Microfluidics for Antibiotic Susceptibility and Toxicity Testing
Abstract
:1. Introduction
2. Microfluidics for Antibiotic Susceptibility Testing
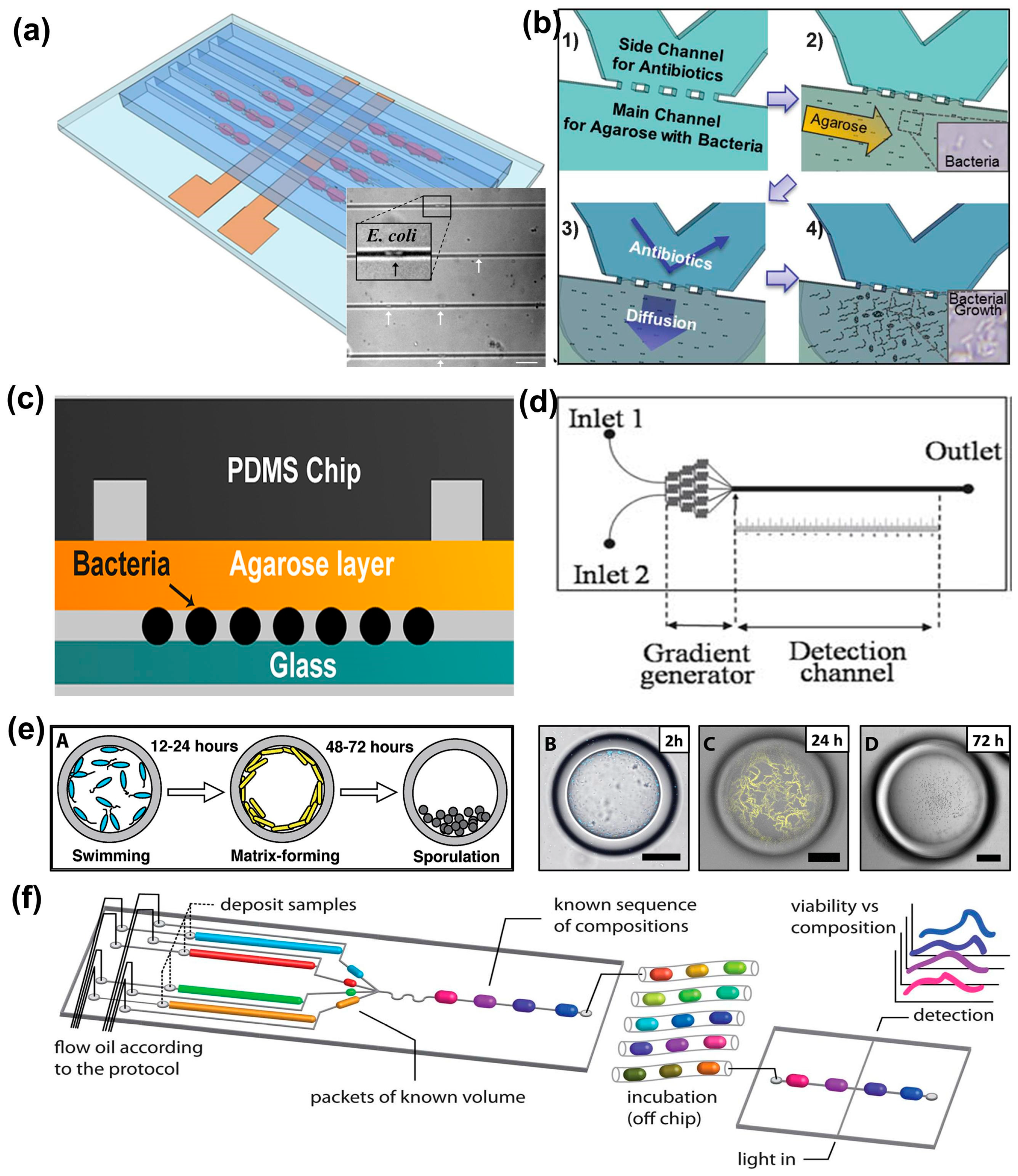
2.1. Bacterial Antibiotic Susceptibility Testing (AST) at the Single-Cell Level
2.2. Bacterial Persisters and Non-Growing but Metabolically Active (NGMA) Bacteria
2.3. Bacterial Biofilm Antibiotic Susceptibility Testing
2.4. Combinatorial Effect of Antibiotics
3. Microfluidics for in Vitro Drug Toxicity Testing
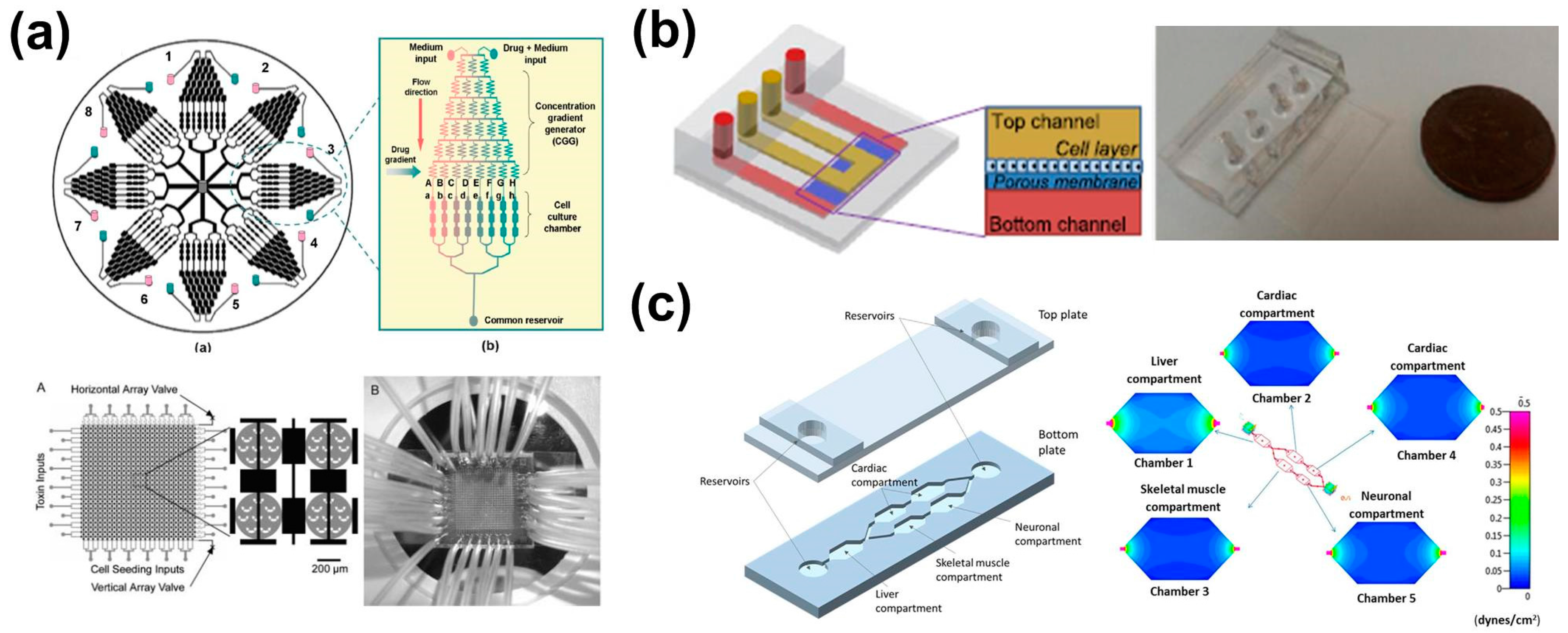
3.1. Cell-on-a-Chip Technology
3.2. Organ-on-a-Chip Technology
4. Conclusions and Outlook
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Towse, A.; Sharma, P. Incentives for R&D for New Antimicrobial Drugs. Int. J. Econ. Bus. 2011, 18, 331–350. [Google Scholar]
- O’Neill, J. Review on antimicrobial resistance. In Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; AMR-Review: London, UK, 2014. [Google Scholar]
- Hamon, M.; Hong, J.W. Systematic evaluation of the efficiencies of proteins and chemicals in pharmaceutical applications. In Microfluidic Technologies for Human Health; Demirci, U., Khdemhosseini, A., Langer, R., Blander, J., Eds.; World Scientific Publishing: Singapore, 2012; pp. 21–45. [Google Scholar]
- Tirella, A.; Marano, M.; Vozzi, F.; Ahluwalia, A. A microfluidic gradient maker for toxicity testing of bupivacaine and lidocaine. Toxicol. in Vitro 2008, 22, 1957–1964. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kim, M.C.; Marquez, M.; Thorsen, T. High-density microfluidic arrays for cell cytotoxicity analysis. Lab Chip 2007, 7, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Brouzes, E.; Medkova, M.; Savenelli, N.; Marran, D.; Twardowski, M.; Hutchison, J.B.; Rothberg, J.M.; Link, D.R.; Perrimon, N.; Samuels, M.L. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA 2009, 106, 14195–14200. [Google Scholar] [CrossRef] [PubMed]
- Caviglia, C.; Zór, K.; Montini, L.; Tilli, V.; Canepa, S.; Melander, F.; Muhammad, H.B.; Carminati, M.; Ferrari, G.; Raiteri, R.; et al. Impedimetric Toxicity Assay in Microfluidics Using Free and Liposome-Encapsulated Anticancer Drugs. Anal. Chem. 2015, 87, 2204–2212. [Google Scholar] [CrossRef] [PubMed]
- Rusconi, R.; Garren, M.; Stocker, R. Microfluidics expanding the frontiers of microbial ecology. Annu. Rev. Biophys. 2014, 43, 65–91. [Google Scholar] [CrossRef] [PubMed]
