Abstract
MicroRNAs (miRNAs) have emerged as a potential class of biomolecules for diagnostic biomarker applications. miRNAs are small non-coding RNA molecules, produced and released by cells in response to various stimuli, that demonstrate remarkable stability in a wide range of biological fluids, in extreme pH fluctuations, and after multiple freeze–thaw cycles. Given these advantages, identification of miRNA-based biomarkers for radiation exposures can contribute to the development of reliable biological dosimetry methods, especially for low-dose radiation (LDR) exposures. In this study, an miRNAome next-generation sequencing (NGS) approach was utilized to identify novel radiation-induced miRNA gene changes within the CGL1 human cell line. Here, irradiations of 10, 100, and 1000 mGy were performed and the samples were collected 1, 6, and 24 h post-irradiation. Corroboration of the miRNAome results with RT-qPCR verification confirmed the identification of numerous radiation-induced miRNA expression changes at all doses assessed. Further evaluation of select radiation-induced miRNAs, including miR-1228-3p and miR-758-5p, as well as their downstream mRNA targets, Ube2d2, Ppp2r2d, and Id2, demonstrated significantly dysregulated reciprocal expression patterns. Further evaluation is needed to determine whether the candidate miRNA biomarkers identified in this study can serve as suitable targets for radiation biodosimetry applications.
Keywords:
miRNA; epigenetics; low-dose radiation; next-generation sequencing; miRNAome; CGL1; biomarkers 1. Introduction
Biodosimetry is the measurement of biological response to radiation. The gold standard in the field of biological dosimetry is the dicentric chromosome assay (DCA), which measures the incidence of dicentric chromosomes [1]. DCA has a predicting error of ±0.5 Gy which is adequate for high-dose radiation (HDR) exposures [2]. However, DCA is unreliable in the low-dose radiation (LDR) range (below 100 mGy) [2]. The cytokinesis-block micronucleus (CBMN) assay is also used for biodosimetry applications. The CBMN assay assesses the formation of micronuclei in binucleated cells derived from peripheral lymphocytes [3]. These micronuclei are formed when chromosomal structures are damaged, forming chromosome fragments that segregate separately during cell division. Unfortunately, both DCA and CBMN are low-throughput methodologies and are ineffective for estimating LDR exposures [4]. This emphasizes the need for new biodosimetry techniques that are reliable, sensitive, high-throughput, and minimally invasive.
microRNAs (miRNAs) are becoming more prevalent within the biomarker field as these biomolecules can be accurately analyzed to predict prognosis while being easily accessible due the fact of its abundance within patient serum [5]. miRNAs also demonstrate remarkable stability in a wide range of biological fluids [6]. In addition, miRNAs are resistant to multiple freeze–thaw cycles and extreme pH fluctuations; therefore, samples can be readily transported [7]. As such, these traits make miRNA key molecules when looking for specific biological markers for radiation exposures.
miRNAs are small non-protein coding RNA molecules approximately 20 nucleotides in length that act to post-transcriptionally modulate the gene expression profile of cells [8]. Two miRNAs with different cellular targets result per miRNA gene and are denoted with either 3p or 5p to indicate their origin. One of these two mature miRNA strands is then bound to Argonaute (AGO), forming a ribonucleoprotein complex known as the RNA-induced silencing complex (RISC) [9]. This complex uses the miRNA to bind to the complementary mRNA target and the AGO protein cleaves the mRNA strand. Despite having two viable miRNAs per gene, 3p and 5p, one of the species is usually more frequently integrated into the RISC complex than the other [10]. This more frequently integrated species is dependent on the particular gene itself and does not relate directly to the 3p or 5p end [11]. Taken together, complementary binding of miRNA to target mRNA prevents its translation and, therefore, reduces downstream protein expression.
Numerous studies have demonstrated that HDR exposure significantly alters miRNA profiles, resulting in downstream changes to various radiation-related molecular pathways including DNA damage repair, apoptosis, and cell cycle arrest [12,13,14]. Overall, the vast majority of the research on radiation and miRNA involves studies examining radiation therapy effects on cancer treatment and are thus HDR focused [15,16,17]. Another limitation of these studies is the lack of next-generation sequencing (NGS)-based miRNAome technology within the context of radiation-induced miRNA profiling [18,19,20]. While many articles investigated the interaction of one or more miRNAs, this selective approach can potentially introduce bias. NGS-based miRNAome profiling would ensure a more comprehensive investigation of the biological system and potentially lead to previously missed novel miRNA interactions resulting from the more focused approach. Taken together, further research is needed to comprehensively profile dose-dependent radiation-induced miRNA that can serve as candidate biomarkers for biodosimetry applications.
The primary aim of this study was to identify novel radiation-induced miRNA biomarkers at various dose ranges spanning LDR and HDR exposures. In this study, the entire miRNAome of the CGL1 cells were analyzed post-radiation exposure to identify dose-dependent radiation-induced miRNA expression changes. The CGL1 cell line is a preneoplastic nontumorigenic model resulting from the hybridization of a normal male skin GM0077 fibroblast cell with the malignant HeLa cervical cancer cell [21,22]. The CGL1 cells are nontumorigenic and demonstrate a normal fibroblast-like phenotype and transcriptome profile [23]. The CGL1 cell model system has been used extensively to investigate the effects of radiation-induced cellular transformation and tumorigenicity [24].
2. Materials and Methods
2.1. Cell Culture
The CGL1 cell line was grown in a humid environment at 37 °C with 5% CO2 [21]. The CGL1 cells were cultured using 1X Minimal Essential media (Corning, Manassas, VA, USA) with the addition of 50 U penicillin–streptomycin (GIBCO, Grand Island, NY, USA) and 5% calf serum (Hyclone, Logan, UT, USA).
2.2. Cell Irradiation
The CGL1 cells were exposed to X-rays in triplicate at 10, 100, or 1000 mGy and collected at 1, 6, and 24 h post-irradiation. Irradiations were performed on an X-RAD 320 irradiation cabinet (Precision X-ray, Madison, CT, USA) operated at 320 kV and 12.5 mA with a 2 mm Al filter. Briefly, cells were grown in a T25 flask (Life Technologies, Carlsbad, CA, USA) and plated to ensure 80% confluence at the desired endpoint. These sample sets consisted of three different timepoints post-irradiation (i.e., 1, 6, and 24 h) to assess the time-dependent expression of the various radiation-induced miRNA. These timepoints allowed for the analysis of acute (1 h), intermediate (6 h), and prolonged effects (24 h). At each of these timepoints, the cells were split into 4 different irradiation groups: the sham irradiation control and 10, 100, and 1000 mGy. This range of dose was selected to evaluate LDR effects (10 and 100 mGy) and a higher dose of radiation (1000 mGy), which is well known to induce DNA damage effects. These samples were irradiated on ice with cold PBS to ensure that no-dose rate effects were observed. The cells were irradiated using three different programs to ensure similar exposure times among the different doses. In short, the 10 mGy dose was irradiated at a dose rate of 5.6 mGy/min, the 100 mGy dose was irradiated at a dose rate of 138 mGy/min, and the 1000 mGy dose was irradiated at a dose rate of 1500 mGy/min. Following irradiation, the samples were re-incubated in media until the desired timepoint, at which point the cells were lysed and collected using 1 mL TRIzol per T-25 flask.
2.3. Total RNA Extraction
Total RNA was extracted using the TRIzol extraction method according to the manufacturer’s instructions. In brief, 0.2 mL of chloroform was added to 1 mL TRIZOL samples and vortexed prior to being centrifuged at 12,000× g for 20 min. The top aqueous layer containing the RNA was transferred into another tube, while the remaining DNA and protein layer were stored for future use. The RNA was precipitated out of the aqueous layer with the addition of 0.3 mL of isopropanol and centrifuged at 12,000× g for 15 min. Afterwards, the supernatant was decanted, and the remaining RNA pellet was washed with 1 mL of 70% ethanol and centrifuged at 7500× g for 5 min. Finally, the supernatant was removed, and the resulting RNA pellet was resuspended in 30 µL of DEPC water. The resulting total RNA samples were assessed to ensure both an adequate concentration and purity using a Nanodrop (Nanodrop One, Thermo Fisher Scientific, Madison, WI, USA), assuring the 260/280 nm absorbance ratio of the samples was greater than 1.8.
2.4. cDNA Synthesis
From the total RNA samples, one of two reverse transcription reactions were performed depending on whether mRNA or miRNA was to be analyzed. In brief, total RNA was first purified of DNA contaminants using a DNAse kit (MilliporeSigma, Oakville, ON, CA) before proceeding to reverse transcription reactions. The key difference between mRNA and miRNA cDNA preparation was that for the analysis of miRNAs, the samples were first subjected to a poly-A tail extension and annealed to an oligo-dT primer with a known sequence at the 3′ end (5′-GCATAGACCTGAATGGCGGTAAGGGTGTGGTAGGCGAGACATTTTTTTTTTTTTTTTTTTT-3′). This additional step before the cDNA synthesis serves to elongate the miRNA and produce a suitable nucleotide length for RT-qPCR analysis, without which RT-qPCR would have been otherwise impossible, as the miRNA are too similar in length to design RT-qPCR primers. For preparation of cDNA from mRNA, random hexamers were used. The resulting products were reverse transcribed using the Promega M-MLV Reverse Transcriptase kit (Madison, WI, USA).
