Identification of Radiation-Induced miRNA Biomarkers Using the CGL1 Cell Model System
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Irradiation
2.3. Total RNA Extraction
2.4. cDNA Synthesis
2.5. miRNAome Profiling via Next-Generation Sequencing
2.6. RT-qPCR Primer Design and Validation
2.7. RT-qPCR
2.8. Statistical Analysis
3. Results
3.1. Identification of Dysregulated miRNA 6 h Post-Irradiation
3.2. Validation of the miRNAome Results Via RT-qPCR Analysis
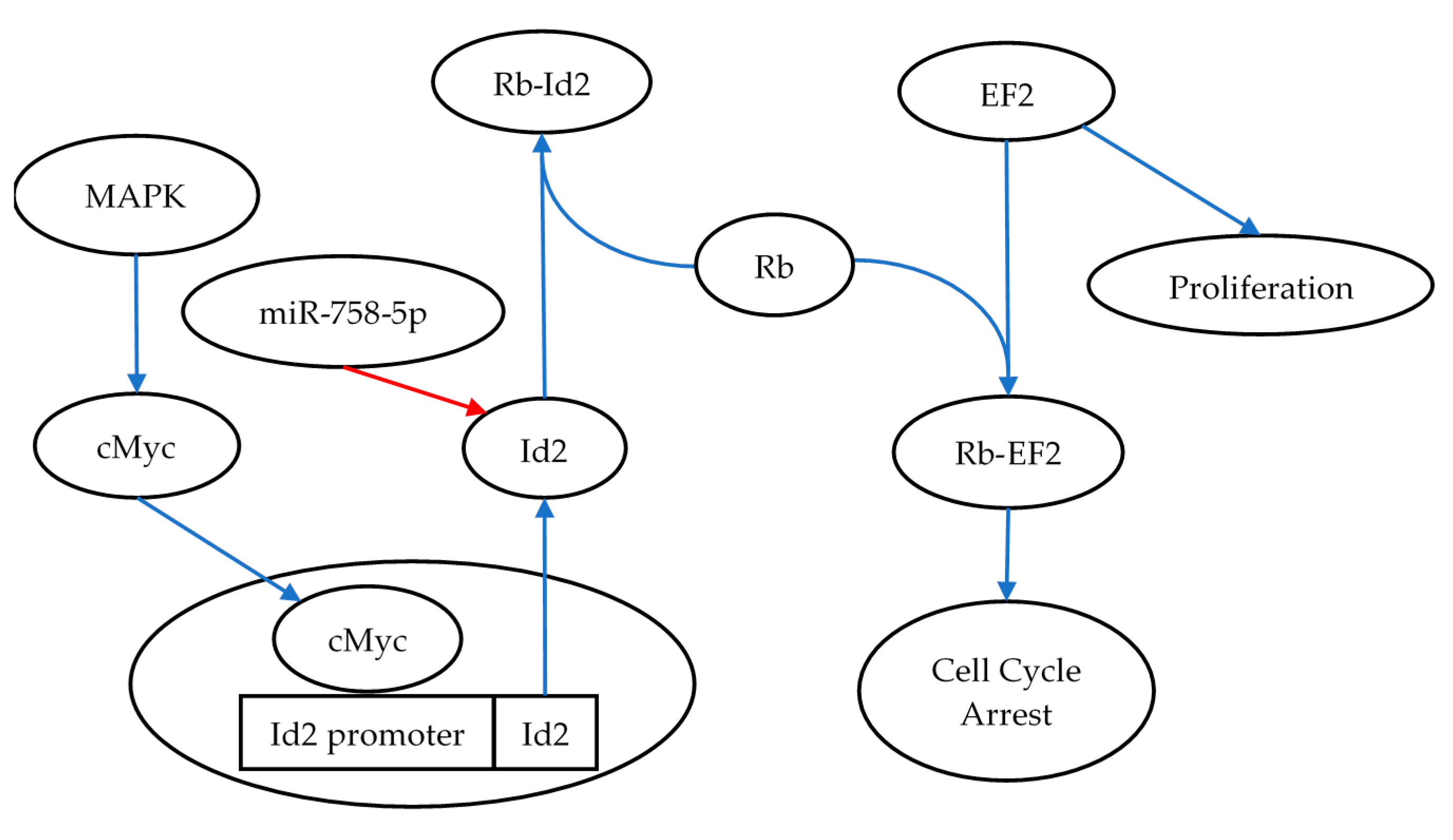
3.3. mRNA Gene Targets of miR-1228-3p and miR-758-5p Showed Reciprocal Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| miRNA | miRNA forward Primer | Tm |
| Oligo-dT adapter | GCATAGACCTGAATGGCGGTAAGGGTGTGGTAGGCGAGACATTTTTTTTTTTTTTTTTTTT | 65 |
| universal reverse | GCATAGACCTGAATGGCGGTA | 60 |
| SNORD48 | TGATGACCCCAGGTAACTCTGA | 60 |
| miR-16-5p | ATTGAAACCTCTAAGAGTGGA | 60 |
| U6 | GTG CTC GCT TCG GCA GCA CAT AT | 60 |
| miR-423-3p | AGCTCGGTCTGAGGCCC | 60 |
| miR-191-5p | CAACGGAATCCCAAAAGCAGC | 60 |
| miR-6134 | UGA GGU GGU AGG AUG UAG A | 60 |
| miR-6082 | GAATACGTCTGGTTGATCCAAAA | 57 |
| miR-208a-5p | AGCTTTTGGCCCGGGTTATAC | 61 |
| miR-7112-3p | CATCACAGCCTTTGGCCCTA | 60 |
| miR-6762-3p | UGGCUGCUUCCCUUGGUCUCCAG | 60 |
| miR-6882-3p | CTCTCCTCTTGCCTGCAGAA | 60 |
| miR-200a-5p | TCTTACCGGACAGTGCTGGA | 61 |
| miR-604 | GCTGCGGAATTCAGGACAAAA | 60 |
| miR-2355-5p | TCCCCAGATACAATGGACAAAA | 57 |
| miR-4692 | UCAGGCAGUGUGGGUAUCAGAU | 60 |
| miR-378e | ACTGGACTTGGAGTCAGGAAA | 59 |
| miR-4535 | GGACCTGGCTGGGACAAAA | 60 |
| miR-1915-3p | CGACGCGGCGGGAAA | 60 |
| miR-1291 | CCTGACTGAAGACCAGCAGTAAA | 60 |
| miR-19a-5p | AGTTTTGCATAGTTGCACTACAAA | 58 |
| miR-5008-3p | TCCCAGGGCCTCGCAAA | 61 |
| miR-4417 | TGGGCTTCCCGGAGGG | 60 |
| miR-1537-3p | AAAACCGUCUAGUUACAGUUGU | 60 |
| miR-3168 | GAGUUCUACAGUCAGAC | 60 |
| miR-5186 | AGAGAUUGGUAGAAAUCAGGU | 60 |
| miR-4680-5p | AGAACTCTTGCAGTCTTAGATGT | 57 |
| miR-554 | AGTCCTGACTCAGCCAGTAAAAA | 60 |
| miR-6845-3p | CCUCUCCUCCCUGUGCCCCAG | 60 |
| miR-6125 | GCGGAGCGGCGGAAAA | 61 |
| miR-3616-3p | CGAGGGCATTTCATGATGCAG | 60 |
| miR-615-3p | UCCGAGCCUGGGUCUCCCUCUU | 60 |
| miR-3152-3p | UGUGUUAGAAUAGGGGCAAUAA | 60 |
| miR-4693-3p | UGAGAGUGGAAUUCACAGUAUUU | 60 |
| miR-4320 | GGGAUUCUGUAGCUUCCU | 60 |
| miR-6783-5p | GGAAAAGTCCTGATCCGGAAAAA | 59 |
| miR-33b-5p | GUGCAUUGCUGUUGCAUUGC | 60 |
| miR-4662a-3p | AAAGAUAGACAAUUGGCUAAAU | 60 |
| miR-8082 | GATGGAGCTGGGAATACTCTGAA | 60 |
| miR-4758-5p | GCCGGTGGGGCTGAAAA | 60 |
| miR-8071 | GGACTGGAGTGGGTGGAAAAA | 60 |
| miR-4477b | AUUAAGGACAUUUGUGAUUGAU | 60 |
| miR-34a-5p | TGGCAGTGTCTTAGCTGGTTG | 60 |
| miR-182-5p | TTTGGCAATGGTAGAACTCACACT | 60 |
| miR-525-3p | AAGGCGCTTCCCTTTAGAGC | 60 |
| hsa-miR-492 | AGGACCTGCGGGACAAGA | 60 |
| hsa-miR-4535 | GTGGACCTGGCTGGGAC | 60 |
| miR-877-5p | GTAGAGGAGATGGCGCAGG | 60 |
| miR-133b | TTTGGTCCCCTTCAACCAGC | 60 |
| miR-205-3p | GATTTCAGTGGAGTGAAGTTC | 60 |
| miR-1 | ACATACTTCTTTATATGCCCAT | 60 |
| miR-19b | AGTTTTGCAGGTTTGCATCCAG | 60 |
| miR-93-5p | AAGTGCTGTTCGTGCAGGTAG | 60 |
| miR-132-5p | ACCGTGGCTTTCGATTGTTAC | 59 |
| miR-671-5p | CTGGAGGGGCTGGAGAAAAA | 60 |
| miR-628-3p | TCTAGTAAGAGTGGCAGTCGAA | 58 |
| miR-125b-1-3p | ACGGGTTAGGCTCTTGGGA | 60 |
| miR-6797-5p | GAAGGGGCTGAGAACAGGAAA | 60 |
| miR-6739-5p | TGGGAAAGAGAAAGAACAAGTAAAA | 57 |
| miR-6823-3p | GCCTCTCCTTCCCTCCAGAAA | 61 |
| miR-449c-3p | GCTAGTTGCACTCCTCTCTGT | 59 |
| miR-328-3p | CCTCTCTGCCCTTCCGTAAA | 59 |
| miR-103a-3p | CAGCATTGTACAGGGCTATGAA | 58 |