- Pulido, M.R.; García-Quintanilla, M.; Martín-Peña, R.; Cisneros, J.M.; McConnell, M.J. Progress on the development of rapid methods for antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013, 68, 2710–2717. [Google Scholar] [CrossRef] [PubMed]
- Dalgaard, P.; Ross, T.; Kamperman, L.; Neumeyer, K.; McMeekin, T.A. Estimation of bacterial growth rates from turbidimetric and viable count data. Int. J. Food Microbiol. 1994, 23, 391–404. [Google Scholar] [CrossRef]
- Shemesh, J.; Ben Arye, T.; Avesar, J.; Kang, J.H.; Fine, A.; Super, M.; Meller, A.; Ingber, D.E.; Levenberg, S. Stationary nanoliter droplet array with a substrate of choice for single adherent/nonadherent cell incubation and analysis. Proc. Natl. Acad. Sci. USA 2014, 111, 11293–11298. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Yoon, S.H.; Sim, H.Y.; Yang, Y.S.; Oh, T.K.; Kim, J.F.; Hong, J.W. Charting Microbial Phenotypes in Multiplex Nanoliter Batch Bioreactors. Anal. Chem. 2013, 85, 5892–5899. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Nugent, E.; Javer, A.; Cicuta, P.; Sclavi, B.; Cosentino Lagomarsino, M.; Dorfman, K.D. Microfluidic chemostat for measuring single cell dynamics in bacteria. Lab Chip 2013, 13, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Rowat, A.C.; Bird, J.C.; Agresti, J.J.; Rando, O.J.; Weitz, D.A. Tracking lineages of single cells in lines using a microfluidic device. Proc. Natl. Acad. Sci. USA 2009, 106, 18149–18154. [Google Scholar] [CrossRef] [PubMed]
- Peitz, I.; van Leeuwen, R. Single-cell bacteria growth monitoring by automated DEP-facilitated image analysis. Lab Chip 2010, 10, 2944–2951. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Gao, J.; Zhang, D.D.; Gau, V.; Liao, J.C.; Wong, P.K. Single Cell Antimicrobial Susceptibility Testing by Confined Microchannels and Electrokinetic Loading. Anal. Chem. 2013, 85, 3971–3976. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Jung, Y.-G.; Kim, J.; Kim, S.; Jung, Y.; Na, H.; Kwon, S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip 2013, 13, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.-G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.-C.; Lee, J.C.; et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef] [PubMed]
- Wong, I.; Atsumi, S.; Huang, W.-C.; Wu, T.-Y.; Hanai, T.; Lam, M.L.; Tang, P.; Yang, J.; Liao, J.C.; Ho, C.M. An agar gel membrane-PDMS hybrid microfluidic device for long term single cell dynamic study. Lab Chip 2010, 10, 2710–2719. [Google Scholar] [CrossRef] [PubMed]