2.5. miRNAome Profiling via Next-Generation Sequencing
An NGS analysis was conducted on the irradiated samples; however, based on various studies in the literature, 6 h post-stimulation was found to be an optimal timepoint, where most miRNA are expected to be expressed and, therefore, of most importance [25]. As such, the 6 h timepoint RNA samples were chosen for miRNAome analysis. The remaining two timepoints, 1 and 24 h, were analyzed subsequently using RT-qPCR and compared to the 6 h results. This allowed for the determination of the time-course for the expression of the dysregulated miRNA. For the sample preparation, the irradiated total RNA samples were treated with the QIAseq miRNA Library kit (QIAGEN Sciences, Germantown, MD, USA), which serves to select smaller RNA within a sample and prepare these RNA sequences for miRNAome profiling. In brief, following the collection of total RNA using the method described earlier, two ligation reactions were accomplished to add adapter sequences to the 5′ and 3′ end of the miRNA. A reverse transcription reaction integrating the UMI adaptors into the resulting cDNA was completed. Finally, a barcode index was added to the end of each of the sample conditions. Samples were then pooled together and sent out for sequencing at the Donnelly Sequencing Centre at the University of Toronto. The samples were sequenced on the NovoSeq6000 platform (Illumina) and consisted of a 75-base length read for a total of 10 million reads per sample. Previous reports have shown that 5 million sequencing reads per sample is adequate for total human miRNAome profiling [26].
2.6. RT-qPCR Primer Design and Validation
Despite the NGS methodology being an excellent technique capable of whole miRNAome profiling, false positives can occur based on statistical probability as well as other contributing factors such as library amplification and sequencing bias [27]. As such, a common practice to verify the integrity of the obtained sequencing results is to utilize a secondary confirmatory technique on randomly selected miRNA targets. Here, randomly selected miRNAs were cross-verified using RT-qPCR analysis.
RT-qPCR primers were designed in-house using the primer blast software offered by the National Center for Biotechnology Information (NCBI). For the design of the mRNA primers, the full-length mRNA transcript sequences were used [28]. However, for the miRNA primers, a modified input sequence was used consisting of the miRNA sequence appended to the oligodT adapter sequence.
The potential primers were screened for the combination of melting temperature, GC content, and lack of self-complementarity to obtain the optimal primer pairs for each gene. These primers were verified across a range of temperatures between 54 and 64 °C and run at various concentrations to ensure their amplification was optimal. The primers for the mRNA gene expression analysis were considered valid if their amplification reaction efficiency was uniform, had an R2 value of >0.99, and an amplification efficiency of 90–110%. For the miRNA, the amplification efficiency was relaxed to 70–130% in order to provide more flexibility than usual due to the limitations when designing miRNA primers; these miRNA primers usually correspond to the miRNA sequence itself with minimal design flexibility. The miRNA cDNA products were qPCR amplified using a universal reverse primer for the known end sequence (5′-GCATAGACCTGAATGGCGGTA-3) common to all of the modified miRNA and a forward primer specific to the miRNA in question. Validated primer information can be found in the appendix (Table A1 for miRNA and Table A2 for mRNA).
2.7. RT-qPCR
The RT-qPCR procedure was performed in 15 µL reaction volume ensuring a final volume of 1X qPCR master mix (LUNA, New England Biolabs, Ipswich, MA, USA) and 600 nM for both primers. The protocol was repeated for 40 cycles: 95 °C for 15 s and 60 °C for 30 s, after which the fluorescence was measured. A melt curve was performed at the end of the 40-cycle run to ensure that the resulting amplicon was unique. Once complete and the raw fluorescence data were obtained, the cycle threshold (Cq) data were analyzed using the QuantStudioTM Design and Analysis Software v1.5.1 (Applied Biosystems). Samples were normalized to the geometric mean of two control housekeeping genes: RPS18/GAPDH for mRNA and SNORD48/U6 for miRNA. The relative expression of the genes was calculated utilizing the ΔΔCT method with the following formula: 2ΔΔCT = 2(ΔCT gene − ΔCT housekeeping genes). The average 2ΔΔCT and standard error of the means (SEMs) were calculated [29].
2.8. Statistical Analysis
For the NGS data, the irradiated samples were first normalized with their sham control using the DEseq2 methodology package previously described [30]. Genes that had a p-value less than 0.05, a gene count greater than 50, and a fold change of at least 1.5 were considered significant.
For the RT-qPCR-based experiments, the different doses within a timepoint were compared to one another by performing a one-way ANOVA followed by Tukey’s post hoc analysis using Jamovi (p-values < 0.05 were considered significant).
3. Results
3.1. Identification of Dysregulated miRNA 6 h Post-Irradiation
The miRNAome analysis of CGL1 cells exposed to 10, 100, and 1000 mGy doses 6 h post-radiation is presented in Table 1. The miRNAome analysis identified a total of 2256 miRNAs within the CGL1 cells. Of these, 38 miRNAs were significantly dysregulated compared to the sham controls, demonstrating radiation-induced expression profiles.

Table 1.
Dysregulated miRNA in CGL1 cells 6 h post-irradiation.
3.2. Validation of the miRNAome Results Via RT-qPCR Analysis
To verify the validity of the significantly dysregulated miRNA identified from the miRNAome results, a secondary quantitative analysis was performed on the irradiated sample sets. Here, the validation of the miRNAome results were corroborated with RT-qPCR analysis. In addition, the 1 and 24 h timepoints were analyzed along with the 6 h timepoint to identify temporal expression patterns. As such, the adapted RT-qPCR technique for the quantification of miRNA described earlier was performed on the samples. The RT-qPCR primers were designed for all of the radiation-induced miRNAs identified in Table 1. However, only 19 of the total primers passed validation due to the limitations in designing primers for the short miRNA sequence (Table A1). From these validated primers, 11 miRNAs were found to be significantly dysregulated as identified via miRNAome and RT-qPCR methodologies across the different experimental conditions: miR-1228-3p, miR-758-5p, miR-502-3p, miR-491-5p, miR-362-5p, miR-3135b, miR-584-5p, miR-143-3p, miR-29a-5p, miR-1292-5p, and miR-370-3p (Figure 1). Of these miRNAs, miR-362-5p, miR-3135b, miR-584-5p, miR-143-3p, miR-29a-5p, and miR-370-3p shared the same temporal expression profile and were significantly upregulated at the 1000 mGy dose at the 1 h timepoint (Figure 1E–K). In fact, all 11 miRNAs, except for miR-1292-5p, were significantly upregulated at 1000 mGy 1 h post-irradiation. Moreover, miR-1292-5p demonstrated dysregulation uniquely at the 10 mGy dose, where it can be shown to be significantly increased at both the 1 and 24 h timepoints (Figure 1J). Another interesting trend was shown with miR-1228-3p, which demonstrated consistent upregulation at 1000 mGy between the 1 and 6 h timepoints (Figure 1A). The remaining miRNAs, miR-758-5p, miR-502-3p, miR-491-5p, shared a similar expression profile, wherein they showed a decrease in expression during the 6 h timepoint at 1000 mGy relative to the sham. Moreover, these miRNAs also demonstrated significant dose-dependent decreases compared to the sham: miR-758-5p showed a decrease at the 10 and 100 mGy doses (Figure 1B), miR-502-3p decreased at the 100 and 1000 mGy doses (Figure 1C), and miR-491-5p was reduced at the 10 and 1000 mGy doses (Figure 1D). Additionally, miR-491-5p also showed a significant increase at the 1000 mGy dose at the 24 h timepoint.
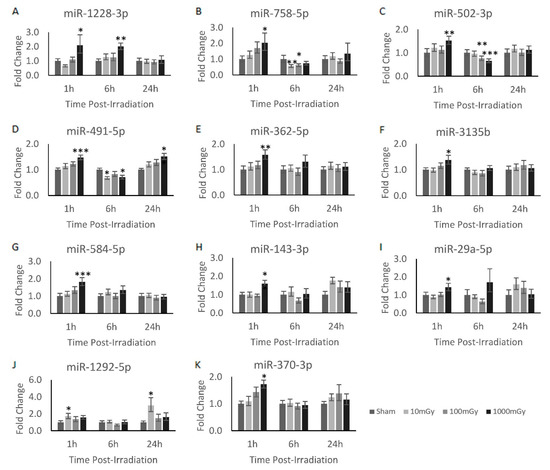
Figure 1.
Radiation-induced miRNA dysregulation in CGL1 cells. Various doses and timepoints were analyzed via RT-qPCR to obtain the temporal expression profile of the analyzed miRNAs in CGL1 cells. Cells were irradiated at 10, 100, and 1000 mGy and collected 1, 6, and 24 h post-irradiation. All irradiation doses were normalized to their own timepoint’s sham condition to obtain the relative fold change. All miRNA shown had at least one dose and timepoint that was considered significant; however, most of the significant findings were shown to be at the 1000 mGy dose. Significance is denoted by an asterisk, where * p < 0.05, ** p < 0.01, and *** p < 0.001.
