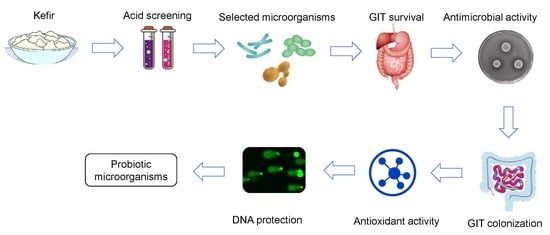
In Vitro Probiotic Properties and DNA Protection Activity of Yeast and Lactic Acid Bacteria Isolated from A Honey-Based Kefir Beverage
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms
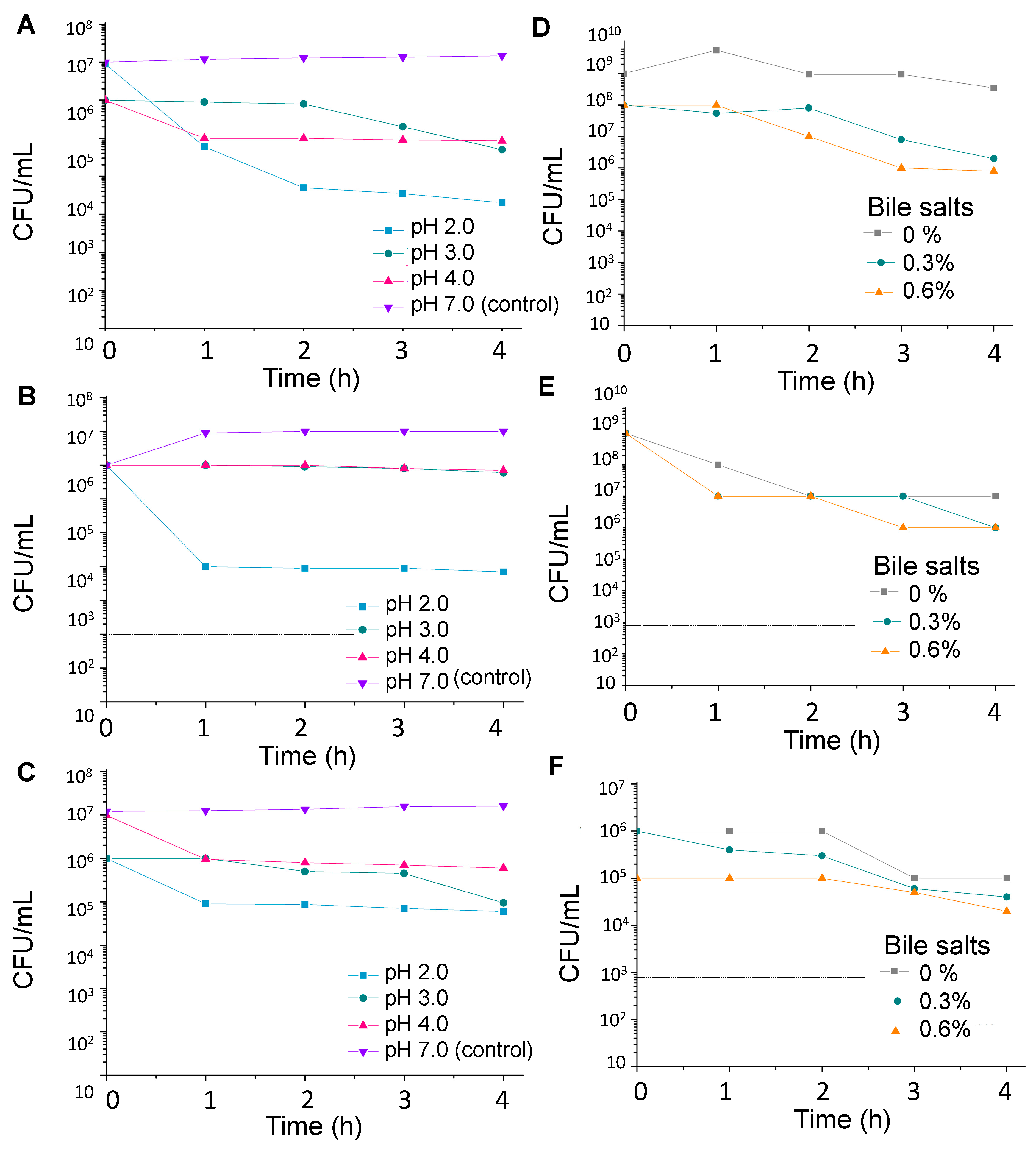
2.2. Stress Tolerance
2.2.1. Acid Tolerance
2.2.2. Resistance to Bile Salts