- Wakamoto, Y.; Dhar, N.; Chait, R.; Schneider, K.; Signorino-Gelo, F.; Leibler, S.; McKinney, J.D. Dynamic Persistence of Antibiotic-Stressed Mycobacteria. Science 2013, 339, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Golchin, S.A.; Stratford, J.; Curry, R.J.; McFadden, J. A microfluidic system for long-term time-lapse microscopy studies of Mycobacteria. Tuberculosis 2012, 92, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qiu, Y.; Glidle, A.; Mcllvenna, D.; Luo, Q.; Cooper, J.; Shi, H.C.; Yin, H. Gradient Microfluidics Enables Rapid Bacterial Growth Inhibition Testing. Anal. Chem. 2014, 86, 3131–3137. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikov, M.; Lee, J.C.; Campbell, J.; Sharon, A.; Sauer-Budge, A.F. A microfluidic platform for rapid, stress-induced antibiotic susceptibility testing of Staphylococcus aureus. Lab Chip 2012, 12, 4523–4532. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; An, Y.; Hjort, K.; Hjort, K.; Sandegren, L.; Wu, Z. Time lapse investigation of antibiotic susceptibility using a microfluidic linear gradient 3D culture device. Lab Chip 2014, 14, 3409–3418. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Sakakihara, S.; Grushnikov, A.; Kikuchi, K.; Noji, H.; Yamaguchi, A.; Iino, R.; Yagi, Y.; Nishino, K. A Microfluidic Channel Method for Rapid Drug-Susceptibility Testing of Pseudomonas aeruginosa. PLoS ONE 2016, 11, e0148797. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.I.; Haswell, S.; Gibson, I. Lab-on-a-chip or Chip-in-a-lab: Challenges of Commercialization Lost in Translation. Procedia Technol. 2015, 20, 54–59. [Google Scholar] [CrossRef]
- Gradientech. Available online: http://gradientech.se/quickmic/ (accessed on 14 September 2016).
- Chen, C.H.; Lu, Y.; Sin, M.L.Y.; Mach, K.E.; Zhang, D.D.; Gau, V.; Liao, J.C.; Wong, P.K. Antimicrobial Susceptibility Testing Using High Surface-to-Volume Ratio Microchannels. Anal. Chem. 2010, 82, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Price, C.S.; Kon, S.E.; Metzger, S. Rapid antibiotic susceptibility phenotypic characterization of Staphylococcus aureus using automated microscopy of small numbers of cells. J. Microbiol. Methods 2014, 98, 50–58. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Mu, X.; Guo, Z.; Hao, H.; Zhang, C.; Zhao, Z.; Wang, Q. A novel microbead-based microfluidic device for rapid bacterial identification and antibiotic susceptibility testing. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2223–2230. [Google Scholar] [CrossRef] [PubMed]
