Abstract
This article outlines a novel data descriptor that provides the Arabic natural language processing community with a dataset dedicated to named entity recognition tasks for diseases. The dataset comprises more than 60 thousand words, which were annotated manually by two independent annotators using the inside–outside (IO) annotation scheme. To ensure the reliability of the annotation process, the inter-annotator agreements rate was calculated, and it scored 95.14%. Due to the lack of research efforts in the literature dedicated to studying Arabic multi-annotation schemes, a distinguishing and a novel aspect of this dataset is the inclusion of six more annotation schemes that will bridge the gap by allowing researchers to explore and compare the effects of these schemes on the performance of the Arabic named entity recognizers. These annotation schemes are IOE, IOB, BIES, IOBES, IE, and BI. Additionally, five linguistic features, including part-of-speech tags, stopwords, gazetteers, lexical markers, and the presence of the definite article, are provided for each record in the dataset.
Dataset: 10.5281/zenodo.3926432
Dataset License: CC-BY
1. Summary
Named entity recognition (NER) is a prominent subfield of natural language processing (NLP). The objective of NER is to recognize specific and predefined entities in a text. In the last decade, Arabic NER has gained considerable interest and focus from the research community due to the popularity of the language, as it is the native tongue for more than 325 million people [1]. While a substantial amount of research work has been dedicated to different domains, such as recognizing people’s names, locations, crimes, organization names, and so on, few research studies have been dedicated to the medical domain. This shortcoming can be attributed to the lack of digital Arabic resources, such as datasets [2,3].
While there are fair amounts of research efforts studying multi-annotation schemes for the task of NER in languages such as English, Spanish, Dutch, Czech [4], Greek [5], Russian [6], and Punjabi [7], the Arabic language suffers from a lack of efforts in this domain. This work is an attempt to rectify this shortcoming by providing the Arabic NLP research community with dataset designated for NER tasks in the medical domain.
The dataset as supplementary was annotated independently by two annotators and the rate of the inter-agreement score was calculated. Another contribution of this work is providing this dataset with different annotation schemes, which will allow the discovery of the effect of using different annotation schemes on NER tasks. Seven well-known annotation schemes were used to annotate the dataset. These schemes are IO, IOE, IOB, BIES, IOBES, IE, and BI.
2. Data Description
This dataset consists of 62,506 records, and each record represents a single word/token from our corpus. Each word is described in terms of six features/columns, which are annotation labels, part-of-speech tags, stopwords, gazetteers, lexical markers, definiteness. Each column is described as follows:
2.1. Annotation Labels
This column determines whether the word of interest is a disease entity or not. Two labels were used to annotate the words. The label I is used to tag disease entities, whereas the label O is used to tag irrelevant words. This annotation mechanism is well known in the literature and is referred to as the IO annotation scheme. However, other annotation schemes are used in the literature, and each has advantages and disadvantages. In this work, we annotated our data using seven different annotations schemes, resulting in seven files, each corresponding to a unique annotation scheme. Further details regarding the annotation process and schemes are given in Section 3.3. In addition, a sample sentence of the dataset is presented with each annotation scheme in Figure 1. The literal translation of the sample sentence is “Leukemia (White blood cells cancer) is considered one of the most common kinds”. It is worth noting that the Arabic language is written from right to left. Therefore, the annotation tags are ordered accordingly.
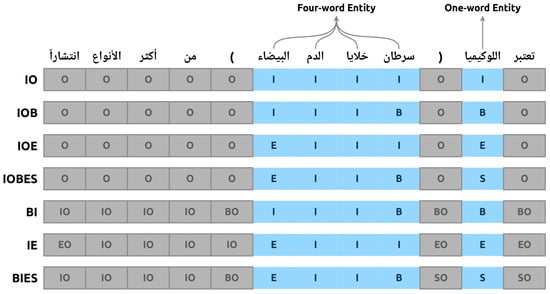
Figure 1.
A sample sentense annotated with seven annotation schemes.
2.2. Part-of-Speech Tags
This column represents the part-of-speech (POS) tags of each word. The POS tags are labels assigned to a word to identify its part of speech (e.g., noun, pronoun, verb, etc.) in a given context. The POS tags are prevalently used in NER tasks due to their ability to reveal the grammatical structure of the sentence. Furthermore, according to [8], a strong correlation exists between POS tags and NER for the Arabic language.
2.3. Stopwords
This column indicates whether the word of interest exists in the predefined list of stopwords. Stopwords are less informative regarding the given task. Usually, the stopwords are primarily conjunctions, prepositions, pronouns, demonstratives, and so on [9]. In the natural language processing literature, lists of stopwords are commonly used for several tasks, including NER tasks. The dataset includes a list of 198 stopwords that have been used in this work.
