RNA molecules [7]. CircRNAs are proposed to be more stable than linear RNAs due to there being no 5′-3′ polarity nor polyadenylated tails. Nowadays, a large amount of circRNAs have been detected in plants (121,971) and animals (32,914), suggesting their importance in transcriptional or post-transcriptional regulation [8]. Expression profiling indicated that some circRNAs exhibit cell and/or tissue-specific expression patterns and are much more abundant than their linear counterparts. They are also evolutionary conserved among different plant species, suggesting the potential physiological functions of these circRNAs [7].
Although the interaction between wheat and Fusarium has been extensively studied for years, the function of circRNAs is still poorly known in common wheat during Fusarium head blight disease development. Herein, we produced a set of high-throughput RNA sequencing reads originating from the CK, 1dpi, 3dpi, and 5dpi wheat samples (Table 1). The reliable circRNAs were identified, and their expression patterns and characters were systemically profiled. This dataset will contribute to the foundation for the characterization of circRNAs in common wheat, specifically for studying circRNAs function analysis during the wheat and Fusarium graminearum interaction.

Table 1.
Statistic of sequencing reads and predicted circRNAs.
2. Data Description
2.1. Identification and Characterization of Wheat circRNAs
The workflow for the sample preparation, circRNA-seq, and bioinformatic analysis is shown in Figure 1. Detailed information on the identified circRNAs and their corresponding expression profiles can be found in Table S1. As shown in Figure 2A,B, among the circRNAs identified by CIRI2, CIRCexplorer2, and find_circ, 2091 circRNAs (1) had two or more junction counts, and two were supported by at least two algorithms. Among these highly confident circRNAs, most (1931, 78.00%) were originally from the exon regions of the genome, 418 circRNAs (19.99%) were originally from the intergenic region, and the remaining 42 circRNAs (2.01%) were originally from intron (Figure 2C). Further analysis showed these circRNAs were generally distributed on all chromosomes (Figure 2D). Nevertheless, according to the genomic density analysis, which is different from the coding genes, circRNA genes were unevenly scattered on chromosomes and more commonly detected at the centre and both ends of chromosomal regions. When considering the length, most circRNAs are shorter than 1600 nucleotide (nt), their average length is ~830 nt, and most parts of long circRNAs originate from the intergenic region (Figure 2E). Most circRNAs (135) were transcribed from Chr5A (chromosomes 5A), followed by Chr2B (116) and Chr3D (115) (Figure 2F). Correlation analyses indicated that the amount of circRNAs was significantly correlated with the length of corresponding chromosomes, suggesting that more circRNAs were produced by longer chromosomes (Figure 2G, R2 = 0.318, t-test ***). According to the alternative circularization analysis, 352 alternative back-splicing circularization events were identified, and they originated from 146 distinct chromosomal loci. To be specific, in the 146 chromosomal loci, 115 loci generated two distinct circRNA isoforms, 18 loci generated three different isoforms, 8 loci generated four isoforms, 2 loci generated five isoforms, 2 loci generated seven isoforms, and 1 locus generated 12 isoforms (Figure 2H). A Wilcoxon rank-sum test revealed that, on the whole, the general expression levels of wheat circRNAs were significantly regulated during Fusarium head blight disease development (p-value < 0.005. Figure 2I).
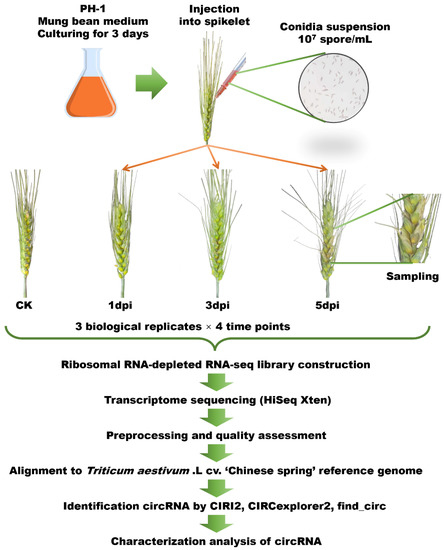
Figure 1.