- Boedicker, J.Q.; Li, L.; Kline, T.R.; Ismagilov, R.F. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip 2008, 8, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Mohan, R.; Mukherjee, A.; Sevgen, S.E.; Sanpitakseree, C.; Lee, J.; Schroeder, C.M.; Kenis, P.J.A. A multiplexed microfluidic platform for rapid antibiotic susceptibility testing. Biosens. Bioelectron. 2013, 49, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Zhao, X. Rapid Identification and Susceptibility Testing of Uropathogenic Microbes via Immunosorbent ATP-Bioluminescence Assay on a Microfluidic Simulator for Antibiotic Therapy. Anal. Chem. 2015, 87, 2410–2418. [Google Scholar] [CrossRef] [PubMed]
- Sinn, I.; Kinnunen, P.; Albertson, T.; McNaughton, B.H.; Newton, D.W.; Burns, M.A.; Kopelman, R. Asynchronous magnetic bead rotation (AMBR) biosensor in microfluidic droplets for rapid bacterial growth and susceptibility measurements. Lab Chip 2011, 11, 2604–2611. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhen, L.; Liu, J.; Wu, J. Rapid Antibiotic Susceptibility Testing in a Microfluidic pH Sensor. Anal. Chem. 2013, 85, 2787–2794. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister Cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Vega, N.M.; Allison, K.R.; Khalil, A.S.; Collins, J.J. Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 2012, 8, 431–433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lambert, G.; Liao, D.; Kim, H.; Robin, K.; Tung, C.K.; Pourmand, N.; Austin, R.H. Acceleration of Emergence of Bacterial Antibiotic Resistance in Connected Microenvironments. Science 2011, 333, 1764–1767. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial Persistence as a Phenotypic Switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef] [PubMed]
- Gefen, O.; Gabay, C.; Mumcuoglu, M.; Engel, G.; Balaban, N.Q. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 6145–6149. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qiu, Y.; Glidle, A.; Cooper, J.; Shi, H.; Yin, H. Single cell growth rate and morphological dynamics revealing an “opportunistic” persistence. Analyst 2014, 139, 3305–3313. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Castro-Camargo, M.; Gerdes, K. (p)ppGpp Controls Bacterial Persistence by Stochastic Induction of Toxin-Antitoxin Activity. Cell. 2013, 154, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Germain, E.; Roghanian, M.; Gerdes, K.; Maisonneuve, E. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. USA 2015, 112, 5171–5176. [Google Scholar] [CrossRef] [PubMed]