2.4. Gazetteers
This column specifies whether the given word is listed in the disease entity gazetteer, which is a dictionary that collects frequently used entities. Gazetteers play a significant role in improving the performance of NE recognizers [10]. However, creating gazetteers from scratch can be a challenging and time-consuming process [11], even though using gazetteers in NER usually improves the precision at the expense of recall.
2.5. Lexical Marker Lists
This column determines whether the word exists in the lexical marker list. Lexical markers, also known as lexical triggers, are words or parts of a word that usually exist in the vicinity of the named entity and can help to recognize an entity. Analyzing the context of NEs can reveal the existence of such markers [12].
2.6. Definiteness (Existence of ‘AL’)
This column features the presence of the definite article ال at the beginning of each word. This article can be translated directly to mean “the” in English. However, in Arabic, this article appears as a prefix for nouns. The column has four possible values, as shown in Table 1.

Table 1.
The possible values for the definiteness column.
3. Methods
This section describes the main methods used to generate the dataset in its final form. The process includes collecting and preprocessing the data, data labeling, and feature engineering.
3.1. Data Collection
King Abdullah Bin Abdulaziz Arabic Health Encyclopedia (KAAHE) [13] was the source for building this dataset. This encyclopedia is considered a reliable provider of health information. The administration and finance of KAAHE are provided by the Ministry of National Guard Health Affairs and King Saud bin Abdulaziz University for Health Sciences in Saudi Arabia. It follows the Executive Regulations of the Electronic Publishing Activity of the Ministry of Media in Saudi Arabia. The content of KAAHE was originally provided by UK National Health Services (NHS). The data consist of 27 Arabic medical articles, totaling around 50,000 words.
3.2. Data Preprocessing
To prepare our raw dataset for further processing, a crucial step to consider is data preprocessing. This step includes data cleansing and tokenization. During the data cleansing step, irrelevant information in the articles, such as hypertext links, images, and so on, was excluded from the dataset. Only the text of interest was included in the body of the dataset. Afterwards, the data were imported into the AMIRA tool [14] to be tokenized, which is a natural language tool devoted to the Arabic language and provides several NLP functionalities, such as the POS tagger, clitic tokenizer, and base phrase chunker. Moreover, AMIRA has several profiles to carry out the tokenization process. In our dataset, all prefixes except the definite article ال were tokenized. The definite article was exempted from tokenization because it is used in later stages to engineer the definiteness column in the final dataset. After the tokenization step, the dataset size increased from around 50,000 words to around 62,500 tokens.
3.3. Data Annotation
Data annotation is the process of tagging the data into predefined categories. It is an important process, especially for supervised learning, and can be done for different types of datasets, such as image, video, and textual datasets. Data annotation plays a crucial role in evaluating the performance of supervised models because they provide ground-truth target labels. According to [15], the process of annotating any dataset should be conducted by at least two independent annotators to make it possible to validate the reliability of the annotation process. Several statistical metrics are used to measure the reliability of annotator labeling, and a well-known measure is Cohen’s kappa metric [16].
Our dataset was annotated independently by two annotators, and each word is classified as either a disease entity or otherwise. To check the reliability of the annotation process, we adopted the Cohen’s kappa statistic. The score of Cohen’s kappa was 95.14%, which indicates a high agreement between the annotators. Given that, many researchers in the literature have argued that the minimum acceptable Cohen’s kappa score is 80% [17]. While the agreement score is high, the researcher analyzed the differences between the two annotated datasets and found that the disparity can primarily be attributed to classifying adjectives in diseases. For example, in the sentence 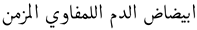 , which is translated as “Chronic Lymphocytic Leukemia,” the adjective chronic was considered part of the NE by the first annotator, while the second annotator decided to exclude it. In the NER literature, several well-known annotation schemes were used to perform the annotation task. We decided to annotate our dataset using seven frequently used annotation schemes, which are listed and described in Table 2.
, which is translated as “Chronic Lymphocytic Leukemia,” the adjective chronic was considered part of the NE by the first annotator, while the second annotator decided to exclude it. In the NER literature, several well-known annotation schemes were used to perform the annotation task. We decided to annotate our dataset using seven frequently used annotation schemes, which are listed and described in Table 2.