Overview of experiment design and analysis pipeline. Wheat spikelet tissues subjected to F. graminearum conidia suspension were harvested at 1, 3, and 5 days post inoculation (dpi) and tissues without inoculation were harvested as control (CK). The circRNA-seq was performed by the Illumina HiSeq Xten platform. Raw reads were quality assessed and filtered, and clean reads were used to identify circRNA by CIRI2, CIRCexplorer, and find_circ. Characterization analyses of circRNAs were also performed.
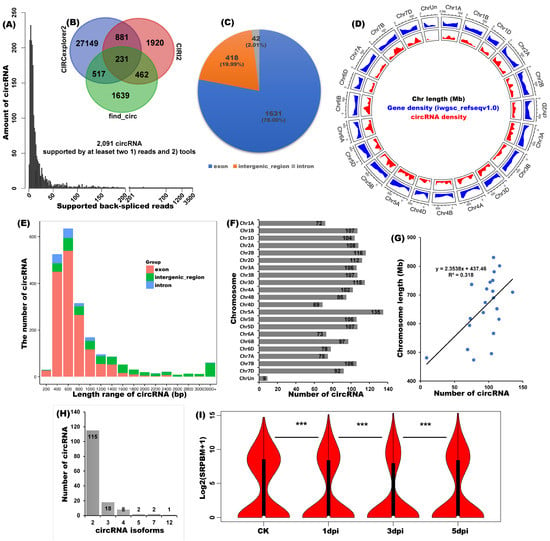
Figure 2.
Characterization of wheat circRNAs. The number of circRNAs is supported by at least (A) two junction reads and (B) two identification tools. (C) Percentage and amount of circRNAs generated by the exon, intergenic, and intron chromosomal reigns. (D) Chromosome distribution and density of circRNA and reference mRNA. (E) Length distribution of circRNA. (F) The number of circRNA in the corresponding chromosome. (G) Correlation analysis between the length of each chromosome and the amount of circRNA in the corresponding chromosome (t-test, p-value < 0.005). (H) Alternative circularization event and the corresponding number of circRNA isoforms. (I) The overall expression patterns of wheat circRNA under CK, 1dpi (days post inoculation), 3dpi, and 5dpi. Log2(SRPBM + 1) values were used to draw the violin plot. *** p < 0.005, Wilcoxon rank-sum test.
2.2. Quality Validation and Analyses
The quality and quantity of total RNA extracted from the control and disease wheat spikelets were estimated by a 1% agarose gel, Nanodrop spectrophotometer, and Agilent Bioanalyzer 2100. The results indicated that the RIN value was larger than 7.5 for all the samples. The quality assessment convinced us that the total RNA met the requirements for constructing the circular RNA sequencing library.
The quality assessment of the circRNA-seq reads was performed using FastQC and multiQC [9] by surveying the mean quality score per sequence and position, as well as the sequence of the GC content and read-length distribution. As shown in Figure 3A, the mean sequence quality of most of the majority reads was greater than the Phred quality score of 30, and all quality scores of the bases per position were larger than a score of 35 (Figure 3B). The GC content distribution of all samples was normal, indicating that the reads were not contaminated during the sequencing process (Figure 3C). As designed, the majority of the reads were 150 bp (Figure 3D). Those indicators validated that the raw reads were of high quality. The alignments of the pre-processed reads resulted in a high mapping rate, and, besides the CK (99.98%), the other samples were all 99.99% (Table 1).
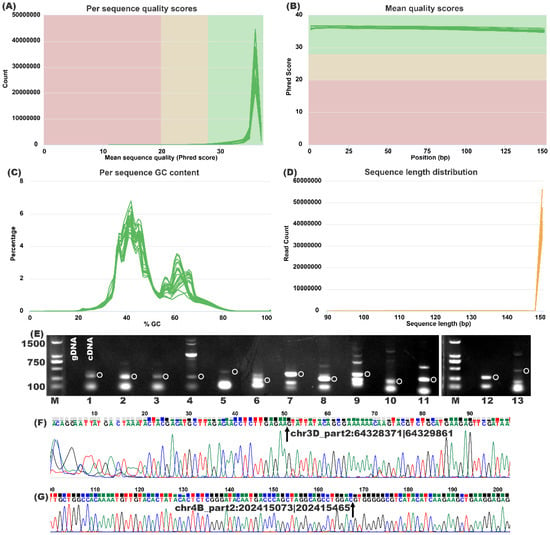
Figure 3.