- Iino, R.; Matsumoto, Y.; Nishino, K.; Yamaguchi, A.; Noji, H. Design of a large-scale femtoliter droplet array for single-cell analysis of drug-tolerant and drug-resistant bacteria. Front. Microbiol. 2013, 4, 300. [Google Scholar] [CrossRef] [PubMed]
- Manina, G.; McKinney, J.D. A single-cell perspective on non-growing but metabolically active (NGMA) bacteria. In Pathogenesis of Mycobacterium Tuberculosis and Its Interaction with the Host Organism; Pieters, J., McKinney, D.J., Eds.; Springer: Berlin, Germany, 2013; pp. 135–161. [Google Scholar]
- Maglica, Ž.; Özdemir, E.; McKinney, J.D. Single-cell tracking reveals antibiotic-induced changes in mycobacterial energy metabolism. MBio 2015, 6, e02236-14. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Microl. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, H.-D.; Chung, S. Microfluidic approaches to bacterial biofilm formation. Molecules 2012, 17, 9818–9834. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Ahmed, I.; Hwang, J.; Seo, Y.; Lee, E.; Choi, J.; Moon, S.; Hong, J.W. A Microfluidic Approach to Investigating a Synergistic Effect of Tobramycin and Sodium Dodecyl Sulfate on Pseudomonas aeruginosa Biofilms. Anal. Sci. 2016, 32, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.P.; Kim, Y.G.; Choi, C.H.; Kim, H.E.; Lee, S.H.; Chang, W.S.; Lee, C.S. In situ monitoring of antibiotic susceptibility of bacterial biofilms in a microfluidic device. Lab Chip 2010, 10, 3296–3299. [Google Scholar] [CrossRef] [PubMed]
- DiCicco, M.; Neethirajan, S. An in vitro microfluidic gradient generator platform for antimicrobial testing. BioChip J. 2014, 8, 282–288. [Google Scholar] [CrossRef]
- Sun, P.; Liu, Y.; Sha, J.; Zhang, Z.; Tu, Q.; Chen, P.; Wang, J. High-throughput microfluidic system for long-term bacterial colony monitoring and antibiotic testing in zero-flow environments. Biosens. Bioelectron. 2011, 26, 1993–1999. [Google Scholar] [CrossRef] [PubMed]
- Webster, T.A.; Sismaet, H.J.; Chan, I.J.; Goluch, E.D. Electrochemically monitoring the antibiotic susceptibility of Pseudomonas aeruginosa biofilms. Analyst 2015, 140, 7195–7201. [Google Scholar] [CrossRef] [PubMed]
- Takagi, R.; Fukuda, J.; Suzuki, H.; Nagata, K.; Nomura, N. Electrochemical microdevice for the determination of the minimum inhibitory concentration of antibiotics. In Proceedings of the 2012 IEEE Sensors, Taipei, Taiwan, 28–31 October 2012; pp. 1–4.