| miR-4721 | CTCCAGGTGACGGTGGAAAAA | 60 |
| miR-589-3p | CAGAACAAATGCCGGTTCCC | 60 |
| miR-769-3p | GGATCTCCGGGGTCTTGGT | 61 |
| miR-27a-3p | CACAGTGGCTAAGTTCCGCA | 61 |
| let-7g-5p | TGAGGTAGTAGTTTGTACAGTT | 60 |
| let-7a-5p | TGAGGTAGTAGGTTGTATAGTT | 60 |
| miR-296-5p | AGGGCCCCCCCTCAATCCTGT | 60 |
| miR-4443 | TTGGAGGCGTGGGTTTT | 60 |
| miR-3120-3p | ACAGCAAGTGTAGACAGGCAA | 60 |
| miR-423-3p | AGCTCGGTCTGAGGCCCCTCAGT | 60 |
| miR-362-5p | CCTTGGAACCTAGGTGTGAGT | 60 |
| miR-148b-5p | AGTTCTGTTATACACTCAGGCAA | 60 |
| miR-495-5p | GAAGTTGCCCATGTTATTTTCG | 60 |
| miR-6835-5p | AGGGGGUAGAAAGUGGCUGAAG | 60 |
| miR-4668-5p | AGGGAAAAAAAAAAGGAUUUGUC | 60 |
| miR-491-5p | GTGGGGAACCCTTCCATGAG | 60 |
| miR-4271 | GGGGGAAGAAAAGGTGGGG | 60 |
| miR-665 | GGAGGCTGAGGCCCCTAAA | 60 |
| miR-193a-5p | TCTTTGCGGGCGAGATGAAA | 60 |
| miR-22-3p | AGCTGCCAGTTGAAGAACTGTA | 60 |
| miR-1228-3p | CACACCTGCCTCGCCC | 60 |
| miR-181a-2-3p | CACTGACCGTTGACTGTACCA | 60 |
| miR-100-5p | ACCCGTAGATCCGAACTTGTG | 60 |
| miR-143-3p | TGAGATGAAGCACTGTAGCTC | 60 |
| miR-10b-5p | ACCCTGTAGAACCGAATTTGTGA | 60 |
| mRNA Primer | Sequence (5′→3′) | Length | Product (bp) | Tm |
| PPP2R2D_F_Hu_1 | GTCAAGGACAGGGCAGACTTC | 21 | 136 | 60 |
| PPP2R2D_R_Hu_2 | AGCTGTTCTCAGCTGTTCTATCA | 23 | ||
| UBE2D2_F_Hu_1 | CGTTTTGCCCGATCCACAAG | 20 | 145 | 60 |
| UBE2D2_R_Hu_1 | GTCCCGTGCCAGATCATTCA | 20 | ||
| IRX2_F_Hu_1 | CACCAAGATGACCCTCACCC | 20 | 119 | 60 |
| IRX2_R_Hu_1 | CGTCCTCGTCCTCATCTTCG | 20 | ||
| ZNF554_F_Hu_1 | CTGCAGTACTGTGCTGATCCA | 21 | 137 | 60 |
| ZNF554_R_Hu_1 | ACGTCTCTGTCCAGCCTTTG | 20 | ||
| NFIA_F_Hu_1 | TGCCGAATCGATTGCAACTTC | 21 | 83 | 60 |
| NFIA_R_Hu_1 | GGCTGGTTCTCAGATTCGCT | 20 | ||
| SOCS6_F_Hu_1 | GGCCGCCTCCGGAAAAT | 17 | 81 | 60 |
| SOCS6_R_Hu_1 | ACATCTGGAGAGGCTGCAAG | 20 | ||
| RABGEF1_F_Hu_1 | GCGGTGACCTGGACCAC | 17 | 82 | 60 |
| RABGEF1_R_Hu_1 | TTCCCAAAGTGATCTCGCCC | 20 | ||
| TJP1_F_Hu_1 | TGGTCTGTTTGCCCACTGTT | 20 | 150 | 60 |
| TJP1_R_Hu_1 | TCTGTACATGCTGGCCAAGG | 20 | ||
| TOR1AIP1_F_Hu_1 | AAGTCCTCTAGTGCAACGCC | 20 | 82 | 60 |
| TOR1AIP1_R_Hu_1 | ATCTTGGCTTGAGGCACTCC | 20 | ||
| ZBTB44_F_Hu_1 | ACGAGTGCAAAACATGTGGC | 20 | 72 | 60 |
| ZBTB44_R_Hu_1 | GGTTCAGACTCCTCAGGTGC | 20 | ||
| Moap1_F_Hu_1 | AGGCCCTTCTCCAGGCAATA | 20 | 70 | 60 |
| Moap1_R_Hu_1 | TGCCATATCCCTTCGTGGTT | 20 | ||
| Csnk2a1_F_Hu_1 | ATCGCCGCCATATTGTCTGT | 20 | 109 | 60 |
| Csnk2a1_R_Hu_1 | CAGCTGGGGGTAAGACCTTG | 20 | ||
| PLAC8_F_Hu_1 | CAGAAGGAGAGCCATGCGTA | 20 | 76 | 60 |
| PLAC8_R_Hu_1 | AACCCACATGTTCTGAGAGGC | 21 | ||
| SLC20A2_F_Hu_1 | TCTCGGCCTAATGTGGTAGGA | 21 | 138 | 60 |
| SLC20A2_R_Hu_1 | CTCCCGATCTGGGAAAGCTG | 20 | ||
| ID2_F_Hu_1 | ATCCTGTCCTTGCAGGCTTC | 20 | 81 | 60 |
| ID2_R_Hu_1 | ACCGCTTATTCAGCCACACA | 20 | ||
| NUFIP2_F_Hu_1 | ATGTCCATTTTGCTTGCCTGG | 21 | 149 | 60 |
| NUFIP2_R_Hu_1 | CCCAATTCAGGTGGGGTCTG | 20 | ||
| PTP4A1_F_Hu_1 | CTGTGAGCTCTTAAGACTTGCTT | 23 | 73 | 58 |
| PTP4A1_R_Hu_1 | CACTGCTGCTGGGAATTATGA | 21 | ||
| TOX4_F_Hu_1 | CTGACGATCACAGGGCCTTC | 20 | 142 | 60 |
| TOX4_R_Hu_1 | GGCCAACCACATCTGAGACA | 20 | ||
| SETD5_F_Hu_1 | CTGTGACAAGTGCAGGGGAA | 20 | 95 | 60 |
| SETD5_R_Hu_1 | CTGTTGCACTGCTATCCCCA | 20 | ||
| PHACTR1_F_Hu_1 | TGTTCATTTGTGCTTGCGGG | 20 | 74 | 60 |
| PHACTR1_R_Hu_1 | CCCTTTCAACAGGACACGGT | 20 | ||
| RTKN_F_Hu_1 | CGAGTGAAGTGTGACTCCGT | 20 | 90 | 60 |
| RTKN_R_Hu_1 | TTCCAGACAGGAAACCAGCC | 20 | ||
| DSG3_F_Hu_1 | GACTCCTTCGGAAAGCAGCA | 20 | 138 | 60 |
| DSG3_R_Hu_1 | GGGGAAGAGCCCCATCATTG | 20 | ||
| CSNK1A1L_F_Hu_1 | CCCTGGGGTTTGCAAATTGT | 20 | 138 | 60 |