3.3. mRNA Gene Targets of miR-1228-3p and miR-758-5p Showed Reciprocal Expression
To better understand the underlying mechanism of action for these various significantly dysregulated miRNAs, their various downstream gene targets were identified based on the predicted interaction using the miRBD database and previously documented in the literature (Table 2) [31]. By examining all these mRNA targets, it can be determined if the change in miRNA expression demonstrates reciprocal effects on the expression of the mRNA targets. Here, reciprocal miRNA and mRNA expression demonstrated biologically relevant interactions [32]. However, for this analysis, it is important to note that miRNAs are typically induced much faster and last much longer than their mRNA counterparts; as such, the various timepoint expressions across mRNA and miRNA may not be a direct temporal relationship [33]. From the analysis of these mRNA targets, three significant miRNA/mRNA reciprocal interactions were identified. Here, miR-1228-3p was upregulated, whereas its mRNA targets, Ube2d2 and Ppp2r2d, were found to be significantly downregulated 6 h post-1000 mGy irradiation (Figure 2A,B). In addition, miR-758-5p was downregulated, while its mRNA target Id2 was found to be significantly upregulated at 24 h post-10/1000 mGy irradiation (Figure 2C).

Table 2.
A list of top mRNA targets for selected radiation-induced miRNA.
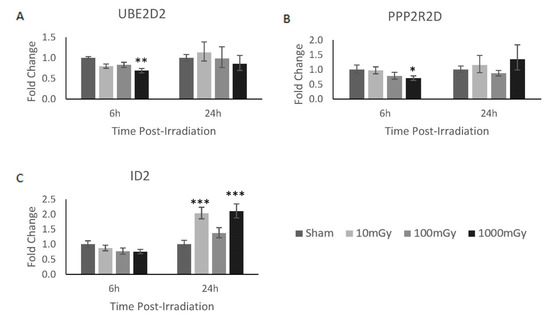
Figure 2.
mRNA gene targets of miR-1228-3p and miR-758-5p showed reciprocal expression. Of the validated possible mRNA interaction shown in Table 2, three mRNA gene targets demonstrated a reciprocal gene expression pattern to their corresponding miRNA. In brief, (A) Ube2d2 and (B) Ppp2r2d demonstrated a reciprocal expression pattern to miR-1228-3p at 6 h, whereas (C) Id2 showed a reciprocal expression pattern to miR-758-5p at 24 h. Significance is denoted with asterisks, where * = p < 0.05, ** = p < 0.01, and *** = p < 0.001.
4. Discussion
The overall miRNAome results showed 38 dysregulated genes across various radiation doses (Table 1). As predicted, an increasing number of genes were shown to be dysregulated when exposed to increasing levels of radiation [34]. To compare radiation dose-dependent expression patterns, 2 miRNAs were dysregulated at 10 mGy, 8 miRNAs were dysregulated at 100 mGy, and 29 miRNAs were dysregulated at 1000 mGy. Of those, 11 miRNAs were validated based on RT-qPCR analysis, and a further 2 miRNAs were shown to have reciprocal response to their predicted mRNA targets (Figure 2).
As mentioned, three mRNA/miRNA target sets were validated from the above results in terms of reciprocal expression patterns. In short, the expression of genes Ube2d2 and Ppp2r2d were inversely related to the expression of miR-1228-3p, whereas Id2 gene expression was inversely related to its target of miR-758-5p. To further explore the role of these radiation-induced mRNA changes within the CGL1 cells, their mechanism of action is discussed below based on the known literature.
Ube2d2 is a component of the protein ubiquitination pathway, a protein modification often associated with protein degradation [35]. The main function of Ube2d2 is to accept ubiquitin from the E1 complex and to catalyze the reaction between ubiquitin and other proteins [35]. Of special interest within our study was its role in the degradation of p53 via its interaction with MDM2 as illustrated in Figure 3 [35]. When paired with the knowledge that p53 is constitutively expressed and degraded via this system, it can be concluded that the levels of p53 within the cell may change in response to the expression of Ube2d2 [31,36]. In short, p53 levels should decrease when Ube2d2 is highly expressed and vice versa. Given that miR-1228-3p inhibits the translation of Ube2d2, radiation-induced expression of miR-1228-3p should therefore reduce Ube2d2-mediated degradation of p53. Therefore, increased expression of miR-1228-3p is expected to result in elevated p53 levels. In this specific case, where miR1228-3p was upregulated following 1 Gy irradiation, it can be inferred that the overall level of active p53 protein is likely elevated via a reduction in its degradation. Therefore, miR-1228-3p is potentially a radiation-induced master regulator of p53 activity and further investigation would be needed to conclude if p53 protein levels are increased in response to radiation-mediated upregulation of miR-1228-3p. Following that assumption, p53-related activities, including cell cycle arrest, DNA damage repair, and apoptosis, would likely be enhanced as a result of increased miR-1228-3p levels [37].
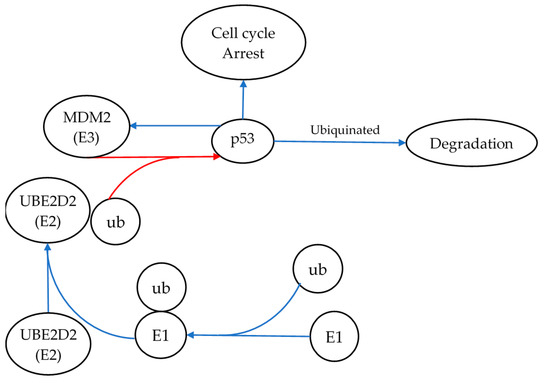
Figure 3.
Ube2d2’s role in p53 degradation, and the signaling pathway demonstrating the role of miR-1228-3p and its target Ube2d2 in regulating the degradation of p53. Here, Ube2d2 is shown transferring ubiquitin (ub) onto p53 via MDM2 and, thus, targeting p53 for degradation. miR-1228-3p inhibits the translation of Ube2d2. Therefore, radiation-induced expression of miR-1228-3p should reduce Ube2d2-mediated degradation of p53. Thus, increased expression of miR-1228-3p is expected to result in elevated p53 levels. The blue arrows represent pathway activation, whereas the red arrows illustrate pathway inhibition. E1 represents ubiquitin-activating enzymes that catalyze the first step in the ubiquitination reaction.
The second validated target of miR-1228-3p is a subunit of the protein phosphatase 2A (PP2A) complex, which is a ubiquitously expressed phosphatase that is generally associated with tumour suppressive activity [38]. The complex is a tri-heteromeric protein in which subunits A and C are responsible for its structural and enzymatic activity, whereas the many types of B subunits are responsible for its specificity and cellular localization [39]. PPP2R2D is a specific B subunit variant (B55 delta) of PP2A that serves to regulate its function and is associated with a key role within the cell cycle by controlling the exit of mitosis via its inhibitory effect on CDK1 [36]. In short, it is highly expressed when the cell is in interphase and found in lower quantities during mitosis [40]. However, a recent finding has also shown Ppp2r2d to have a potential oncogenic role within the cell, as it has repeatedly been shown to be upregulated in gastrointestinal cancer cells and its inhibition is deleterious to the cells [41]. This study demonstrated that the Ppp2r2d subunit was correlated with higher levels of p-mTOR protein, thus potentially affecting the cell proliferative state; however, a mechanism for this correlation could not be identified [41]. This discovery aligns with previous knowledge demonstrating that while PP2A acts primarily as a tumour suppressor, certain subunits, including Ppp2r2d, show the ability to positively regulate signalling pathways such as the MAPK signalling cascade [42]. These findings indicate that a reduction in Ppp2r2d via miR-1228-3p may lead to a decrease in proliferative signals after exposure to 1 Gy of radiation. This coincides with the expected radiation-induced stress response discussed with Ube2d2, where the cell initially undergoes cell cycle arrest post-radiation to allow progression of various repair pathways.
In addition to miR-1228-3p, this study identified reciprocal expression of miR-758-5p and Id2 (Inhibitor of DNA Binding 2). ID2 is a transcriptional regulator that is capable of binding to other transcription factors and preventing their binding to DNA, thus negatively regulating their activity [43]. As such, it can affect a diverse set of signalling pathways, which may be of interest in the context of radiation biology including proliferation, cell cycle arrest, and apoptosis pathways [44]. One such study showed results consistent with those presented here, where Id2 demonstrated an increase in expression following 24 h post-irradiation [45]. This study established that Id2 had the ability to reverse the cell cycle arrest induced by gamma irradiation, demonstrated a protective role when cells were exposed to irradiation, and promoted cell proliferation [45]. Taken together these finding may indicate that subsequent decreases in miR-758-5p at later timepoints post-irradiation trigger an increase in ID2, which promotes the return of the cell to normal functions following the irradiation-induced cell cycle arrest (Figure 4).
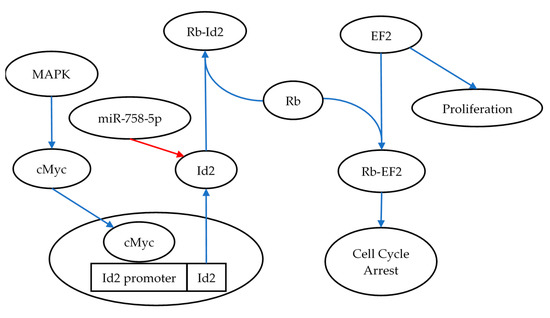
Figure 4.
Id2’s role in reversing cell cycle arrest, illustrating the role of miR-758-5p in regulating Id2 and the consequence of upregulating Id2 via the activation of the MAPK signaling pathway. As part of the recovery process following radiation exposure, Id2 is expressed and binds to Rb. This interaction prevents binding to its other binding partner EF2, thus inducing cell proliferation [46]. The blue arrows in the figure represent pathway activation, whereas the red arrows illustrate pathway inhibition.