- Chang, C.B.; Wilking, J.N.; Kim, S.H.; Shum, H.C.; Weitz, D.A. Monodisperse Emulsion Drop Microenvironments for Bacterial Biofilm Growth. Small 2015, 11, 3954–3961. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for Emerging Pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Suh, S.-J.; Hamon, M.; Hong, J.W. Determination of antibiotic EC50 using a zero-flow microfluidic chip based growth phenotype assay. Biotechnol. J. 2015, 10, 1783–1791. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Taylor, D.; Agrawal, N.; Wang, H.; Kim, H.; Han, A.; Rege, K.; Jayaraman, A. A programmable microfluidic cell array for combinatorial drug screening. Lab Chip 2012, 12, 1813–1822. [Google Scholar] [CrossRef] [PubMed]
- Kilinc, D.; Schwab, J.; Rampini, S.; Ikpekha, O.W.; Thampi, A.; Blasiak, A.; Li, P.; Schwamborn, R.; Kolch, W.; Matallanas, D.; et al. A microfluidic dual gradient generator for conducting cell-based drug combination assays. Integr. Biol. 2016, 8, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Köhler, J.M. Droplet-based microfluidics for microtoxicological studies. Eng. Life Sci. 2015, 15, 306–317. [Google Scholar] [CrossRef]
- Churski, K.; Kaminski, T.S.; Jakiela, S.; Kamysz, W.; Baranska-Rybak, W.; Weibel, D.B.; Garstecki, P. Rapid screening of antibiotic toxicity in an automated microdroplet system. Lab Chip 2012, 12, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Kursten, D.; Schneider, S.; Knauer, A.; Günther, P.M.; Köhler, J.M. Uncovering toxicological complexity by multi-dimensional screenings in microsegmented flow: Modulation of antibiotic interference by nanoparticles. Lab Chip 2012, 12, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Goldhan, J.; Martin, K.; Köhler, J.M. Investigation of mixture toxicity of widely used drugs caffeine and ampicillin in the presence of an ACE inhibitor on bacterial growth using droplet-based microfluidic technique. Green Process. Synth. 2013, 2, 591–601. [Google Scholar] [CrossRef]
- Mingeot-Leclercq, M.P.; Tulkens, P.M. Aminoglycosides: Nephrotoxicity. Antimicrob. Agents Chemother. 1999, 43, 1003–1012. [Google Scholar] [PubMed]
- Westphal, J.F.; Vetter, D.; Brogard, J.M. Hepatic side-effects of antibiotics. J. Antimicrob. Chemother. 1994, 33, 387–401. [Google Scholar] [CrossRef]
- Ye, N.; Qin, J.; Shi, W.; Liu, X.; Lin, B. Cell-based high content screening using an integrated microfluidic device. Lab Chip 2007, 7, 1696–1704. [Google Scholar] [CrossRef] [PubMed]
- Hamon, M.; Jambovane, S.; Bradley, L.; Khademhosseini, A.; Hong, J.W. Cell-Based Dose Responses from Open-Well Microchambers. Anal. Chem. 2013, 85, 5249–5254. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.J.; Ghorashian, N.; Gaige, T.A.; Hung, P.J. Microfluidic System for Automated Cell-based Assays. J. Lab. Autom. 2007, 12, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Ostafe, R.; Prodanovic, R.; Ung, W.L.; Weitz, D.A.; Fischer, R. A high-throughput cellulase screening system based on droplet microfluidics. Biomicrofluidics 2014, 8, 041102. [Google Scholar] [CrossRef] [PubMed]
- Chokkalingam, V.; Tel, J.; Wimmers, F.; Liu, X.; Semenov, S.; Thiele, J.; Figdor, C.G.; Huck, W.T. Probing cellular heterogeneity in cytokine-secreting immune cells using droplet-based microfluidics. Lab Chip 2013, 13, 4740–4744. [Google Scholar] [CrossRef] [PubMed]
- Pompano, R.R.; Liu, W.; Du, W.; Ismagilov, R.F. Microfluidics Using Spatially Defined Arrays of Droplets in One, Two, and Three Dimensions. Annu. Rev. Anal. Chem. 2011, 4, 59–81. [Google Scholar] [CrossRef] [PubMed]
- Malsch, D.; Gleichmann, N.; Kielpinski, M.; Mayer, G.; Henkel, T.; Mueller, D.; van Steijn, V.; Kleijn, C.R.; Kreutzer, M.T. Dynamics of droplet formation at T-shaped nozzles with elastic feed lines. Microfluid. Nanofluid. 2010, 8, 497–507. [Google Scholar] [CrossRef]