| CSNK1A1L_R_Hu_1 | TCTTCACAGGTAAGCAGGCG | 20 | ||
| CD36_F_Hu_1 | CCACACACTGGGATCTGACA | 20 | 132 | 60 |
| CD36_R_Hu_1 | TCTGCAGGAAAGTCCTACACTG | 22 | ||
| ZBTB20_F_Hu_1 | TCCTGACAAATGCTAGAACGGA | 22 | 86 | 60 |
| ZBTB20_R_Hu_1 | CCACCCGGCTGAGTAATCTC | 20 | ||
| CBX5_F_Hu_1 | GGGAGGCCCCTCCTGTTAG | 19 | 72 | 60 |
| CBX5_R_Hu_1 | AAGACTAAGGCCACCAGGTC | 20 | ||
| CTTN_F_Hu_1 | GACAAATGTGCCCTTGGCTG | 20 | 111 | 60 |
| CTTN_R_Hu_1 | CTGCCTCTCCGACTGAACAC | 20 | ||
| NCOA2_F_Hu_1 | CCCTCCCTCTACCACAGTCA | 20 | 87 | 60 |
| NCOA2_R_Hu_1 | CAGAGTCCTCTGAGAAGGCG | 20 | ||
| ZNF281_F_Hu_1 | ATGACCACCATGGCACTGAG | 20 | 70 | 60 |
| ZNF281_R_Hu_1 | TCTGGCTTTGGCCTTTTTGC | 20 | ||
| TNPO1_F_Hu_1 | TGCCCGGCCGTTTGAAG | 17 | 121 | 60 |
| TNPO1_R_Hu_1 | GCTCGTCAGGTTTCCACTCA | 20 | ||
| HNRNPD_F_Hu_1 | GCCATTCAAACTCCTCCCCA | 20 | 89 | 60 |
| HNRNPD_R_Hu_1 | GTCCCAGCTAAGGCCTCCTA | 20 | ||
| SBNO1_F_Hu_1 | CAATGCCTACCCCGTCAGTT | 20 | 72 | 60 |
| SBNO1_R_Hu_1 | CTGCTTCGGTCTCCAAACCT | 20 | ||
| DIPK2A_F_Hu_1 | GTGGGTGTGAGACATCCTAGC | 21 | 74 | 60 |
| DIPK2A_R_Hu_1 | CACGACAAGTGGGGTCTGC | 19 | ||
| MCUB_F_Hu_1 | GGAGGATGCTCCAGAGGGG | 19 | 119 | 60 |
| MCUB_R_Hu_1 | CTTCACACGCAAAACCTGGG | 20 | ||
| NOL4L_F_Hu_1 | GCCAAAACCAAGACGGTGAC | 20 | 124 | 6 |
| NOL4L_R_Hu_1 | CCCAGAACTGGAACTTGCCT | 20 | ||
| CAMTA1_F_Hu_1 | GAAAACAAGCCGGAAGAGCG | 20 | 81 | 60 |
| CAMTA1_R_Hu_1 | ATAGGTGGCACGGTGTTGAG | 202 | ||
| cdk6_F_Hu_1 | CTGCAGGGAAAGAAAAGTGCAA | 22 | 95 | 60 |
| cdk6_R_Hu_1 | CTCCTCGAAGCGAAGTCCTC | 20 | ||
| MTA3_F_Hu_1 | GTCCTCCCCCTCCGCTC | 17 | 131 | 60 |
| MTA3_R_Hu_1 | CTGGGGACTGCCCAATTCAT | 20 | ||
| TRIM50_F_Hu_1 | GGCATCTAACTGGAGCGACA | 20 | 85 | 60 |
| TRIM50_R_Hu_1 | CCAAGCCATCCACACTCACT | 20 | ||
| SGIP1_F_Hu_1 | TGGAATTCCTTCAGGCGGAC | 20 | 73 | 60 |
| SGIP1_R_Hu_1 | ACGATTCCAGGTCCCAGCTA | 20 | ||
| PLAGL2_F_Hu_1 | ACAATGCACCGCACAATGG | 19 | 138 | 60 |
| PLAGL2_R_Hu_1 | CCTCCAACGCAGCTTTCAGA | 20 | ||
| PRKACB_F_Hu_1 | GCTAGCAGTAAGAGCTGGTGT | 21 | 75 | 60 |
| PRKACB_R_Hu_1 | TGAACCTGGCAAGGAGCAAA | 20 | ||
| LUC7L3_F_Hu_1 | GGTCAATGGGACCAGTGAAGA | 21 | 147 | 60 |
| LUC7L3_R_Hu_1 | CGCTGCACTGTCAAACAGTAA | 21 | ||
| EDEM1_F_Hu_1 | ACAACTACATGGCTCACGCC | 20 | 91 | 60 |
| EDEM1_R_Hu_1 | AGATTTGAAGGGTCCCCGC | 19 | ||
| CYP1B1_F_Hu_1 | GCTGTGAGGAAACCTCGACT | 20 | 121 | 60 |
| CYP1B1_R_Hu_1 | GAGTCTCTTGGCGTCGTCAG | 20 | ||
| RMI1_F_Hu_1 | GCGGTTCCTGTCCTTACAGT | 19 | 86 | 60 |
| RMI1_R_Hu_1 | ACTGCTCAGAAATGGCCCTG | 20 | ||
| MRPL35_F_Hu_1 | TGCAAAGAAATTGGGTCTGTGT | 22 | 141 | 60 |
| MRPL35_R_Hu_1 | TGAAGGGCCACCCTTAAACC | 20 | ||
| PIK3C2B_F_Hu_1 | CCACCATAGAGATGGCGTCC | 20 | 134 | 60 |
| PIK3C2B_R_Hu_1 | TGGGCGCCTGATTCTTCTAC | 20 | ||
| Cyld_F_Hu_1 | CCCCCTTTCTAGGGTGAGGA | 20 | 98 | 60 |
| Cyld_R_Hu_1 | TTCAGCAACGTGGTGTCCAT | 20 | ||
| GAS7_F_Hu_1 | AGCCAACGAGTCTCTGCTTC | 20 | 79 | 60 |
| GAS7_R_Hu_1 | GCCGTCTCTGGGGTGC | 16 | ||
| LRRC27_F_Hu_1 | CTCGCCAGCGCTTCAGT | 17 | 102 | 60 |
| LRRC27_R_Hu_1 | TAGGAGCTGCTTCCCTCCAT | 20 | ||
| FMNL3_F_Hu_1 | GAGTCGGGACTCGGGGAG | 18 | 130 | 60 |
| FMNL3_R_Hu_1 | CTCTCCAGGTTGCCCATCG | 9 | ||
| TTC21B_F_Hu_1 | TGCGTCTTCCTTTAGGCTGC | 20 | 87 | 60 |
| TTC21B_R_Hu_1 | TCTTCAATTCCTGCGAGTCCA | 21 | ||
| CASTOR3_F_Hu_1 | AGCTTTTCCAGACCAGGCAT | 20 | 76 | 60 |
| CASTOR3_R_Hu_1 | CTAGGGGCTGATGTGCCAAA | 20 | ||
| XPO7_F_Hu_1 | GCAATCACAGACGTCACAAGG | 21 | 126 | 60 |
| XPO7_R_Hu_1 | TGCTTCAATGAGGAAGGCTGT | 21 | ||
| PPM1A_F_Hu_1 | CTGCTCCGGACCTAGAGGAT | 20 | 124 | 60 |
| PPM1A_R_Hu_1 | CAGCCTTGCATGCTGCTTAG | 20 | ||
| DNM1L_F_Hu_1 | TCACCCGGAGACCTCTCATT | 20 | 91 | 60 |