Taken together, miR-1228-3p and miR-758-5p appear to be the most promising radiation-induced miRNAs identified from this study. However, all of the radiation-induced miRNAs presented in Table 1 from the miRNAome analysis are potential targets to pursue further as possible biomarkers for LDR exposures. Other promising results include miR-4443, which was shown to be dysregulated at 10 and 100 mGy doses and may potentially show high sensitivity to radiation. In fact, all miRNAs found within the 10 and 100 mGy LDR range are novel radiation-induced miRNAs and should be further assessed for biodosimetry applications of LDR exposures.
The results from this study demonstrate that a single miRNA may not be sufficient to serve as a biomarker for a broad range of doses that spans LDR and HDR exposures. This study points to the use of dose-range specific miRNAs that can be assessed together to develop biodosimetry biomarkers to accurately identify a broad range of radiation dose exposures. In this scenario, a multiplex RT-qPCR design encompassing various dose-range specific primers for radiation-induced miRNAs may serve as a reliable approach for biodosimetry applications.
In addition, as shown in Figure 1, most of the miRNAs were dysregulated at the 1 h timepoint, whereas minimal lasting miRNA dysregulation was identified at the 24 h timepoint. This would potentially be a problem when determining the applicability of miRNA-based biodosimeter biomarkers, as the temporal window for detection should be as large as possible. Therefore, miRNAs, such as miR-1292-5p and miR-491-5p, that demonstrated prolonged dysregulation at 24 h post-irradiation may be of note. However, it is possible that the radiation-induced miRNAs may have been secreted as exosomes, contributing to the lack of elevated expression at 24 h post-irradiation with the cell.
Additionally, this study indicates that mRNA targets may also serve as potential radiation biomarkers. Here, we identified reduced expression of miR-758-5p and an upregulation of its mRNA target Id2 24 h post-radiation. Interestingly, Id2 expression has been previously identified in the literature as a radiation-induced biomarker [43].
This study provides a technical framework for identifying radiation-induced miRNA biomarkers using NGS-based miRNAome technology. Similar studies need to be performed in various cell types and in vivo models to elucidate reliable radiation-induced miRNA biomarkers. In addition, it is important to verify whether the radiation-induced miRNAs are also detectable in the extracellular environment in exosome fractions [47]. Given that serum and urine samples are likely the ideal choice for sampling radiation exposures, identification of released miRNA is crucial for developing reliable miRNA biomarkers for biodosimetry application.
5. Conclusions
In conclusion, this study identified numerous potential candidate radiation-induced miRNAs at various dose ranges. The results of the miRNAome study identified 38 radiation-induced miRNA that were dysregulated within the irradiated samples. From there, a total of 11 miRNA were further verified via RT-qPCR-based miRNA expression analysis. Here, two miRNAs demonstrated reciprocal gene expression with its predicted mRNA targets: miRNAs 1228-3p and 758-5p and their corresponding mRNA targets Ube2d2/Ppp2r2d and Id2, respectively. The most promising was the interaction of the miR-1228-3p target Ube2d2 and its role in p53 degradation. This finding suggests that radiation-induced expression of miR-1228-3p may promote higher levels of p53. Elevated p53 suggests a stress response typically expected after radiation exposures.
Altogether, the results discussed in this paper revealed novel potential miRNA biomarkers within the CGL1 cell line using miRNAome sequencing. Although this study requires additional investigation to validate its findings, the data presented here represents a good foundation for future investigations.
Author Contributions
Conceptualization, J.P., T.C.T. and S.T.; Formal analysis, J.P., C.D.M. and S.T.; Funding acquisition, C.T., N.K., S.J.L., D.R.B., T.C.T. and S.T.; Investigation, J.P. and S.T.; Methodology, J.P., C.D.M., T.C.T. and S.T.; Supervision, T.C.T. and S.T.; Writing—original draft, J.P. and S.T.; Writing—review and editing, J.P., C.D.M., C.T., N.K., S.J.L., D.R.B., T.C.T. and S.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) Collaborative Research and Development (CRD) in partnership with Bruce Power and the Nuclear Innovation Institute. The research was also supported by a Northern Ontario School of Medicine Faculty Association Research Development Award.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon a reasonable request.
Conflicts of Interest
D.R.B. was previously employed by the company Bruce Power.
Appendix A

Table A1.
RT-qPCR primers for miRNA gene expression profiling.
Table A1.
RT-qPCR primers for miRNA gene expression profiling.
| miRNA | miRNA forward Primer | Tm |
| Oligo-dT adapter | GCATAGACCTGAATGGCGGTAAGGGTGTGGTAGGCGAGACATTTTTTTTTTTTTTTTTTTT | 65 |