- Khan, S.A.; Duraiswamy, S. Microfluidic emulsions with dynamic compound drops. Lab Chip 2009, 9, 1840–1842. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Iwai, S.; Araki, S.; Sakakihara, S.; Iino, R.; Noji, H. Large-scale femtoliter droplet array for digital counting of single biomolecules. Lab Chip 2012, 12, 4986–4991. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gu, Y.; Le Roux, R.B.; Matthews, S.M.; Bratton, D.; Yunus, K.; Fisher, A.C.; Huck, W.T.S. The electrochemical detection of droplets in microfluidic devices. Lab Chip 2008, 8, 1937–1942. [Google Scholar] [CrossRef] [PubMed]
- Ahn, B.; Lee, K.; Louge, R.; Oh, K.W. Concurrent droplet charging and sorting by electrostatic actuation. Biomicrofluidics 2009, 3, 044102. [Google Scholar] [CrossRef] [PubMed]
- Granieri, L.; Baret, J.-C.; Griffiths, A.D.; Merten, C.A. High-Throughput Screening of Enzymes by Retroviral Display Using Droplet-Based Microfluidics. Chem. Biol. 2010, 17, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Ji, X.-H.; Liu, K.; He, R.-X.; Zhao, L.-B.; Guo, Z.-X.; Liu, W.; Guo, S.-S.; Zhao, X.-Z. Droplet electric separator microfluidic device for cell sorting. Appl. Phys. Lett. 2010, 96, 193701. [Google Scholar] [CrossRef]
- Tan, Y.-C.; Ho, Y.L.; Lee, A.P. Droplet coalescence by geometrically mediated flow in microfluidic channels. Microfluid. Nanofluid. 2007, 3, 495–499. [Google Scholar] [CrossRef]
- Niu, X.; Gulati, S.; Edel, J.B.; deMello, A.J. Pillar-induced droplet merging in microfluidic circuits. Lab Chip 2008, 8, 1837–1841. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, C.; Li, C.M. On-demand microfluidic droplet trapping and fusion for on-chip static droplet assays. Lab Chip 2009, 9, 1504–1506. [Google Scholar] [CrossRef] [PubMed]
- Baroud, C.N.; Gallaire, F.; Dangla, R. Dynamics of microfluidic droplets. Lab Chip 2010, 10, 2032–2045. [Google Scholar] [CrossRef] [PubMed]
- Fair, R.B. Digital microfluidics: Is a true lab-on-a-chip possible? Microfluid. Nanofluid. 2007, 3, 245–281. [Google Scholar] [CrossRef]
- Schumacher, J.T.; Grodrian, A.; Lemke, K.; Roemer, R.; Metze, J. System development for generating homogeneous cell suspensions and transporting them in microfluidic devices. Eng. Life Sci. 2008, 8, 49–55. [Google Scholar] [CrossRef]
- Arias, I.; Wolkoff, A.; Boyer, J.; Shafritz, D.; Fausto, N.; Alter, H.; Cohen, D. The Liver: Biology and Pathobiology, 5th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Yeon, J.H.; Park, J.K. A microfluidic platform for 3-dimensional cell culture and cell-based assays. Biomed. Microdevices 2007, 9, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Ziolkowska, K.; Stelmachowska, A.; Kwapiszewski, R.; Chudy, M.; Dybko, A.; Brzózka, Z. Long-term three-dimensional cell culture and anticancer drug activity evaluation in a microfluidic chip. Biosens. Bioelectron. 2013, 40, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Au, S.H.; Chamberlain, M.D.; Mahesh, S.; Sefton, M.V.; Wheeler, A.R. Hepatic organoids for microfluidic drug screening. Lab Chip 2014, 14, 3290–3299. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.-J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.Y.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Leslie, D.C.; Matthews, B.D.; Fraser, J.P.; Jurek, S.; Hamilton, G.A.; Thorneloe, K.S.; McAlexander, M.A.; Ingber, D.E. A Human Disease Model of Drug Toxicity-Induced Pulmonary Edema in a Lung-on-a-Chip Microdevice. Sci. Transl. Med. 2012, 4, 159ra147. [Google Scholar] [CrossRef] [PubMed]