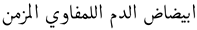 , which is translated as “Chronic Lymphocytic Leukemia,” the adjective chronic was considered part of the NE by the first annotator, while the second annotator decided to exclude it. In the NER literature, several well-known annotation schemes were used to perform the annotation task. We decided to annotate our dataset using seven frequently used annotation schemes, which are listed and described in Table 2.
, which is translated as “Chronic Lymphocytic Leukemia,” the adjective chronic was considered part of the NE by the first annotator, while the second annotator decided to exclude it. In the NER literature, several well-known annotation schemes were used to perform the annotation task. We decided to annotate our dataset using seven frequently used annotation schemes, which are listed and described in Table 2.
Table 2.
A description of annotation schemes and their tags.
The process of annotating our datasets using the aforementioned schemes is illustrated in Figure 2. The annotation step was performed based on the preprocessed data. At the beginning, each annotator independently labeled the dataset manually using the IO annotation scheme due to its simplicity. However, the IO scheme cannot determine the boundaries of consecutive entities. Therefore, the data were annotated automatically using the IOB scheme except for the consecutive entities that the annotators marked manually. Afterwards, the annotation process was automatically performed for the rest of the schemes using a Python script especially developed for this purpose. The script relies on identifying the named entities boundaries to be able to generate the subsequent annotation schemes. Based on the specific rules designated for each annotation scheme, the script generates the required scheme. For example, the code listing shown in Listing 1 highlights some of the important rules and steps performed for generating the IOBES annotation scheme.
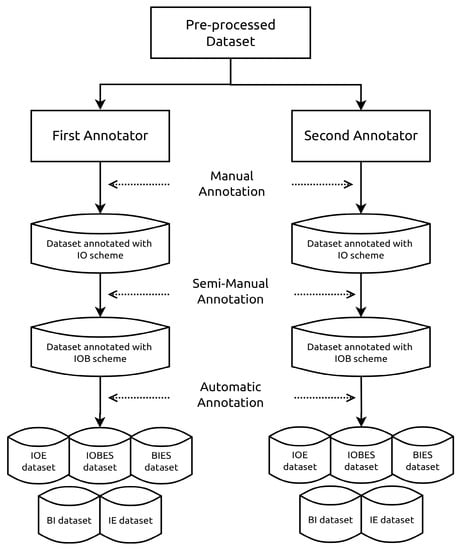
Figure 2.
The dataset annotation process.
| Listing 1: Code snippet of the IOBES scheme script. |
def generate_IOBES(dataset): # make a new copy of the new_dataset = dataset.copy() # loop over every record in the new dataset for i, row in enumerate(new_dataset): # Check if the current token is a single entity if ( row[LABEL] == "I" and dataset.iloc[i - 1][LABEL] == "O" and dataset.iloc[i + 1][LABEL] == "O" ): new_dataset.at[i, LABEL] = "S" # check if the current token is the beginning of a multi-token entity if row[LABEL] == "I" and dataset.iloc[i - 1][LABEL] == "O": new_dataset.at[i, LABEL] = "B" # check if the current token is the beginning of a multi-token entity if row[LABEL] == "I" and dataset.iloc[i + 1][LABEL] == "O": new_dataset.at[i, LABEL] = "E" # return the newly generated dataset return new_dataset |
3.4. Feature Engineering
In this section, we describe the method used to derive the features/columns that were mentioned in Section 2. The MADAMIRA tool was used to obtain the POS tags and the definiteness columns. MADAMIRA [18] is an Arabic morphological analyzer developed by combining two previous NLP tools: MADA [19] and AMIRA [14].
Regarding the lexical markers, stopwords, and gazetteers columns, several statistical methods were used to analyze the dataset, such as frequency, concordance, and n-gram analyses. These methods are essential to exploring and understanding the context in which the entities exist. Therefore, these tools allowed us to derive these lexical marks, stopwords, and gazetteers.
Frequency analysis is considered one of the most prominent techniques that is used to study any corpora. Analyzing the words in the dataset based on their frequency can indicate the most important keywords present in the dataset. However, the most frequent words are considered stopwords which are less informative. We implemented this technique to derive the stopwords list used in this work. After that, we removed the stopwords from the list which revealed the informative keywords in this domain and assisted in the gazetteers creation process. Then, the concordance analysis was carried out based on these keywords. The concordance analysis allows us to explore the context of a given word/named entity and the structure in which it frequently appears in. It aided in the extraction of the most common verbs, nouns that appear in the surrounding context which have the potential of being a lexical marker. Another statistical technique that was used in this work is n-gram analysis. The probability of a given set of words appearing together in the text, is the basis idea in n-gram analysis. This technique has several applications even outside of NLP and it gives us an additional perspective to understand and discover the structure of the text. This technique was helpful in this work specifically for recognizing disease entities as they are usually composed of more than one word appearing with each other.