| DNM1L_R_Hu_1 | TCTGCTTCCACCCCATTTTCT | 21 | ||
| KDM3B_F_Hu_1 | CTGCGCACTCGAGCCTG | 17 | 114 | 60 |
| KDM3B_R_Hu_1 | CCAGGAGTGTTGCTTCCAGT | 20 | ||
| RBP1_F_Hu_1 | CCGCTACAATGGATCCTCCC | 20 | 96 | 60 |
| RBP1_R_Hu_1 | GGAAATGAGCGCCCTCCG | 18 | ||
| FAAP20_F_Hu_1 | GGGTCCCCTTCTCCACTGTA | 20 | 78 | 60 |
| FAAP20_R_Hu_1 | CTGGCAGGAGCTGGAGATG | 20 | ||
| PTEN_F_Hu_1 | CTGCAGAAAGACTTGAAGGCG | 21 | 70 | 58 |
| PTEN_R_Hu_1 | TGCTTTGAATCCAAAAACCTTACT | 24 | ||
| GOLPH3_F_Hu_1 | TGTTTCCTCATGACTGCCCC | 20 | 80 | 60 |
| GOLPH3_R_Hu_1 | CGATCCGGGTTTCCGTGTTA | 20 | ||
| ADAMTS3_F_Hu_1 | CAAGCATTCTCCGCGCTAAC | 20 | 147 | 60 |
| ADAMTS3_R_Hu_1 | GGAGCGAGAAGGTGCTGTAA | 20 | ||
| KCTD9_F_Hu_1 | CCCAAGAACGGAAAGGTGGT | 20 | 88 | 60 |
| KCTD9_R_Hu_1 | TGGTGGCTTTTATGCCGAGT | 20 | ||
| FBN2_F_Hu_1 | GTTTTCTGCCAGTCATCCAGC | 21 | 142 | 60 |
| FBN2_R_Hu_1 | AGCTGCTTTGGCTTCGATCT | 20 | ||
| SEC63_F_Hu_1 | GGACATAAATAGGGCAATCCACT | 23 | 119 | 58 |
| SEC63_R_Hu_1 | CCCTCTCACTCCTGGGTTTT | 20 | ||
| ZFX_F_Hu_1 | ACCCTAGTGGAGTGTTGGCT | 20 | 123 | 60 |
| ZFX_R_Hu_1 | TGAACCACTGAAGGGAGTCG | 20 | ||
| DAPK1_F_Hu_1 | TTCGGAGTGTGAGGAGGACA | 20 | 149 | 60 |
| DAPK1_R_Hu_1 | GGGAACACAGCTAGGGAGTG | 20 | ||
| SMIM13_F_Hu_1 | AGTGGGTGAAAATTCCCGCT | 20 | 94 | 60 |
| SMIM13_R_Hu_1 | CCCTGGTAAACACTCAGCCC | 20 | ||
| NAP1L5_F_Hu_1 | CTCCTAGACCTCTGCGGCTT | 20 | 147 | 62 |
| NAP1L5_R_Hu_1 | GCTGTCACAGTCTCCACCCT | 20 | ||
| CSDE1_F_Hu_1 | CGCTGAGCTGTTGGGTATGA | 20 | 78 | 60 |
| CSDE1_R_Hu_1 | ACGAGGTTTGTTCCTTGCCT | 20 | ||
| set_F_Hu_1 | AGTCTCAGTGTTCAGCCTGC | 20 | 78 | 60 |
| set_R_Hu_1 | GGCCATGCTGTTAGGGAAGT | 20 | ||
| OLFM4_F_Hu_1 | AAATGCTCGAGAGTTGCGGA | 20 | 133 | 60 |
| OLFM4_R_Hu_1 | CACAGCAATCGTGTTGGTGG | 20 | ||
| USP6NL_F_Hu_1 | TGGAAGGGAAACAATGGGGC | 20 | 144 | 60 |
| USP6NL_R_Hu_1 | CATGTCCTCAGTACGGTCCC | 20 | ||
| AVPR1A_F_Hu_1 | TGGGCGCCTTTCTTCATCAT | 20 | 75 | 60 |
| AVPR1A_R_Hu_1 | AGGGTTTTCCGATTCGGTCC | 20 | ||
| HSD11B1_F_Hu_1 | TGCCTGCTTAGGAGGTTGTAG | 21 | 90 | 60 |
| HSD11B1_R_Hu_1 | AAAAGCCATCCGACAGGGAG | 20 | ||
| GALNT15_F_Hu_1 | ACCCAGATGGCTTATTGCCT | 20 | 126 | 60 |
| GALNT15_R_Hu_1 | TACCAGCCAGGGACTGAGTT | 20 | ||
| GBP5_F_Hu_1 | AGTTCTGCTTGACACCGAGG | 20 | 149 | 60 |
| GBP5_R_Hu_1 | GCAGTAGGTCGATAGCACCC | 20 | ||
| CD200_F_Hu_1 | TCCAGGAGCAAGGATGGAGA | 20 | 76 | 60 |
| CD200_R_Hu_1 | GACCCAAACCAGGCTGTAGG | 20 | ||
| PPM1A_F_Hu_1 | CTGCTCCGGACCTAGAGGAT | 20 | 124 | 60 |
| PPM1A_R_Hu_1 | CAGCCTTGCATGCTGCTTAG | 20 | ||
| NPHP1_F_Hu_1 | CCCACTTCTCCACTCCACAC | 20 | 76 | 60 |
| NPHP1_R_Hu_1 | AACTTTCCACCGTGCAGTCT | 20 | ||
| Rock_F_Hu_1 | CCCTTTGCTTTCGCCTTTCC | 20 | 150 | 60 |
| Rock1_R_Hu_1 | GAGGTGCTTCAGTCTAGCGG | 20 | ||
| mmp14_F_Hu_1 | GGGTCTTCGTTGCTCAGTCA | 20 | 145 | 60 |
| mmp14_R_Hu_1 | AACATTCGAGAGGCACAGGG | 20 | ||
| kcne2_F_Hu_1 | ACGGGAACACTCCAATGACC | 20 | 105 | 60 |
| kcne2_R_Hu_1 | TGGATGGTGGCCTTCGATTC | 20 | ||
| WWP1_F_Hu_1 | TGCTACTTTTAGCAAACTGGGC | 22 | 128 | 58 |
| WWP1_R_Hu_1 | TTAAGAAGTCAGTTCCATGGCT | 22 | ||
| cdk16_F_Hu_1 | TTGGGCCGTTGGCTGTTC | 18 | 70 | 60 |
| cdk16_R_Hu_1 | GGCTCGCGGCACAGAG | 16 | ||
| SEMA6A_F_Hu_1 | CTTACAACACAGTGTATGGGCA | 22 | 81 | 60 |
| SEMA6A_R_Hu_1 | CATACCCCTCTTGAGCCGTC | 20 | ||
| CHRNB2_F_Hu_1 | GAAAGTTCGGCTCCCTTCCA | 20 | 115 | 60 |
| CHRNB2_R_Hu_1 | GCCATCATAGGAGACCACGG | 20 | ||
| POGK_F_Hu_1 | TGGGAAGTTTTGACGGAGCA | 20 | 113 | 60 |
| POGK_R_Hu_1 | CGGGTGATCATGTCTGGCTT | 20 | ||
| KSR2_F_Hu_1 | AAAGCACTCCAAACCGTGGA | 20 | 133 | 60 |
| KSR2_R_Hu_1 | ACTCTTTACACACCGGCTCC | 20 | ||
| SAMD4B_F_Hu_1 | CCTTCCTACTGGGCAGATGAG | 21 | 101 | 60 |