- Mahler, G.J.; Esch, M.B.; Glahn, R.P.; Shuler, M.L. Characterization of a Gastrointestinal Tract Microscale Cell Culture Analog Used to Predict Drug Toxicity. Biotechnol. Bioeng. 2009, 104, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Torisawa, Y.-S.; Spina, C.S.; Mammoto, T.; Mammoto, A.; Weaver, J.C.; Tat, T.; Collins, J.J.; Ingber, D.E. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro. Nat. Methods 2014, 11, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Yeon, J.H.; Na, D.; Choi, K.; Ryu, S.-W.; Choi, C.; Park, J.K. Reliable permeability assay system in a microfluidic device mimicking cerebral vasculatures. Biomed. Microdevices 2012, 14, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Inamdar, N.K.; Borenstein, J.T. Microfluidic cell culture models for tissue engineering. Curr. Opin. Biotechnol. 2011, 22, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Balis, U.J.; Yarmush, M.L.; Toner, M. Effect of cell-cell interactions in preservation of cellular phenotype: Cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999, 13, 1883–1900. [Google Scholar] [PubMed]
- Aref, A.R.; Huang, R.Y.J.; Yu, W.; Chua, K.N.; Sun, W.; Tu, T.Y.; Bai, J.; Sim, W.J.; Zervantonakis, I.K.; Thiery, J.P. Screening therapeutic EMT blocking agents in a three-dimensional microenvironment. Integr. Biol. 2013, 5, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Hattersley, S.M.; Greenman, J.; Haswell, S.J. Study of ethanol induced toxicity in liver explants using microfluidic devices. Biomed. Microdevices 2011, 13, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Sasha Cai, L.; Byoung choul, C.K.; Cameron, Y.; Labuz, J.M.; Leung, B.; Takayama, S. Pharmacokinetic profile that reduces nephrotoxicity of gentamicin in a perfused kidney-on-a-chip. Biofabrication 2016, 8, 015021. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.G.; Shuler, M.L. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol. Bioeng. 2016, 113, 2213–2227. [Google Scholar] [CrossRef] [PubMed]
- Oleaga, C.; Bernabini, C.; Smith, A.S.T.; Srinivasan, B.; Jackson, M.; McLamb, W.; Platt, V.; Bridges, R.; Cai, Y.; Santhanam, N.; et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci. Rep. 2016, 6, 20030. [Google Scholar] [CrossRef] [PubMed]
| Growth Monitoring Methods | Description | AST Time [Reference] | Advantage/Limitation | |
|---|---|---|---|---|
| Direct | Optical imaging | Number of cells | 2 h [28], 4 h [29], 5 h [15], 1 h [16], 3 h [25] | All clinic isolates |
| Area of cells | 4 h [17], 4 h [22] | |||
| Greyscale imaging of cells | 2.5~4 h [24] | |||
| Indirect | Fluorescence | Fluorescent signal of bacteria-microbead complex | 4~8 h [30] | Immunoassay is required |
| GFP expression of bacterial strains | 7.5 h [31], 2~4 h [32] | Only molecularly engineered strains | ||
| Bioluminescence | ATP bioluminescence of bacteria-antibodies complex | 3~6 h [33] | Immunoassay is required | |
| Magnetics | Magnetic beads rotation rate which is inversely proportional to bacterial mass | 30 min [34] | Immunoassay & external rotational magnetic field are required | |
| pH | pH changes due to the accumulation of metabolic products | 2 h [35] | All clinic isolates | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, J.; Hamon, M.; Jambovane, S. Microfluidics for Antibiotic Susceptibility and Toxicity Testing. Bioengineering 2016, 3, 25. https://doi.org/10.3390/bioengineering3040025
Dai J, Hamon M, Jambovane S. Microfluidics for Antibiotic Susceptibility and Toxicity Testing. Bioengineering. 2016; 3(4):25. https://doi.org/10.3390/bioengineering3040025
Chicago/Turabian StyleDai, Jing, Morgan Hamon, and Sachin Jambovane. 2016. "Microfluidics for Antibiotic Susceptibility and Toxicity Testing" Bioengineering 3, no. 4: 25. https://doi.org/10.3390/bioengineering3040025
APA StyleDai, J., Hamon, M., & Jambovane, S. (2016). Microfluidics for Antibiotic Susceptibility and Toxicity Testing. Bioengineering, 3(4), 25. https://doi.org/10.3390/bioengineering3040025