| universal reverse | GCATAGACCTGAATGGCGGTA | 60 |
| SNORD48 | TGATGACCCCAGGTAACTCTGA | 60 |
| miR-16-5p | ATTGAAACCTCTAAGAGTGGA | 60 |
| U6 | GTG CTC GCT TCG GCA GCA CAT AT | 60 |
| miR-423-3p | AGCTCGGTCTGAGGCCC | 60 |
| miR-191-5p | CAACGGAATCCCAAAAGCAGC | 60 |
| miR-6134 | UGA GGU GGU AGG AUG UAG A | 60 |
| miR-6082 | GAATACGTCTGGTTGATCCAAAA | 57 |
| miR-208a-5p | AGCTTTTGGCCCGGGTTATAC | 61 |
| miR-7112-3p | CATCACAGCCTTTGGCCCTA | 60 |
| miR-6762-3p | UGGCUGCUUCCCUUGGUCUCCAG | 60 |
| miR-6882-3p | CTCTCCTCTTGCCTGCAGAA | 60 |
| miR-200a-5p | TCTTACCGGACAGTGCTGGA | 61 |
| miR-604 | GCTGCGGAATTCAGGACAAAA | 60 |
| miR-2355-5p | TCCCCAGATACAATGGACAAAA | 57 |
| miR-4692 | UCAGGCAGUGUGGGUAUCAGAU | 60 |
| miR-378e | ACTGGACTTGGAGTCAGGAAA | 59 |
| miR-4535 | GGACCTGGCTGGGACAAAA | 60 |
| miR-1915-3p | CGACGCGGCGGGAAA | 60 |
| miR-1291 | CCTGACTGAAGACCAGCAGTAAA | 60 |
| miR-19a-5p | AGTTTTGCATAGTTGCACTACAAA | 58 |
| miR-5008-3p | TCCCAGGGCCTCGCAAA | 61 |
| miR-4417 | TGGGCTTCCCGGAGGG | 60 |
| miR-1537-3p | AAAACCGUCUAGUUACAGUUGU | 60 |
| miR-3168 | GAGUUCUACAGUCAGAC | 60 |
| miR-5186 | AGAGAUUGGUAGAAAUCAGGU | 60 |
| miR-4680-5p | AGAACTCTTGCAGTCTTAGATGT | 57 |
| miR-554 | AGTCCTGACTCAGCCAGTAAAAA | 60 |
| miR-6845-3p | CCUCUCCUCCCUGUGCCCCAG | 60 |
| miR-6125 | GCGGAGCGGCGGAAAA | 61 |
| miR-3616-3p | CGAGGGCATTTCATGATGCAG | 60 |
| miR-615-3p | UCCGAGCCUGGGUCUCCCUCUU | 60 |
| miR-3152-3p | UGUGUUAGAAUAGGGGCAAUAA | 60 |
| miR-4693-3p | UGAGAGUGGAAUUCACAGUAUUU | 60 |
| miR-4320 | GGGAUUCUGUAGCUUCCU | 60 |
| miR-6783-5p | GGAAAAGTCCTGATCCGGAAAAA | 59 |
| miR-33b-5p | GUGCAUUGCUGUUGCAUUGC | 60 |
| miR-4662a-3p | AAAGAUAGACAAUUGGCUAAAU | 60 |
| miR-8082 | GATGGAGCTGGGAATACTCTGAA | 60 |
| miR-4758-5p | GCCGGTGGGGCTGAAAA | 60 |
| miR-8071 | GGACTGGAGTGGGTGGAAAAA | 60 |
| miR-4477b | AUUAAGGACAUUUGUGAUUGAU | 60 |
| miR-34a-5p | TGGCAGTGTCTTAGCTGGTTG | 60 |
| miR-182-5p | TTTGGCAATGGTAGAACTCACACT | 60 |
| miR-525-3p | AAGGCGCTTCCCTTTAGAGC | 60 |
| hsa-miR-492 | AGGACCTGCGGGACAAGA | 60 |
| hsa-miR-4535 | GTGGACCTGGCTGGGAC | 60 |
| miR-877-5p | GTAGAGGAGATGGCGCAGG | 60 |
| miR-133b | TTTGGTCCCCTTCAACCAGC | 60 |
| miR-205-3p | GATTTCAGTGGAGTGAAGTTC | 60 |
| miR-1 | ACATACTTCTTTATATGCCCAT | 60 |
| miR-19b | AGTTTTGCAGGTTTGCATCCAG | 60 |
| miR-93-5p | AAGTGCTGTTCGTGCAGGTAG | 60 |
| miR-132-5p | ACCGTGGCTTTCGATTGTTAC | 59 |
| miR-671-5p | CTGGAGGGGCTGGAGAAAAA | 60 |
| miR-628-3p | TCTAGTAAGAGTGGCAGTCGAA | 58 |
| miR-125b-1-3p | ACGGGTTAGGCTCTTGGGA | 60 |
| miR-6797-5p | GAAGGGGCTGAGAACAGGAAA | 60 |
| miR-6739-5p | TGGGAAAGAGAAAGAACAAGTAAAA | 57 |
| miR-6823-3p | GCCTCTCCTTCCCTCCAGAAA | 61 |
| miR-449c-3p | GCTAGTTGCACTCCTCTCTGT | 59 |
| miR-328-3p | CCTCTCTGCCCTTCCGTAAA | 59 |
| miR-103a-3p | CAGCATTGTACAGGGCTATGAA | 58 |
| miR-4721 | CTCCAGGTGACGGTGGAAAAA | 60 |
| miR-589-3p | CAGAACAAATGCCGGTTCCC | 60 |
| miR-769-3p | GGATCTCCGGGGTCTTGGT | 61 |
| miR-27a-3p | CACAGTGGCTAAGTTCCGCA | 61 |
| let-7g-5p | TGAGGTAGTAGTTTGTACAGTT | 60 |
| let-7a-5p | TGAGGTAGTAGGTTGTATAGTT | 60 |
| miR-296-5p | AGGGCCCCCCCTCAATCCTGT | 60 |
| miR-4443 | TTGGAGGCGTGGGTTTT | 60 |
| miR-3120-3p | ACAGCAAGTGTAGACAGGCAA | 60 |
| miR-423-3p | AGCTCGGTCTGAGGCCCCTCAGT | 60 |
| miR-362-5p | CCTTGGAACCTAGGTGTGAGT | 60 |
| miR-148b-5p | AGTTCTGTTATACACTCAGGCAA | 60 |
| miR-495-5p | GAAGTTGCCCATGTTATTTTCG | 60 |
| miR-6835-5p | AGGGGGUAGAAAGUGGCUGAAG | 60 |
| miR-4668-5p | AGGGAAAAAAAAAAGGAUUUGUC | 60 |
| miR-491-5p | GTGGGGAACCCTTCCATGAG | 60 |
| miR-4271 | GGGGGAAGAAAAGGTGGGG | 60 |
| miR-665 | GGAGGCTGAGGCCCCTAAA | 60 |
| miR-193a-5p | TCTTTGCGGGCGAGATGAAA | 60 |
| miR-22-3p | AGCTGCCAGTTGAAGAACTGTA | 60 |
| miR-1228-3p | CACACCTGCCTCGCCC | 60 |
| miR-181a-2-3p | CACTGACCGTTGACTGTACCA | 60 |
| miR-100-5p | ACCCGTAGATCCGAACTTGTG | 60 |
| miR-143-3p | TGAGATGAAGCACTGTAGCTC | 60 |
| miR-10b-5p | ACCCTGTAGAACCGAATTTGTGA | 60 |
A total list of miRNA primers tested during the course of the secondary validation step of the obtained NGS results. The sequences and their optimal melting temperature can be found above.

Table A2.
RT-qPCR primers for mRNA gene expression profiling.
Table A2.
RT-qPCR primers for mRNA gene expression profiling.
| mRNA Primer | Sequence (5′→3′) | Length | Product (bp) | Tm |
| PPP2R2D_F_Hu_1 | GTCAAGGACAGGGCAGACTTC | 21 | 136 | 60 |
| PPP2R2D_R_Hu_2 | AGCTGTTCTCAGCTGTTCTATCA | 23 | ||
| UBE2D2_F_Hu_1 | CGTTTTGCCCGATCCACAAG | 20 | 145 | 60 |
| UBE2D2_R_Hu_1 | GTCCCGTGCCAGATCATTCA | 20 | ||
| IRX2_F_Hu_1 | CACCAAGATGACCCTCACCC | 20 | 119 | 60 |
| IRX2_R_Hu_1 | CGTCCTCGTCCTCATCTTCG | 20 | ||
| ZNF554_F_Hu_1 | CTGCAGTACTGTGCTGATCCA | 21 | 137 | 60 |
| ZNF554_R_Hu_1 | ACGTCTCTGTCCAGCCTTTG | 20 | ||
| NFIA_F_Hu_1 | TGCCGAATCGATTGCAACTTC | 21 | 83 | 60 |
| NFIA_R_Hu_1 | GGCTGGTTCTCAGATTCGCT | 20 | ||
| SOCS6_F_Hu_1 | GGCCGCCTCCGGAAAAT | 17 | 81 | 60 |
| SOCS6_R_Hu_1 | ACATCTGGAGAGGCTGCAAG | 20 | ||
| RABGEF1_F_Hu_1 | GCGGTGACCTGGACCAC | 17 | 82 | 60 |
| RABGEF1_R_Hu_1 | TTCCCAAAGTGATCTCGCCC | 20 | ||
| TJP1_F_Hu_1 | TGGTCTGTTTGCCCACTGTT | 20 | 150 | 60 |
| TJP1_R_Hu_1 | TCTGTACATGCTGGCCAAGG | 20 | ||
| TOR1AIP1_F_Hu_1 | AAGTCCTCTAGTGCAACGCC | 20 | 82 | 60 |