| SAMD4B_R_Hu_1 | CCAGAAGTGGACATGGGGTA | 20 | ||
| OPTC_F_Hu_1 | TGTCTTCAACCTGGCCTGTC | 20 | 70 | 60 |
| OPTC_R_Hu_1 | AGTACACAAGCCCATCCAGG | 20 | ||
| CPNE5_F_Hu_1 | TGTGTCCAACGGTGGTGTC | 19 | 113 | 60 |
| CPNE5_R_Hu_1 | CCAGCTTGTTGGCACAGAAC | 20 | ||
| FOXP4_F_Hu_1 | AGCTGATTTGCTGCAGGGAT | 20 | 100 | 60 |
| FOXP4_R_Hu_1 | GAAGGACACCTGGGAATGGG | 20 | ||
| MSN_F_Hu_1 | GCCCAAAACGATCAGTGTGC | 20 | 88 | 60 |
| MSN_R_Hu_1 | AAATAGCTGCTTCCCGGTGG | 20 | ||
| IGF2_F_Hu_1 | TCGCCGAACCAAAGTGGATTA | 21 | 148 | 60 |
| IGF2_R_Hu_1 | GTGGGAGAGACAGAGTGAACG | 21 | ||
| Smad3_F_Hu_1 | CCGGGGGTTGGACTTTCCT | 19 | 70 | 60 |
| Smad3_R_Hu_1 | CAGAAGTTTGGGTTTCCGCA | 20 | ||
| Bcl2l1_F_Hu_1 | AGGCGGATTTGAATCTCTTTCTCT | 24 | 129 | 60 |
| Bcl2l1_R_Hu_1 | GGGCTCAACCAGTCCATTGT | 20 | ||
| Egfr_F_Hu_1 | GACAGGCCACCTCGTCG | 17 | 106 | 60 |
| Egfr_R_Hu_1 | CCGGCTCTCCCGATCAATAC | 20 | ||
| Notch3_F_Hu_1 | TCTAGGTAAGGTGGGGAGTGG | 21 | 70 | 60 |
| Notch3_R_Hu_1 | TGGGAGCTCAAGTTAGCCCT | 20 | ||
| Mmp9 _F_Hu_1 | GTACTCGACCTGTACCAGCG | 20 | 92 | 60 |
| Mmp9 _R_Hu_1 | AGAAGCCCCACTTCTTGTCG | 20 | ||
| Igf2bp1_F_Hu_1 | AGCTCCTTTATGCAGGCTCC | 20 | 111 | 60 |
| lgf2bp1_R_Hu_1 | CCGGGAGAGCTGTTTGATGT | 20 | ||
| Wnt3a_F_Hu_1 | CTCCTCCCTGGAGCTAGTGT | 20 | 138 | 60 |
| Wnt3a_R_Hu_1 | AATCTGTAGCCCCGCCTCTG | 20 | ||
| capns1_F_Hu_1 | CGGACGCTGCGGGAG | 15 | 72 | 60 |
| capns1_R_Hu_1 | TCACTGCGCCGCACAC | 16 |
References
- Rothkamm, K.; Beinke, C.; Romm, H.; Badie, C.; Balagurunathan, Y.; Barnard, S.; Bernard, N.; Boulay-Greene, H.; Brengues, M.; de Amicis, A. Comparison of established and emerging biodosimetry assays. Radiat. Res. 2013, 180, 111–119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, Y.; Jin, Y.W.; Wilkins, R.C.; Jang, S. Validation of the dicentric chromosome assay for radiation biological dosimetry in South Korea. J. Radiat. Res. 2019, 60, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Vral, A.; Fenech, M.; Thierens, H. The micronucleus assay as a biological dosimeter of in vivo ionising radiation exposure. Mutagenesis 2011, 26, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.M.; Prasanna, P.G.; Grace, M.B.; Wathen, L.K.; Wallace, R.L.; Koerner, J.F.; Coleman, C.N. Assessment of biodosimetry methods for a mass-casualty radiological incident: Medical response and management considerations. Health Phys. 2013, 105, 540–554. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Li, J.R.; Tong, C.Y.; Sung, T.J.; Kang, T.Y.; Zhou, X.J.; Liu, C.C. CMEP: A database for circulating microRNA expression profiling. Bioinformatics 2019, 35, 3127–3132. [Google Scholar] [CrossRef]
- Enelund, L.; Nielsen, L.N.; Cirera, S. Evaluation of microRNA Stability in Plasma and Serum from Healthy Dogs. Microrna 2017, 6, 42–52. [Google Scholar] [CrossRef]
- Glinge, C.; Clauss, S.; Boddum, K.; Jabbari, R.; Jabbari, J.; Risgaard, B.; Tomsits, P.; Hildebrand, B.; Kääb, S.; Wakili, R.; et al. Stability of Circulating Blood-Based MicroRNAs—Pre-Analytic Methodological Considerations. PLoS ONE 2017, 12, e0167969. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Hu, H.Y.; Yan, Z.; Xu, Y.; Hu, H.; Menzel, C.; Zhou, Y.H.; Chen, W.; Khaitovich, P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genom. 2009, 10, 413. [Google Scholar] [CrossRef]
- Tharmalingam, S.; Sreetharan, S.; Brooks, A.L.; Boreham, D.R. Re-evaluation of the linear no-threshold (LNT) model using new paradigms and modern molecular studies. Chem. Biol. Interact. 2019, 301, 54–67. [Google Scholar] [CrossRef]