Quality control of raw sequencing reads and circRNAs. (A) Per sequence quality scores. The x-axis depicts the mean value of sequence quality scores, and the y-axis represents the read counts. (B) Mean quality scores of each sequence position. The x-axis depicts the position, and the y-axis depicts the Phred1.9 score. (C) The GC content of sequencing reads. The x-axis depicts the GC content, and the y-axis represents the percentage of reads. (D) Sequence length distribution. The x-axis represents the length of sequencing reads, and the y-axis depicts the count of a sequencing read. (E) PCR and (F,G) Sanger confirmation of circRNAs. The gDNA and cDNA were both used as templates for PCR validation. The circular indicates the designed amplicon hugging the back-splicing site. 1: chr1A_part1:261131674|261134157; 2: chr1D_part1:416533069|416534514; 3: chr2A_part1:41738558|41741294; 4: chr2D_part1:344564233|344568330; 5: chr2D_part1:34672266|34676866; 6: chr3B_part2:286756868|286758204; 7: chr3D_part2:64328371|64329861; 8: chr4B_part2:202415073|202415465; 9: chr5A_part1:6666752|6667506; 10: chr5A_part2:178282187|178282717; 11: chr5B_part1:170596562|170597173; 12: chr6A_part1:114227778|114230210; 13: chr6A_part1:212312197|212313644. Arrow indicates the back-splicing site.
2.3. Experimental Validation of cicrRNAs
For the further validation of circRNAs, divergent primers (Table S2) were designed to carry out the divergent polymerase chain reaction (PCR) [10]. As shown in Figure 3E, the PCR products spanning the back-splicing sites with the designed length were successfully amplified. The Sanger sequencing results further demonstrated the back-splicing sites and validated the existence of identified wheat circRNAs.
3. Methods
3.1. Plant Treatment and Materials Collection
Wheat (Triticum aestivum L. cultivar ‘Yangmai 158’, preserved by Lixiahe Institute of Agriculture Sciences, Jiangsu, China) was normally grown in the field for 160 days. When the wheat plants grew in the flowering period, the spikelet was inoculated with a conidia suspension according to methods reported by Zhang et al. [11]. Briefly, the F. graminearum strain, PH-1 (preserved by Lixiahe Institute of Agriculture Sciences), was cultured in a PDA (potato dextrose agar) medium for 3–4 days, then the vigorous hyphae growing at the edge of the colony were transferred into 3% sterile mung and cultured at 25 °C at 180 RPM for 3 days. The conidia were collected by centrifugation, suspended with sterile water (1.0 × 107 spores/mL), and injected into spikelets. The spikelet tissues were collected after inoculation for 1, 3, and 5 days, and a spikelet without inoculation was used as a control. Each sample contained three biological replications. The samples were immediately frozen in liquid nitrogen and preserved at −80 °C until they were used.
3.2. RNA Isolation, Library Preparation, and Sequencing
Total RNA was extracted using a Trizol reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. The quality and quantity of the total RNA extracted from the control and disease wheat spikelets were firstly determined by a 1% agarose gel and Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The RNA integrity number (RIN) of the total RNA was further visually evaluated by using the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The total RNA was digested with a Ribo-Zero rRNA Removal Kit (Epicentre, Madison, WI, USA) to obtain rRNA-depleted RNA, which was then digested with RNase R (Epicentre, Madison, WI, USA) to remove the linear RNAs. The remaining RNA was used for the library construction using the NEB Next®Ultra™ Directional RNA Library Prep Kit (NEB, Ipswich, MA, USA). The strand-specific pair-end 150 bp reads were generated using the Illumina Xten platform at Biomarker Technologies Co., Ltd. (Beijing, China).