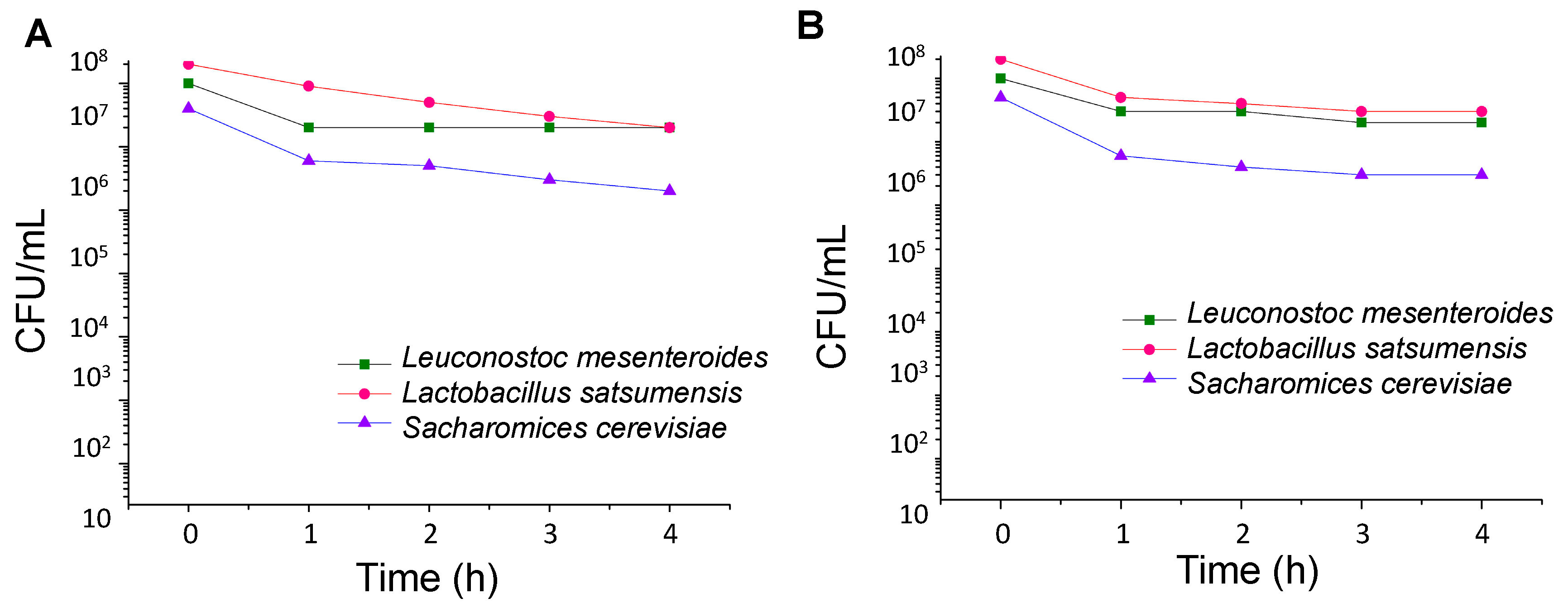
2.2.3. Survival in Simulated Gastrointestinal Tract
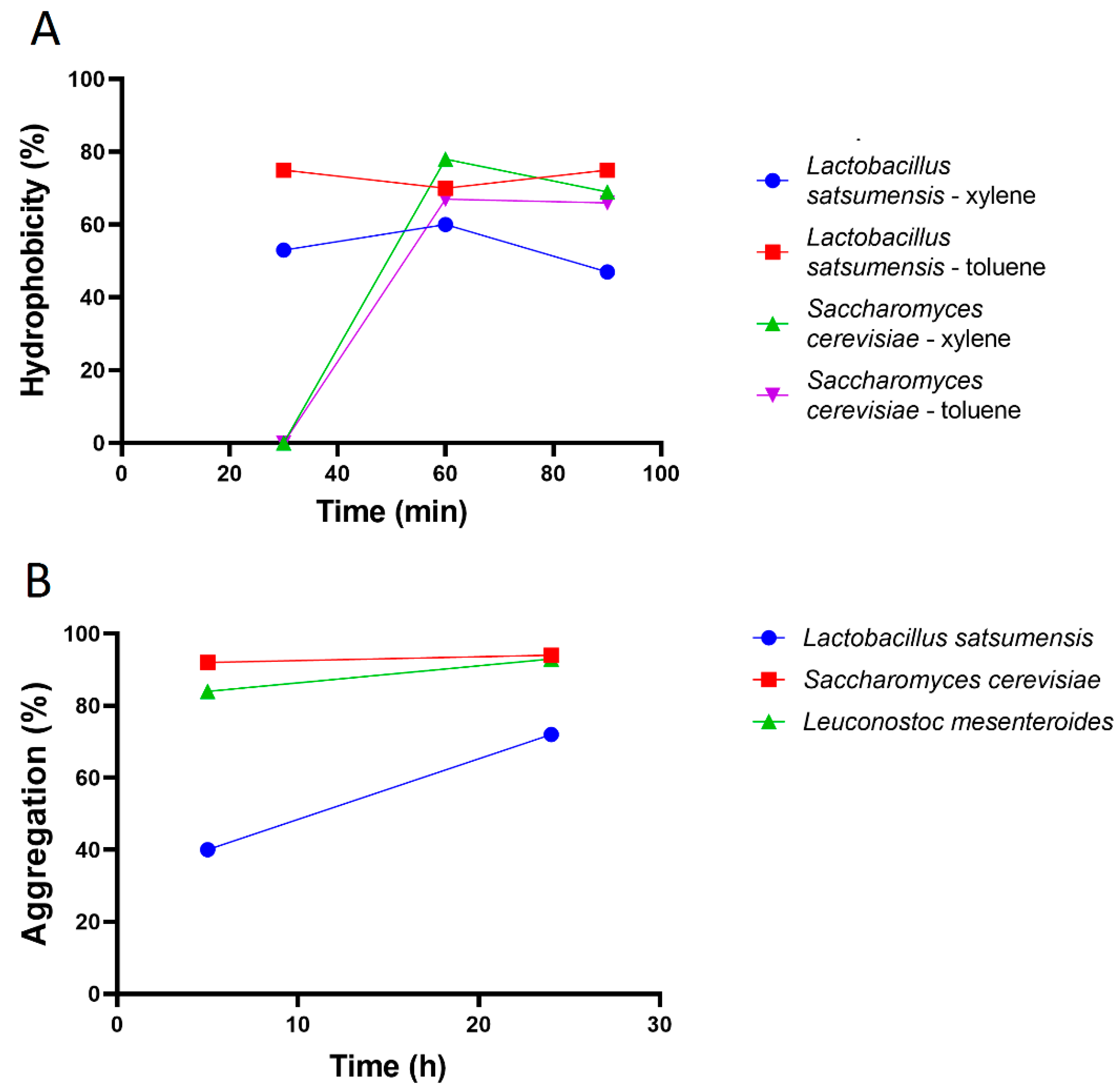
2.3. Adhesion Ability
2.3.1. Hydrophobicity
2.3.2. Aggregation