- Tharmalingam, S.; Sreetharan, S.; Kulesza, A.V.; Boreham, D.R.; Tai, T.C. Low-Dose Ionizing Radiation Exposure, Oxidative Stress and Epigenetic Programing of Health and Disease. Radiat. Res. 2017, 188, 525–538. [Google Scholar] [CrossRef]
- Puukila, S.; Tharmalingam, S.; Al-Khayyat, W.; Peterson, J.; Hooker, A.M.; Muise, S.; Boreham, D.R.; Dixon, D.L. Transcriptomic Response in the Spleen after Whole-Body Low-Dose X-ray Irradiation. Radiat. Res. 2021, 196, 66–73. [Google Scholar] [CrossRef]
- Mao, A.; Zhao, Q.; Zhou, X.; Sun, C.; Si, J.; Zhou, R.; Gan, L.; Zhang, H. MicroRNA-449a enhances radiosensitivity by downregulation of c-Myc in prostate cancer cells. Sci. Rep. 2016, 6, 27346. [Google Scholar] [CrossRef]
- Duan, X.M.; Liu, X.N.; Li, Y.X.; Cao, Y.Q.; Silayiding, A.; Zhang, R.K.; Wang, J.P. MicroRNA-498 promotes proliferation, migration, and invasion of prostate cancer cells and decreases radiation sensitivity by targeting PTEN. Kaohsiung J. Med. Sci. 2019, 35, 659–671. [Google Scholar] [CrossRef]
- Körner, C.; Keklikoglou, I.; Bender, C.; Wörner, A.; Münstermann, E.; Wiemann, S. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J. Biol. Chem. 2013, 288, 8750–8761. [Google Scholar] [CrossRef]
- Zaleska, K.; Przybyła, A.; Kulcenty, K.; Wichtowski, M.; Mackiewicz, A.; Suchorska, W.; Murawa, D. Wound fluids affect miR-21, miR-155 and miR-221 expression in breast cancer cell lines, and this effect is partially abrogated by intraoperative radiation therapy treatment. Oncol. Lett. 2017, 14, 4029–4036. [Google Scholar] [CrossRef]
- Chaudhry, M.A.; Sachdeva, H.; Omaruddin, R.A. Radiation-induced micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol. 2010, 29, 553–561. [Google Scholar] [CrossRef]
- Maia, D.; de Carvalho, A.C.; Horst, M.A.; Carvalho, A.L.; Scapulatempo-Neto, C.; Vettore, A.L. Expression of miR-296-5p as predictive marker for radiotherapy resistance in early-stage laryngeal carcinoma. J. Transl. Med. 2015, 13, 262. [Google Scholar] [CrossRef]
- Stanbridge, E.J.; Flandermeyer, R.R.; Daniels, D.W.; Nelson-Rees, W.A. Specific chromosome loss associated with the expression of tumorigenicity in human cell hybrids. Somatic. Cell Genet. 1981, 7, 699–712. [Google Scholar] [CrossRef]
- Stanbridge, E.J.; Wilkinson, J. Dissociation of anchorage independence form tumorigenicity in human cell hybrids. Int. J. Cancer 1980, 26, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pirkkanen, J.; Tharmalingam, S.; Morais, I.H.; Lam-Sidun, D.; Thome, C.; Zarnke, A.M.; Benjamin, L.V.; Losch, A.C.; Borgmann, A.J.; Sinex, H.C.; et al. Transcriptomic profiling of gamma ray induced mutants from the CGL1 human hybrid cell system reveals novel insights into the mechanisms of radiation-induced carcinogenesis. Free Radic. Biol. Med. 2019, 145, 300–311. [Google Scholar] [CrossRef]
- Pirkkanen, J.S.; Boreham, D.R.; Mendonca, M.S. The CGL1 (HeLa × Normal Skin Fibroblast) Human Hybrid Cell Line: History of Ionizing Radiation Induced Effects on Neoplastic Transformation and Novel Future Directions in SNOLAB. Radiat. Res. 2017, 188, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, M.A.; Omaruddin, R.A.; Brumbaugh, C.D.; Tariq, M.A.; Pourmand, N. Identification of radiation-induced microRNA transcriptome by next-generation massively parallel sequencing. J. Radiat. Res. 2013, 54, 808–822. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.D.; Liu, G.; Luo, L.; Xiao, J.; Gerrein, J.; Juan-Guardela, B.; Tedrow, J.; Alekseyev, Y.O.; Yang, I.V.; Correll, M.; et al. Assessment of microRNA differential expression and detection in multiplexed small RNA sequencing data. RNA 2015, 21, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Abnizova1, I.; Boekhorst, R.; Orlov, Y.L. Computational Errors and Biases in Short Read Next Generation Sequencing. J. Proteomics Bioinform. 2017, 10, 1. [Google Scholar] [CrossRef]