| TOR1AIP1_R_Hu_1 | ATCTTGGCTTGAGGCACTCC | 20 | ||
| ZBTB44_F_Hu_1 | ACGAGTGCAAAACATGTGGC | 20 | 72 | 60 |
| ZBTB44_R_Hu_1 | GGTTCAGACTCCTCAGGTGC | 20 | ||
| Moap1_F_Hu_1 | AGGCCCTTCTCCAGGCAATA | 20 | 70 | 60 |
| Moap1_R_Hu_1 | TGCCATATCCCTTCGTGGTT | 20 | ||
| Csnk2a1_F_Hu_1 | ATCGCCGCCATATTGTCTGT | 20 | 109 | 60 |
| Csnk2a1_R_Hu_1 | CAGCTGGGGGTAAGACCTTG | 20 | ||
| PLAC8_F_Hu_1 | CAGAAGGAGAGCCATGCGTA | 20 | 76 | 60 |
| PLAC8_R_Hu_1 | AACCCACATGTTCTGAGAGGC | 21 | ||
| SLC20A2_F_Hu_1 | TCTCGGCCTAATGTGGTAGGA | 21 | 138 | 60 |
| SLC20A2_R_Hu_1 | CTCCCGATCTGGGAAAGCTG | 20 | ||
| ID2_F_Hu_1 | ATCCTGTCCTTGCAGGCTTC | 20 | 81 | 60 |
| ID2_R_Hu_1 | ACCGCTTATTCAGCCACACA | 20 | ||
| NUFIP2_F_Hu_1 | ATGTCCATTTTGCTTGCCTGG | 21 | 149 | 60 |
| NUFIP2_R_Hu_1 | CCCAATTCAGGTGGGGTCTG | 20 | ||
| PTP4A1_F_Hu_1 | CTGTGAGCTCTTAAGACTTGCTT | 23 | 73 | 58 |
| PTP4A1_R_Hu_1 | CACTGCTGCTGGGAATTATGA | 21 | ||
| TOX4_F_Hu_1 | CTGACGATCACAGGGCCTTC | 20 | 142 | 60 |
| TOX4_R_Hu_1 | GGCCAACCACATCTGAGACA | 20 | ||
| SETD5_F_Hu_1 | CTGTGACAAGTGCAGGGGAA | 20 | 95 | 60 |
| SETD5_R_Hu_1 | CTGTTGCACTGCTATCCCCA | 20 | ||
| PHACTR1_F_Hu_1 | TGTTCATTTGTGCTTGCGGG | 20 | 74 | 60 |
| PHACTR1_R_Hu_1 | CCCTTTCAACAGGACACGGT | 20 | ||
| RTKN_F_Hu_1 | CGAGTGAAGTGTGACTCCGT | 20 | 90 | 60 |
| RTKN_R_Hu_1 | TTCCAGACAGGAAACCAGCC | 20 | ||
| DSG3_F_Hu_1 | GACTCCTTCGGAAAGCAGCA | 20 | 138 | 60 |
| DSG3_R_Hu_1 | GGGGAAGAGCCCCATCATTG | 20 | ||
| CSNK1A1L_F_Hu_1 | CCCTGGGGTTTGCAAATTGT | 20 | 138 | 60 |
| CSNK1A1L_R_Hu_1 | TCTTCACAGGTAAGCAGGCG | 20 | ||
| CD36_F_Hu_1 | CCACACACTGGGATCTGACA | 20 | 132 | 60 |
| CD36_R_Hu_1 | TCTGCAGGAAAGTCCTACACTG | 22 | ||
| ZBTB20_F_Hu_1 | TCCTGACAAATGCTAGAACGGA | 22 | 86 | 60 |
| ZBTB20_R_Hu_1 | CCACCCGGCTGAGTAATCTC | 20 | ||
| CBX5_F_Hu_1 | GGGAGGCCCCTCCTGTTAG | 19 | 72 | 60 |
| CBX5_R_Hu_1 | AAGACTAAGGCCACCAGGTC | 20 | ||
| CTTN_F_Hu_1 | GACAAATGTGCCCTTGGCTG | 20 | 111 | 60 |
| CTTN_R_Hu_1 | CTGCCTCTCCGACTGAACAC | 20 | ||
| NCOA2_F_Hu_1 | CCCTCCCTCTACCACAGTCA | 20 | 87 | 60 |
| NCOA2_R_Hu_1 | CAGAGTCCTCTGAGAAGGCG | 20 | ||
| ZNF281_F_Hu_1 | ATGACCACCATGGCACTGAG | 20 | 70 | 60 |
| ZNF281_R_Hu_1 | TCTGGCTTTGGCCTTTTTGC | 20 | ||
| TNPO1_F_Hu_1 | TGCCCGGCCGTTTGAAG | 17 | 121 | 60 |
| TNPO1_R_Hu_1 | GCTCGTCAGGTTTCCACTCA | 20 | ||
| HNRNPD_F_Hu_1 | GCCATTCAAACTCCTCCCCA | 20 | 89 | 60 |
| HNRNPD_R_Hu_1 | GTCCCAGCTAAGGCCTCCTA | 20 | ||
| SBNO1_F_Hu_1 | CAATGCCTACCCCGTCAGTT | 20 | 72 | 60 |
| SBNO1_R_Hu_1 | CTGCTTCGGTCTCCAAACCT | 20 | ||
| DIPK2A_F_Hu_1 | GTGGGTGTGAGACATCCTAGC | 21 | 74 | 60 |
| DIPK2A_R_Hu_1 | CACGACAAGTGGGGTCTGC | 19 | ||
| MCUB_F_Hu_1 | GGAGGATGCTCCAGAGGGG | 19 | 119 | 60 |
| MCUB_R_Hu_1 | CTTCACACGCAAAACCTGGG | 20 | ||
| NOL4L_F_Hu_1 | GCCAAAACCAAGACGGTGAC | 20 | 124 | 6 |
| NOL4L_R_Hu_1 | CCCAGAACTGGAACTTGCCT | 20 | ||
| CAMTA1_F_Hu_1 | GAAAACAAGCCGGAAGAGCG | 20 | 81 | 60 |
| CAMTA1_R_Hu_1 | ATAGGTGGCACGGTGTTGAG | 202 | ||
| cdk6_F_Hu_1 | CTGCAGGGAAAGAAAAGTGCAA | 22 | 95 | 60 |
| cdk6_R_Hu_1 | CTCCTCGAAGCGAAGTCCTC | 20 | ||
| MTA3_F_Hu_1 | GTCCTCCCCCTCCGCTC | 17 | 131 | 60 |
| MTA3_R_Hu_1 | CTGGGGACTGCCCAATTCAT | 20 | ||
| TRIM50_F_Hu_1 | GGCATCTAACTGGAGCGACA | 20 | 85 | 60 |
| TRIM50_R_Hu_1 | CCAAGCCATCCACACTCACT | 20 | ||
| SGIP1_F_Hu_1 | TGGAATTCCTTCAGGCGGAC | 20 | 73 | 60 |
| SGIP1_R_Hu_1 | ACGATTCCAGGTCCCAGCTA | 20 | ||
| PLAGL2_F_Hu_1 | ACAATGCACCGCACAATGG | 19 | 138 | 60 |
| PLAGL2_R_Hu_1 | CCTCCAACGCAGCTTTCAGA | 20 | ||
| PRKACB_F_Hu_1 | GCTAGCAGTAAGAGCTGGTGT | 21 | 75 | 60 |
| PRKACB_R_Hu_1 | TGAACCTGGCAAGGAGCAAA | 20 | ||
| LUC7L3_F_Hu_1 | GGTCAATGGGACCAGTGAAGA | 21 | 147 | 60 |
| LUC7L3_R_Hu_1 | CGCTGCACTGTCAAACAGTAA | 21 | ||
| EDEM1_F_Hu_1 | ACAACTACATGGCTCACGCC | 20 | 91 | 60 |
| EDEM1_R_Hu_1 | AGATTTGAAGGGTCCCCGC | 19 | ||
| CYP1B1_F_Hu_1 | GCTGTGAGGAAACCTCGACT | 20 | 121 | 60 |
| CYP1B1_R_Hu_1 | GAGTCTCTTGGCGTCGTCAG | 20 | ||
| RMI1_F_Hu_1 | GCGGTTCCTGTCCTTACAGT | 19 | 86 | 60 |
| RMI1_R_Hu_1 | ACTGCTCAGAAATGGCCCTG | 20 | ||
| MRPL35_F_Hu_1 | TGCAAAGAAATTGGGTCTGTGT | 22 | 141 | 60 |
| MRPL35_R_Hu_1 | TGAAGGGCCACCCTTAAACC | 20 | ||
| PIK3C2B_F_Hu_1 | CCACCATAGAGATGGCGTCC | 20 | 134 | 60 |
| PIK3C2B_R_Hu_1 | TGGGCGCCTGATTCTTCTAC | 20 | ||
| Cyld_F_Hu_1 | CCCCCTTTCTAGGGTGAGGA | 20 | 98 | 60 |
| Cyld_R_Hu_1 | TTCAGCAACGTGGTGTCCAT | 20 | ||
| GAS7_F_Hu_1 | AGCCAACGAGTCTCTGCTTC | 20 | 79 | 60 |
| GAS7_R_Hu_1 | GCCGTCTCTGGGGTGC | 16 | ||
| LRRC27_F_Hu_1 | CTCGCCAGCGCTTCAGT | 17 | 102 | 60 |
| LRRC27_R_Hu_1 | TAGGAGCTGCTTCCCTCCAT | 20 | ||
| FMNL3_F_Hu_1 | GAGTCGGGACTCGGGGAG | 18 | 130 | 60 |
| FMNL3_R_Hu_1 | CTCTCCAGGTTGCCCATCG | 9 | ||
| TTC21B_F_Hu_1 | TGCGTCTTCCTTTAGGCTGC | 20 | 87 | 60 |
| TTC21B_R_Hu_1 | TCTTCAATTCCTGCGAGTCCA | 21 | ||
| CASTOR3_F_Hu_1 | AGCTTTTCCAGACCAGGCAT | 20 | 76 | 60 |
| CASTOR3_R_Hu_1 | CTAGGGGCTGATGTGCCAAA | 20 | ||
| XPO7_F_Hu_1 | GCAATCACAGACGTCACAAGG | 21 | 126 | 60 |
| XPO7_R_Hu_1 | TGCTTCAATGAGGAAGGCTGT | 21 | ||
| PPM1A_F_Hu_1 | CTGCTCCGGACCTAGAGGAT | 20 | 124 | 60 |
| PPM1A_R_Hu_1 | CAGCCTTGCATGCTGCTTAG | 20 | ||
| DNM1L_F_Hu_1 | TCACCCGGAGACCTCTCATT | 20 | 91 | 60 |
| DNM1L_R_Hu_1 | TCTGCTTCCACCCCATTTTCT | 21 | ||
| KDM3B_F_Hu_1 | CTGCGCACTCGAGCCTG | 17 | 114 | 60 |