3.3. CircRNA Prediction and Annotation
To guarantee the quality of the sequencing data, the raw reads were trimmed using Cutadapt (v1.9) with a quality score cutoff of 30 and a minimum length of 30 bp [12]. The remaining 1.04 billion high-quality reads were used to predict circRNA (Table 1). In order to detect the reliable circRNAs, we used three identification tools: CIRI2 [13], which employs the BWA [14] as an external aligner; find_circ [15], employing bowtie2 [16] as an aligner; CIRCexplorer2 [17], employing STAR [18] as an aligner (Figure 1). The default parameters were used during the circRNA detection. We sought to generate a high-confidence set of circRNAs expressed in wheat with the following rules: (1) supported by at least two tools; (2) at least two junction reads spanning the back-splicing site. The expression levels of reliable circRNAs were normalized as SRPBM (SRPBM = number of circular reads/number of mapped reads (units in billion)/read length). Several commonly used databases, including Nr, GO, KEGG, KOG/COG, Swiss-Prot, and Pfam, were used to perform circRNAs function annotation [10]. A Veen diagram was drawn using an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/, accessed on 1 August 2022). The circos and violin diagrams were drawn using the correspondence R packages [7]. Data were analyzed using the R v4.1.2 statistical package (https://cran.r-project.org, accessed on 1 August 2022). The circlize v0.4.14 [19], ggplot2 v3.3.5 [20], and vioplot v0.3.7 (https://github.com/TomKellyGenetics/vioplot, accessed on 1 August 2022) packages were used to draw the circular, histogram, and violin plots.
3.4. CircRNA Validation
For circRNAs validation, total RNA was extracted from the wheat spikelet tissues using the TRizol reagent (Invitrogen, Carlsbad, CA, USA), and then they were subjected to a reverse transcription reaction with the PrimeScritpt RT reagent Kit (Vazyme, Nanjing, China) to synthesize the first cDNA strand [21]. The divergent PCR primers were designed according to the “out-facing” strategy to ensure the amplifications were from the circle template [22]. The primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Sanger sequencing was performed by BGI Co., Ltd. (Wuhan, Hubei, China) to validate the presence of the back-spliced junction sites. The primers for circRNA validation were designed using the Primer Premier 5 software (PREMIER Biosoft International, Palo Alto, CA, USA) and are listed in Supplementary Materials Table S2 [23].
4. Conclusions
In this study, circRNA-seq was used to determine the changes in circRNAs among the control (CK) and 1, 3, and 5 days post-Fusarium graminearum inoculation (dpi) samples. In total, 2091 high-confidence circRNAs were detected. These circRNAs showed distinct expression patterns among the CK, 1dpi, 3dpi and 5dpi samples. They were widely scattered across all chromosomes, and the longer chromosomes produced more circRNAs. The wheat circRNAs were unevenly distributed, and more circRNAs were commonly found at the centre and both ends of the chromosomes. Most circRNAs originated from the exon. Our results provide valuable clues for identifying F. graminearum infection-responsive circRNAs and lay the foundation for further functional characterization of wheat circRNA participating in the Fusarium head blight disease response.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/data7090121/s1. Table S1: Detailed information on circRNAs; Table S2: Primers for circRNA validation.