- Hulley, E.N.; Tharmalingam, S.; Zarnke, A.; Boreham, D.R. Development and validation of probe-based multiplex real-time PCR assays for the rapid and accurate detection of freshwater fish species. PLoS ONE 2019, 14, e0210165. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Wang, B.D.; Ceniccola, K.; Yang, Q.; Andrawis, R.; Patel, V.; Ji, Y.; Rhim, J.; Olender, J.; Popratiloff, A.; Latham, P.; et al. Identification and Functional Validation of Reciprocal microRNA-mRNA Pairings in African American Prostate Cancer Disparities. Clin. Cancer Res. 2015, 21, 4970–4984. [Google Scholar] [CrossRef]
- Cifuentes-Bernal, A.M.; Pham, V.V.; Xiaomei, L.; Lin, L.; Jiuyong, L.; Thuc, D.L. A pseudotemporal causality approach to identifying miRNA–mRNA interactions during biological processes. Bioinformatics 2021, 37, 807–814. [Google Scholar] [CrossRef]
- Santivasi, W.L.; Xia, F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid Redox. Signal. 2014, 21, 251–259. [Google Scholar] [CrossRef]
- Saville, M.K.; Sparks, A.; Xirodimas, D.P.; Wardrop, J.; Stevenson, L.F.; Bourdon, J.C.; Woods, Y.L.; Lane, D.P. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J. Biol. Chem. 2004, 279, 42169–42181. [Google Scholar] [CrossRef]
- Pant, V.; Lozano, G. Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev. 2014, 28, 1739–1751. [Google Scholar] [CrossRef]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef]
- Junttila, M.R.; Puustinen, P.; Niemelä, M.; Ahola, R.; Arnold, H.; Böttzauw, T.; Ala-aho, R.; Nielsen, C.; Ivaska, J.; Taya, Y.; et al. CIP2A inhibits PP2A in human malignancies. Cell 2007, 130, 51–62. [Google Scholar] [CrossRef]
- Cho, U.S.; Xu, W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 2007, 445, 53–57. [Google Scholar] [CrossRef]
- Mochida, S.; Ikeo, S.; Gannon, J.; Hunt, T. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 2009, 28, 2777–2785. [Google Scholar] [CrossRef]
- Yu, S.; Li, L.; Wu, Q.; Dou, N.; Li, Y.; Gao, Y. PPP2R2D, a regulatory subunit of protein phosphatase 2A, promotes gastric cancer growth and metastasis via mechanistic target of rapamycin activation. Int. J. Oncol. 2018, 52, 2011–2020. [Google Scholar] [CrossRef]
- Adams, D.G.; Coffee, R.L., Jr.; Zhang, H.; Pelech, S.; Strack, S.; Wadzinski, B.E. Positive regulation of Raf1-MEK1/2-ERK1/2 signaling by protein serine/threonine phosphatase 2A holoenzymes. J. Biol. Chem. 2005, 280, 42644–42654. [Google Scholar] [CrossRef]
- Baghdoyan, S.; Lamartine, J.; Castel, D.; Pitaval, A.; Roupioz, Y.; Franco, N.; Duarte, M.; Martin, M.T.; Gidrol, X. Id2 reverses cell cycle arrest induced by {gamma}-irradiation in human HaCaT keratinocytes. J. Biol. Chem. 2005, 280, 15836–15841. [Google Scholar] [CrossRef] [PubMed]
- Fukuma, M.; Okita, H.; Hata, J.I.; Umezawa, A. Upregulation of Id2, an oncogenic helix-loop-helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene 2003, 22, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Losada, M.; González, R.; Peropadre, A.; Gil-Gálvez, A.; Tena, J.J.; Baonza, A.; Estella, C. Coordination between cell proliferation and apoptosis after DNA damage in Drosophila. Cell Death Differ. 2022, 29, 832–845. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 2007, 9, 654–659. [Google Scholar] [CrossRef]