| KDM3B_R_Hu_1 | CCAGGAGTGTTGCTTCCAGT | 20 | ||
| RBP1_F_Hu_1 | CCGCTACAATGGATCCTCCC | 20 | 96 | 60 |
| RBP1_R_Hu_1 | GGAAATGAGCGCCCTCCG | 18 | ||
| FAAP20_F_Hu_1 | GGGTCCCCTTCTCCACTGTA | 20 | 78 | 60 |
| FAAP20_R_Hu_1 | CTGGCAGGAGCTGGAGATG | 20 | ||
| PTEN_F_Hu_1 | CTGCAGAAAGACTTGAAGGCG | 21 | 70 | 58 |
| PTEN_R_Hu_1 | TGCTTTGAATCCAAAAACCTTACT | 24 | ||
| GOLPH3_F_Hu_1 | TGTTTCCTCATGACTGCCCC | 20 | 80 | 60 |
| GOLPH3_R_Hu_1 | CGATCCGGGTTTCCGTGTTA | 20 | ||
| ADAMTS3_F_Hu_1 | CAAGCATTCTCCGCGCTAAC | 20 | 147 | 60 |
| ADAMTS3_R_Hu_1 | GGAGCGAGAAGGTGCTGTAA | 20 | ||
| KCTD9_F_Hu_1 | CCCAAGAACGGAAAGGTGGT | 20 | 88 | 60 |
| KCTD9_R_Hu_1 | TGGTGGCTTTTATGCCGAGT | 20 | ||
| FBN2_F_Hu_1 | GTTTTCTGCCAGTCATCCAGC | 21 | 142 | 60 |
| FBN2_R_Hu_1 | AGCTGCTTTGGCTTCGATCT | 20 | ||
| SEC63_F_Hu_1 | GGACATAAATAGGGCAATCCACT | 23 | 119 | 58 |
| SEC63_R_Hu_1 | CCCTCTCACTCCTGGGTTTT | 20 | ||
| ZFX_F_Hu_1 | ACCCTAGTGGAGTGTTGGCT | 20 | 123 | 60 |
| ZFX_R_Hu_1 | TGAACCACTGAAGGGAGTCG | 20 | ||
| DAPK1_F_Hu_1 | TTCGGAGTGTGAGGAGGACA | 20 | 149 | 60 |
| DAPK1_R_Hu_1 | GGGAACACAGCTAGGGAGTG | 20 | ||
| SMIM13_F_Hu_1 | AGTGGGTGAAAATTCCCGCT | 20 | 94 | 60 |
| SMIM13_R_Hu_1 | CCCTGGTAAACACTCAGCCC | 20 | ||
| NAP1L5_F_Hu_1 | CTCCTAGACCTCTGCGGCTT | 20 | 147 | 62 |
| NAP1L5_R_Hu_1 | GCTGTCACAGTCTCCACCCT | 20 | ||
| CSDE1_F_Hu_1 | CGCTGAGCTGTTGGGTATGA | 20 | 78 | 60 |
| CSDE1_R_Hu_1 | ACGAGGTTTGTTCCTTGCCT | 20 | ||
| set_F_Hu_1 | AGTCTCAGTGTTCAGCCTGC | 20 | 78 | 60 |
| set_R_Hu_1 | GGCCATGCTGTTAGGGAAGT | 20 | ||
| OLFM4_F_Hu_1 | AAATGCTCGAGAGTTGCGGA | 20 | 133 | 60 |
| OLFM4_R_Hu_1 | CACAGCAATCGTGTTGGTGG | 20 | ||
| USP6NL_F_Hu_1 | TGGAAGGGAAACAATGGGGC | 20 | 144 | 60 |
| USP6NL_R_Hu_1 | CATGTCCTCAGTACGGTCCC | 20 | ||
| AVPR1A_F_Hu_1 | TGGGCGCCTTTCTTCATCAT | 20 | 75 | 60 |
| AVPR1A_R_Hu_1 | AGGGTTTTCCGATTCGGTCC | 20 | ||
| HSD11B1_F_Hu_1 | TGCCTGCTTAGGAGGTTGTAG | 21 | 90 | 60 |
| HSD11B1_R_Hu_1 | AAAAGCCATCCGACAGGGAG | 20 | ||
| GALNT15_F_Hu_1 | ACCCAGATGGCTTATTGCCT | 20 | 126 | 60 |
| GALNT15_R_Hu_1 | TACCAGCCAGGGACTGAGTT | 20 | ||
| GBP5_F_Hu_1 | AGTTCTGCTTGACACCGAGG | 20 | 149 | 60 |
| GBP5_R_Hu_1 | GCAGTAGGTCGATAGCACCC | 20 | ||
| CD200_F_Hu_1 | TCCAGGAGCAAGGATGGAGA | 20 | 76 | 60 |
| CD200_R_Hu_1 | GACCCAAACCAGGCTGTAGG | 20 | ||
| PPM1A_F_Hu_1 | CTGCTCCGGACCTAGAGGAT | 20 | 124 | 60 |
| PPM1A_R_Hu_1 | CAGCCTTGCATGCTGCTTAG | 20 | ||
| NPHP1_F_Hu_1 | CCCACTTCTCCACTCCACAC | 20 | 76 | 60 |
| NPHP1_R_Hu_1 | AACTTTCCACCGTGCAGTCT | 20 | ||
| Rock_F_Hu_1 | CCCTTTGCTTTCGCCTTTCC | 20 | 150 | 60 |
| Rock1_R_Hu_1 | GAGGTGCTTCAGTCTAGCGG | 20 | ||
| mmp14_F_Hu_1 | GGGTCTTCGTTGCTCAGTCA | 20 | 145 | 60 |
| mmp14_R_Hu_1 | AACATTCGAGAGGCACAGGG | 20 | ||
| kcne2_F_Hu_1 | ACGGGAACACTCCAATGACC | 20 | 105 | 60 |
| kcne2_R_Hu_1 | TGGATGGTGGCCTTCGATTC | 20 | ||
| WWP1_F_Hu_1 | TGCTACTTTTAGCAAACTGGGC | 22 | 128 | 58 |
| WWP1_R_Hu_1 | TTAAGAAGTCAGTTCCATGGCT | 22 | ||
| cdk16_F_Hu_1 | TTGGGCCGTTGGCTGTTC | 18 | 70 | 60 |
| cdk16_R_Hu_1 | GGCTCGCGGCACAGAG | 16 | ||
| SEMA6A_F_Hu_1 | CTTACAACACAGTGTATGGGCA | 22 | 81 | 60 |
| SEMA6A_R_Hu_1 | CATACCCCTCTTGAGCCGTC | 20 | ||
| CHRNB2_F_Hu_1 | GAAAGTTCGGCTCCCTTCCA | 20 | 115 | 60 |
| CHRNB2_R_Hu_1 | GCCATCATAGGAGACCACGG | 20 | ||
| POGK_F_Hu_1 | TGGGAAGTTTTGACGGAGCA | 20 | 113 | 60 |
| POGK_R_Hu_1 | CGGGTGATCATGTCTGGCTT | 20 | ||
| KSR2_F_Hu_1 | AAAGCACTCCAAACCGTGGA | 20 | 133 | 60 |
| KSR2_R_Hu_1 | ACTCTTTACACACCGGCTCC | 20 | ||
| SAMD4B_F_Hu_1 | CCTTCCTACTGGGCAGATGAG | 21 | 101 | 60 |
| SAMD4B_R_Hu_1 | CCAGAAGTGGACATGGGGTA | 20 | ||
| OPTC_F_Hu_1 | TGTCTTCAACCTGGCCTGTC | 20 | 70 | 60 |
| OPTC_R_Hu_1 | AGTACACAAGCCCATCCAGG | 20 | ||
| CPNE5_F_Hu_1 | TGTGTCCAACGGTGGTGTC | 19 | 113 | 60 |
| CPNE5_R_Hu_1 | CCAGCTTGTTGGCACAGAAC | 20 | ||
| FOXP4_F_Hu_1 | AGCTGATTTGCTGCAGGGAT | 20 | 100 | 60 |
| FOXP4_R_Hu_1 | GAAGGACACCTGGGAATGGG | 20 | ||
| MSN_F_Hu_1 | GCCCAAAACGATCAGTGTGC | 20 | 88 | 60 |
| MSN_R_Hu_1 | AAATAGCTGCTTCCCGGTGG | 20 | ||
| IGF2_F_Hu_1 | TCGCCGAACCAAAGTGGATTA | 21 | 148 | 60 |
| IGF2_R_Hu_1 | GTGGGAGAGACAGAGTGAACG | 21 | ||
| Smad3_F_Hu_1 | CCGGGGGTTGGACTTTCCT | 19 | 70 | 60 |
| Smad3_R_Hu_1 | CAGAAGTTTGGGTTTCCGCA | 20 | ||
| Bcl2l1_F_Hu_1 | AGGCGGATTTGAATCTCTTTCTCT | 24 | 129 | 60 |
| Bcl2l1_R_Hu_1 | GGGCTCAACCAGTCCATTGT | 20 | ||
| Egfr_F_Hu_1 | GACAGGCCACCTCGTCG | 17 | 106 | 60 |
| Egfr_R_Hu_1 | CCGGCTCTCCCGATCAATAC | 20 | ||
| Notch3_F_Hu_1 | TCTAGGTAAGGTGGGGAGTGG | 21 | 70 | 60 |
| Notch3_R_Hu_1 | TGGGAGCTCAAGTTAGCCCT | 20 | ||
| Mmp9 _F_Hu_1 | GTACTCGACCTGTACCAGCG | 20 | 92 | 60 |
| Mmp9 _R_Hu_1 | AGAAGCCCCACTTCTTGTCG | 20 | ||
| Igf2bp1_F_Hu_1 | AGCTCCTTTATGCAGGCTCC | 20 | 111 | 60 |
| lgf2bp1_R_Hu_1 | CCGGGAGAGCTGTTTGATGT | 20 | ||
| Wnt3a_F_Hu_1 | CTCCTCCCTGGAGCTAGTGT | 20 | 138 | 60 |
| Wnt3a_R_Hu_1 | AATCTGTAGCCCCGCCTCTG | 20 | ||
| capns1_F_Hu_1 | CGGACGCTGCGGGAG | 15 | 72 | 60 |
| capns1_R_Hu_1 | TCACTGCGCCGCACAC | 16 |
A total list of mRNA primers tested, consisting of both predicted mRNA targets for the studied miRNA and the genes chosen for use in the DNA damage panel. The sequences of both primer pairs and their optimal melting temperature can be found above.