Author Contributions
Conceptualization, J.Y., D.M., D.G., and Y.Z.; Methodology, J.Y.; Validation, J.Y., D.M., and D.G.; Formal Analysis, J.Y.; Investigation, J.Y., X.H., D.M., Z.F., and D.G.; Resources, D.G.; Writing—Original Draft, J.Y.; Writing, J.Y.; Review and Editing, J.Y., D.M., Z.F., D.G., and Y.Z.; Funding Acquisition, D.M. and D.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Seed Industry Revitalization Project of Jiangsu Province, grant number JBGS(2021)047. The APC was funded by JBGS(2021)047.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yin, J.; Yan, J.; Hou, L.; Jiang, L.; Xian, W.; Guo, Q. Identification and functional deciphering suggested the regulatory roles of long intergenic ncRNAs (lincRNAs) in increasing grafting pepper resistance to Phytophthora capsici. BMC Genom. 2021, 22, 868. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Lu, W.; Zhong, C.; Zhou, R.; Xu, J.; Liu, W.; Gou, X.; Wang, Q.; Yin, J.; Xu, C.; et al. The 25–26 nt small RNAs in Phytophthora parasitica are associated with efficient silencing of homologous endogenous genes. Front. Microbiol. 2017, 8, 773. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. Piwi-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, T.; Xu, K.; Zhang, W.; Wang, X.; Quan, J.; Jin, W.; Zhang, M.; Fan, G.; Wang, M.-B.; et al. The tRNA-derived small RNAs regulate gene expression through triggering sequence-specific degradation of target transcripts in the oomycete pathogen Phytophthora sojae. Front. Plant Sci. 2016, 7, 1938. [Google Scholar] [CrossRef]
- Zhu, Y.-X.; Gong, H.-J.; Yin, J.-L. Role of silicon in mediating salt tolerance in plants: A review. Plants 2019, 8, 147. [Google Scholar] [CrossRef]
- Hutchins, E.; Reiman, R.; Winarta, J.; Beecroft, T.; Richholt, R.; De Both, M.; Shahbander, K.; Carlson, E.; Janss, A.; Siniard, A.; et al. Extracellular circular RNA profiles in plasma and urine of healthy, male college athletes. Sci. Data 2021, 8, 276. [Google Scholar] [CrossRef]
- Yin, J.; Liu, Y.; Lu, L.; Zhang, J.; Chen, S.; Wang, B. Comparison of tolerant and susceptible cultivars revealed the roles of circular RNAs in rice responding to salt stress. Plant Growth Regul. 2022, 96, 243–254. [Google Scholar] [CrossRef]
- Song, Y.; Bu, C.; Chen, P.; Liu, P.; Zhang, D. Miniature inverted repeat transposable elements cis-regulate circular RNA expression and promote ethylene biosynthesis, reducing heat tolerance in Populus tomentosa. J. Exp. Bot. 2021, 72, 1978–1994. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. Multiqc: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Zhu, Y.; Jia, J.; Yang, L.; Xia, Y.; Zhang, H.-L.; Jia, J.-B.; Zhou, R.; Nie, P.; Yin, J.; Ma, D.; et al. Identification of cucumber circular RNAs responsive to salt stress. BMC Plant Biol. 2019, 19, 164. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Zhu, Y.; Ma, D.; Xu, W.; Zhou, J.; Yan, H.; Yang, L.; Yin, J. Screening, identification, and optimization of fermentation conditions of an antagonistic endophyte to wheat head blight. Agronomy 2019, 9, 476. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, J.; Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2017, 1, 8. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Zhang, X.-O.; Dong, R.; Zhang, Y.; Zhang, J.-L.; Luo, Z.; Zhang, J.; Chen, L.-L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Ginestet, C. Ggplot2: Elegant graphics for data analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Xiao, Y.; Wen, Y.; Li, K.; Ma, Z.; Yang, L.; Zhu, Y.; Yin, J. Genome-wide characterization and function analysis uncovered roles of wheat LIMs in responding to adverse stresses and TaLIM8-4d function as a susceptible gene. Plant Genome 2022, 16, e20246. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zhu, Y.X.; Zhao, J.; Fang, Z.W.; Wang, S.P.; Yin, J.L.; Chu, Z.H.; Ma, D.F. Transcriptome-wide identification and characterization of potato circular RNAs in response to Pectobacterium carotovorum subspecies Brasiliense infection. Int. J. Mol. Sci. 2018, 19, 71. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Jiang, X.; Zhang, J.; He, Y.; Zhu, X.; Zhou, X.; Gong, H.; Yin, J.; Liu, Y. Silicon confers cucumber resistance to salinity stress through regulation of proline and cytokinins. Plant Physiol. Biochem. 2020, 156, 209–220. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).