| Dose (mGy) | miRNA | Fold Change | p-Value |
|---|---|---|---|
| 10 | miR-3120-3p | 11.3 | 0.042 |
| miR-4443 | 1.9 | 0.048 | |
| 100 | miR-4443 | 2.3 | 0.009 |
| miR-362-5p | 2.1 | 0.034 | |
| miR-148b-5p | 2.0 | 0.043 | |
| miR-423-3p | 1.9 | 0.016 | |
| miR-500b-5p | 1.7 | 0.040 | |
| miR-502-3p | 1.6 | 0.040 | |
| miR-125b-1-3p | 1.6 | 0.041 | |
| miR-495-5p | -1.7 | 0.040 | |
| 1000 | miR-3168 | 8.8 | 0.0008 |
| miR-671-5p | 5.3 | 0.003 | |
| miR-6835-5p | 4.2 | 0.001 | |
| miR-5694 | 4.1 | 0.009 | |
| miR-491-5p | 3.9 | 0.005 | |
| miR-2054 | 3.6 | 0.003 | |
| miR-4668-5p | 3.5 | 0.002 | |
| miR-6069 | 3.4 | 0.019 | |
| miR-23a-5p | 3.3 | 0.003 | |
| miR-3135b | 2.8 | 0.015 | |
| miR-22-3p | 2.7 | 0.022 | |
| miR-29a-5p | 2.7 | 0.045 | |
| miR-665 | 2.5 | 0.007 | |
| miR-296-3p | 2.1 | 0.0004 | |
| miR-6813-3p | 2.1 | 0.008 | |
| miR-1292-5p | 2.0 | 0.011 | |
| miR-4271 | 1.9 | 0.006 | |
| miR-1228-3p | 1.8 | 0.043 | |
| miR-193a-5p | 1.7 | 0.007 | |
| miR-370-3p | 1.7 | 0.020 | |
| miR-758-5p | 1.7 | 0.044 | |
| miR-584-5p | -2.1 | 0.021 | |
| miR-598-3p | -2.2 | 0.050 | |
| miR-449c-3p | -2.4 | 0.042 | |
| miR-181a-2-3p | -2.5 | 0.022 | |
| miR-10b-5p | -;2.7 | 0.035 | |
| miR-143-3p | -2.8 | 0.034 | |
| miR-889-3p | -2.8 | 0.041 | |
| miR-100-5p | -2.9 | 0.028 |
| miRNA | mRNA Targets |
|---|---|
| miR-362-5p | Rbm27, Trim50, Sgip1, Plagl2, Prkacb, Luc7l3, Edem1, Cyp1b1, Rmi1, Mrpl35, Pik3c2b, Cyld, Gas7 |
| miR-491-5p | Sema6a, Chrnb2, Pogk, Ksr2, Samd4b, Optc, Cpne5, Foxp4, Msn, Igf2, Smad3, Bcl2l1, Egfr, Notch3, Mmp9, Igf2bp1, Wnt3a, Capns1 |
| miR-495-5p | Cttn, Ncoa2, Znf281, Tnpo1, Hnrnpd, Sbno1, Dipk2a, Mcub, Nol4l, Camta1, Cdk6, Mta3 |
| miR-502-3p | Adamts3, Kctd9, Fbn2, Sec63, Zfx, Dapk1, Smim13, Napil5, Csde1, Set, Olfm4 |
| miR-584-5p | Usp6nl, Avpr1a, Hsd11b1, Galnt15, Gbp5, Cd200, Ppm1a, Nphp1, Rock1, Mmp14, Kcne2, Wwp1, Cdk16 |
| miR-758-5p | Slc20a2, Id2, Nufip2, Ptp4a1, Tox4, Setd5, Phactr1, Rtkn, Dsg3, Csnk1a1l, Cd36, Zbtb20, Cbx5 |
| miR-1228-3p | Ppp2r2d, Ube2d2, Irx2, Znf554, Nfia, Socs6, Rabgef1, Tjp1, Tor1aip1, Zbtb44, Moap1, Csnk2a1, Plac8 |
| miR-3135b | Lrrc27, Fmnl3, Ttc21b, Castor3, Xpo7, Ppm1a, Dnm1l, Kdm3b, Rbp1, Faap20, Pten, Golph3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peterson, J.; McTiernan, C.D.; Thome, C.; Khaper, N.; Lees, S.J.; Boreham, D.R.; Tai, T.C.; Tharmalingam, S. Identification of Radiation-Induced miRNA Biomarkers Using the CGL1 Cell Model System. Bioengineering 2022, 9, 214. https://doi.org/10.3390/bioengineering9050214
Peterson J, McTiernan CD, Thome C, Khaper N, Lees SJ, Boreham DR, Tai TC, Tharmalingam S. Identification of Radiation-Induced miRNA Biomarkers Using the CGL1 Cell Model System. Bioengineering. 2022; 9(5):214. https://doi.org/10.3390/bioengineering9050214
Chicago/Turabian StylePeterson, Jayden, Christopher D. McTiernan, Christopher Thome, Neelam Khaper, Simon J. Lees, Douglas R. Boreham, Tze Chun Tai, and Sujeenthar Tharmalingam. 2022. "Identification of Radiation-Induced miRNA Biomarkers Using the CGL1 Cell Model System" Bioengineering 9, no. 5: 214. https://doi.org/10.3390/bioengineering9050214
APA StylePeterson, J., McTiernan, C. D., Thome, C., Khaper, N., Lees, S. J., Boreham, D. R., Tai, T. C., & Tharmalingam, S. (2022). Identification of Radiation-Induced miRNA Biomarkers Using the CGL1 Cell Model System. Bioengineering, 9(5), 214. https://doi.org/10.3390/bioengineering9050214