References
- Rothkamm, K.; Beinke, C.; Romm, H.; Badie, C.; Balagurunathan, Y.; Barnard, S.; Bernard, N.; Boulay-Greene, H.; Brengues, M.; de Amicis, A. Comparison of established and emerging biodosimetry assays. Radiat. Res. 2013, 180, 111–119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, Y.; Jin, Y.W.; Wilkins, R.C.; Jang, S. Validation of the dicentric chromosome assay for radiation biological dosimetry in South Korea. J. Radiat. Res. 2019, 60, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Vral, A.; Fenech, M.; Thierens, H. The micronucleus assay as a biological dosimeter of in vivo ionising radiation exposure. Mutagenesis 2011, 26, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.M.; Prasanna, P.G.; Grace, M.B.; Wathen, L.K.; Wallace, R.L.; Koerner, J.F.; Coleman, C.N. Assessment of biodosimetry methods for a mass-casualty radiological incident: Medical response and management considerations. Health Phys. 2013, 105, 540–554. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Li, J.R.; Tong, C.Y.; Sung, T.J.; Kang, T.Y.; Zhou, X.J.; Liu, C.C. CMEP: A database for circulating microRNA expression profiling. Bioinformatics 2019, 35, 3127–3132. [Google Scholar] [CrossRef]
- Enelund, L.; Nielsen, L.N.; Cirera, S. Evaluation of microRNA Stability in Plasma and Serum from Healthy Dogs. Microrna 2017, 6, 42–52. [Google Scholar] [CrossRef]
- Glinge, C.; Clauss, S.; Boddum, K.; Jabbari, R.; Jabbari, J.; Risgaard, B.; Tomsits, P.; Hildebrand, B.; Kääb, S.; Wakili, R.; et al. Stability of Circulating Blood-Based MicroRNAs—Pre-Analytic Methodological Considerations. PLoS ONE 2017, 12, e0167969. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Hu, H.Y.; Yan, Z.; Xu, Y.; Hu, H.; Menzel, C.; Zhou, Y.H.; Chen, W.; Khaitovich, P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genom. 2009, 10, 413. [Google Scholar] [CrossRef]
- Tharmalingam, S.; Sreetharan, S.; Brooks, A.L.; Boreham, D.R. Re-evaluation of the linear no-threshold (LNT) model using new paradigms and modern molecular studies. Chem. Biol. Interact. 2019, 301, 54–67. [Google Scholar] [CrossRef]
- Tharmalingam, S.; Sreetharan, S.; Kulesza, A.V.; Boreham, D.R.; Tai, T.C. Low-Dose Ionizing Radiation Exposure, Oxidative Stress and Epigenetic Programing of Health and Disease. Radiat. Res. 2017, 188, 525–538. [Google Scholar] [CrossRef]
- Puukila, S.; Tharmalingam, S.; Al-Khayyat, W.; Peterson, J.; Hooker, A.M.; Muise, S.; Boreham, D.R.; Dixon, D.L. Transcriptomic Response in the Spleen after Whole-Body Low-Dose X-ray Irradiation. Radiat. Res. 2021, 196, 66–73. [Google Scholar] [CrossRef]
- Mao, A.; Zhao, Q.; Zhou, X.; Sun, C.; Si, J.; Zhou, R.; Gan, L.; Zhang, H. MicroRNA-449a enhances radiosensitivity by downregulation of c-Myc in prostate cancer cells. Sci. Rep. 2016, 6, 27346. [Google Scholar] [CrossRef]
- Duan, X.M.; Liu, X.N.; Li, Y.X.; Cao, Y.Q.; Silayiding, A.; Zhang, R.K.; Wang, J.P. MicroRNA-498 promotes proliferation, migration, and invasion of prostate cancer cells and decreases radiation sensitivity by targeting PTEN. Kaohsiung J. Med. Sci. 2019, 35, 659–671. [Google Scholar] [CrossRef]
- Körner, C.; Keklikoglou, I.; Bender, C.; Wörner, A.; Münstermann, E.; Wiemann, S. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J. Biol. Chem. 2013, 288, 8750–8761. [Google Scholar] [CrossRef]
- Zaleska, K.; Przybyła, A.; Kulcenty, K.; Wichtowski, M.; Mackiewicz, A.; Suchorska, W.; Murawa, D. Wound fluids affect miR-21, miR-155 and miR-221 expression in breast cancer cell lines, and this effect is partially abrogated by intraoperative radiation therapy treatment. Oncol. Lett. 2017, 14, 4029–4036. [Google Scholar] [CrossRef]
- Chaudhry, M.A.; Sachdeva, H.; Omaruddin, R.A. Radiation-induced micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol. 2010, 29, 553–561. [Google Scholar] [CrossRef]
- Maia, D.; de Carvalho, A.C.; Horst, M.A.; Carvalho, A.L.; Scapulatempo-Neto, C.; Vettore, A.L. Expression of miR-296-5p as predictive marker for radiotherapy resistance in early-stage laryngeal carcinoma. J. Transl. Med. 2015, 13, 262. [Google Scholar] [CrossRef]
- Stanbridge, E.J.; Flandermeyer, R.R.; Daniels, D.W.; Nelson-Rees, W.A. Specific chromosome loss associated with the expression of tumorigenicity in human cell hybrids. Somatic. Cell Genet. 1981, 7, 699–712. [Google Scholar] [CrossRef]
- Stanbridge, E.J.; Wilkinson, J. Dissociation of anchorage independence form tumorigenicity in human cell hybrids. Int. J. Cancer 1980, 26, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pirkkanen, J.; Tharmalingam, S.; Morais, I.H.; Lam-Sidun, D.; Thome, C.; Zarnke, A.M.; Benjamin, L.V.; Losch, A.C.; Borgmann, A.J.; Sinex, H.C.; et al. Transcriptomic profiling of gamma ray induced mutants from the CGL1 human hybrid cell system reveals novel insights into the mechanisms of radiation-induced carcinogenesis. Free Radic. Biol. Med. 2019, 145, 300–311. [Google Scholar] [CrossRef]
- Pirkkanen, J.S.; Boreham, D.R.; Mendonca, M.S. The CGL1 (HeLa × Normal Skin Fibroblast) Human Hybrid Cell Line: History of Ionizing Radiation Induced Effects on Neoplastic Transformation and Novel Future Directions in SNOLAB. Radiat. Res. 2017, 188, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, M.A.; Omaruddin, R.A.; Brumbaugh, C.D.; Tariq, M.A.; Pourmand, N. Identification of radiation-induced microRNA transcriptome by next-generation massively parallel sequencing. J. Radiat. Res. 2013, 54, 808–822. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.D.; Liu, G.; Luo, L.; Xiao, J.; Gerrein, J.; Juan-Guardela, B.; Tedrow, J.; Alekseyev, Y.O.; Yang, I.V.; Correll, M.; et al. Assessment of microRNA differential expression and detection in multiplexed small RNA sequencing data. RNA 2015, 21, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Abnizova1, I.; Boekhorst, R.; Orlov, Y.L. Computational Errors and Biases in Short Read Next Generation Sequencing. J. Proteomics Bioinform. 2017, 10, 1. [Google Scholar] [CrossRef]
- Hulley, E.N.; Tharmalingam, S.; Zarnke, A.; Boreham, D.R. Development and validation of probe-based multiplex real-time PCR assays for the rapid and accurate detection of freshwater fish species. PLoS ONE 2019, 14, e0210165. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Wang, B.D.; Ceniccola, K.; Yang, Q.; Andrawis, R.; Patel, V.; Ji, Y.; Rhim, J.; Olender, J.; Popratiloff, A.; Latham, P.; et al. Identification and Functional Validation of Reciprocal microRNA-mRNA Pairings in African American Prostate Cancer Disparities. Clin. Cancer Res. 2015, 21, 4970–4984. [Google Scholar] [CrossRef]
- Cifuentes-Bernal, A.M.; Pham, V.V.; Xiaomei, L.; Lin, L.; Jiuyong, L.; Thuc, D.L. A pseudotemporal causality approach to identifying miRNA–mRNA interactions during biological processes. Bioinformatics 2021, 37, 807–814. [Google Scholar] [CrossRef]
- Santivasi, W.L.; Xia, F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid Redox. Signal. 2014, 21, 251–259. [Google Scholar] [CrossRef]
- Saville, M.K.; Sparks, A.; Xirodimas, D.P.; Wardrop, J.; Stevenson, L.F.; Bourdon, J.C.; Woods, Y.L.; Lane, D.P. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J. Biol. Chem. 2004, 279, 42169–42181. [Google Scholar] [CrossRef]
- Pant, V.; Lozano, G. Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev. 2014, 28, 1739–1751. [Google Scholar] [CrossRef]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef]
- Junttila, M.R.; Puustinen, P.; Niemelä, M.; Ahola, R.; Arnold, H.; Böttzauw, T.; Ala-aho, R.; Nielsen, C.; Ivaska, J.; Taya, Y.; et al. CIP2A inhibits PP2A in human malignancies. Cell 2007, 130, 51–62. [Google Scholar] [CrossRef]
- Cho, U.S.; Xu, W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 2007, 445, 53–57. [Google Scholar] [CrossRef]
- Mochida, S.; Ikeo, S.; Gannon, J.; Hunt, T. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 2009, 28, 2777–2785. [Google Scholar] [CrossRef]
- Yu, S.; Li, L.; Wu, Q.; Dou, N.; Li, Y.; Gao, Y. PPP2R2D, a regulatory subunit of protein phosphatase 2A, promotes gastric cancer growth and metastasis via mechanistic target of rapamycin activation. Int. J. Oncol. 2018, 52, 2011–2020. [Google Scholar] [CrossRef]
- Adams, D.G.; Coffee, R.L., Jr.; Zhang, H.; Pelech, S.; Strack, S.; Wadzinski, B.E. Positive regulation of Raf1-MEK1/2-ERK1/2 signaling by protein serine/threonine phosphatase 2A holoenzymes. J. Biol. Chem. 2005, 280, 42644–42654. [Google Scholar] [CrossRef]
- Baghdoyan, S.; Lamartine, J.; Castel, D.; Pitaval, A.; Roupioz, Y.; Franco, N.; Duarte, M.; Martin, M.T.; Gidrol, X. Id2 reverses cell cycle arrest induced by {gamma}-irradiation in human HaCaT keratinocytes. J. Biol. Chem. 2005, 280, 15836–15841. [Google Scholar] [CrossRef] [PubMed]
- Fukuma, M.; Okita, H.; Hata, J.I.; Umezawa, A. Upregulation of Id2, an oncogenic helix-loop-helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene 2003, 22, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Losada, M.; González, R.; Peropadre, A.; Gil-Gálvez, A.; Tena, J.J.; Baonza, A.; Estella, C. Coordination between cell proliferation and apoptosis after DNA damage in Drosophila. Cell Death Differ. 2022, 29, 832–845. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 2007, 9, 654–659. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).