Supplementary Materials
The following are available at https://www.mdpi.com/2306-5729/5/3/60/s1.
Author Contributions
Conceptualization, N.A. and S.A.; methodology, N.A. and S.A.; software, N.A.; resources, S.A.; writing—original draft preparation, N.A. and S.A.; writing—review and editing, N.A. and S.A.; visualization, N.A. and S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- McLoughlin, L. Colloquial Arabic (Levantine); Routledge: London, UK, 2009. [Google Scholar]
- Alanazi, S. A Named Entity Recognition System Applied to Arabic Text in the Medical Domain. Ph.D. Thesis, Staffordshire University, Stoke-on-Trent, UK, 2017. [Google Scholar]
- Shaalan, K.; Raza, H. Arabic named entity recognition from diverse text types. In International Conference on Natural Language Processing; Springer: New York, NY, USA, 2008; pp. 440–451. [Google Scholar]
- Konkol, M.; Konopík, M. Segment representations in named entity recognition. In International Conference on Text, Speech, and Dialogue; Springer: New York, NY, USA, 2015; pp. 61–70. [Google Scholar]
- Demiros, I.; Boutsis, S.; Giouli, V.; Liakata, M.; Papageorgiou, H.; Piperidis, S. Named Entity Recognition in Greek Texts. Ph.D. Thesis, Aristotle University of Thessaloniki, Thessaloniki, Greece, 2019. [Google Scholar]
- Mozharova, V.A.; Loukachevitch, N.V. Combining knowledge and CRF-based approach to named entity recognition in Russian. In International Conference on Analysis of Images, Social Networks and Texts; Springer: Cham, Switzerland, 2016; pp. 185–195. [Google Scholar]
- Ahmad, M.T.; Malik, M.K.; Shahzad, K.; Aslam, F.; Iqbal, A.; Nawaz, Z.; Bukhari, F. Named Entity Recognition and Classification for Punjabi Shahmukhi. ACM Trans. Asian Low-Resour. Lang. Inform. Process. (TALLIP) 2020, 19, 1–13. [Google Scholar] [CrossRef]
- Algahtani, S.M. Arabic Named Entity Recognition: A Corpus-Based Study. Ph.D. Thesis, University of Manchester, Manchester, UK, 2012. [Google Scholar]
- Elsebai, A.; Meziane, F.; Belkredim, F.Z. A rule based persons names Arabic extraction system. Commun. IBIMA 2009, 11, 53–59. [Google Scholar]
- Torisawa, K. Exploiting Wikipedia as external knowledge for named entity recognition. In Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL), Prague, Czech Republic, 28–30 June 2007; pp. 698–707. [Google Scholar]
- Alruily, M. Using Text Mining to Identify Crime Patterns from Arabic Crime News Report Corpus; De Montfort University: Leicester, UK, 2012. [Google Scholar]
- Shaalan, K. A survey of arabic named entity recognition and classification. Comput. Linguist. 2014, 40, 469–510. [Google Scholar] [CrossRef]
- King Abdullah Bin Abdulaziz Arabic Health Encyclopedia. Available online: https://kaahe.org/ (accessed on 30 April 2020).
- Diab, M. Second generation AMIRA tools for Arabic processing: Fast and robust tokenization, POS tagging, and base phrase chunking. In Proceedings of the 2nd International Conference on Arabic Language Resources and Tools (MEDAR), Cairo, Egypt, 22–23 April 2009; p. 198. [Google Scholar]
- Hovy, E.; Lavid, J. Towards a ‘science’ of corpus annotation: A new methodological challenge for corpus linguistics. Int. J. Transl. 2010, 22, 13–36. [Google Scholar]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Pasha, A.; Al-Badrashiny, M.; Diab, M.T.; El Kholy, A.; Eskander, R.; Habash, N.; Pooleery, M.; Rambow, O.; Roth, R. Madamira: A fast, comprehensive tool for morphological analysis and disambiguation of arabic. LREC 2014, 14, 1094–1101. [Google Scholar]
- Habash, N.; Rambow, O.; Roth, R. MADA+ TOKAN: A toolkit for Arabic tokenization, diacritization, morphological disambiguation, POS tagging, stemming and lemmatization. In Proceedings of the 2nd International Conference on Arabic Language Resources and Tools (MEDAR), Cairo, Egypt, 22–23 April 2009; p. 62. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).