2.4. Anti-Pathogenic Activity
2.4.1. Antimicrobial
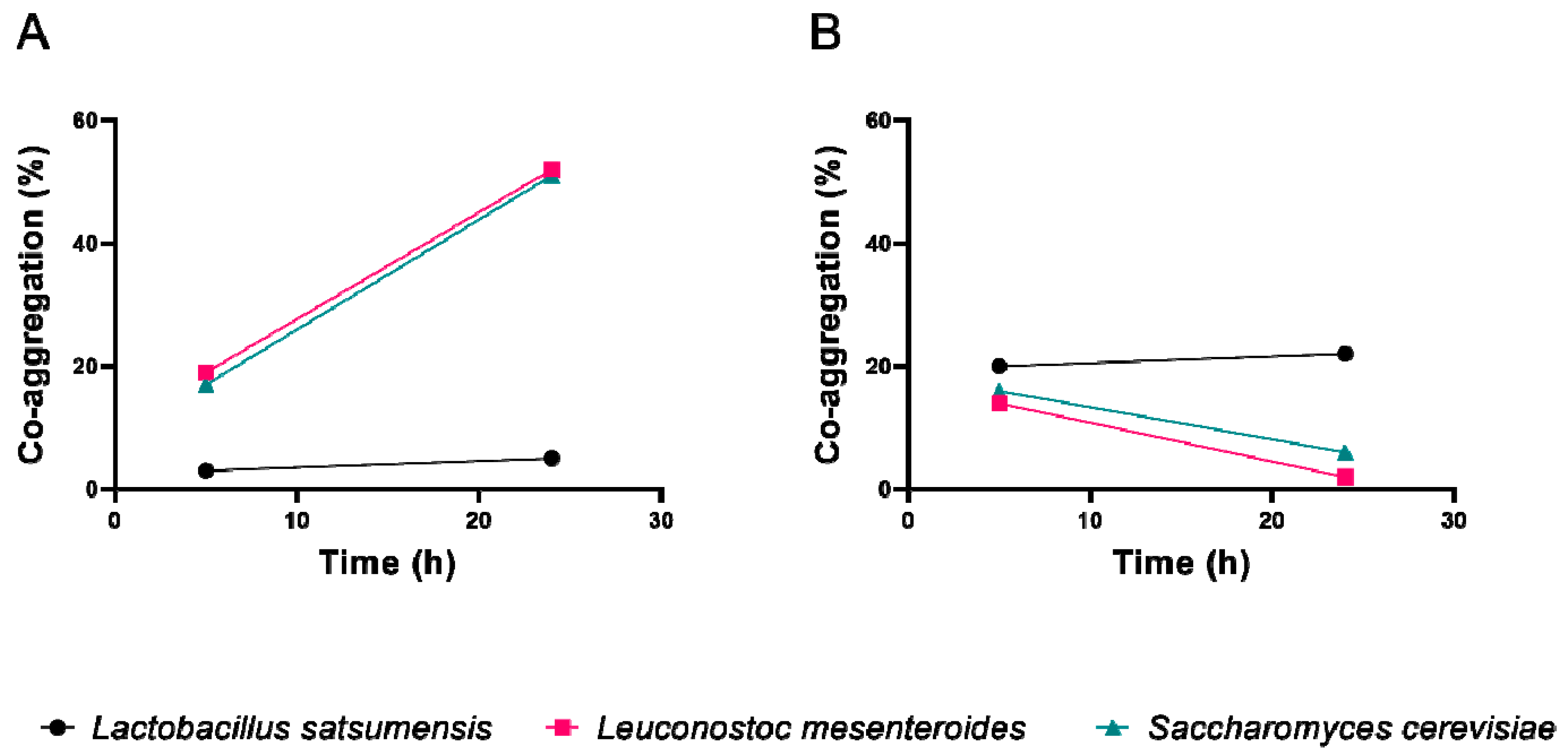
2.4.2. Co-Aggregation
2.5. Functional Characteristics
2.5.1. Antioxidant Activity by the DPPH Method
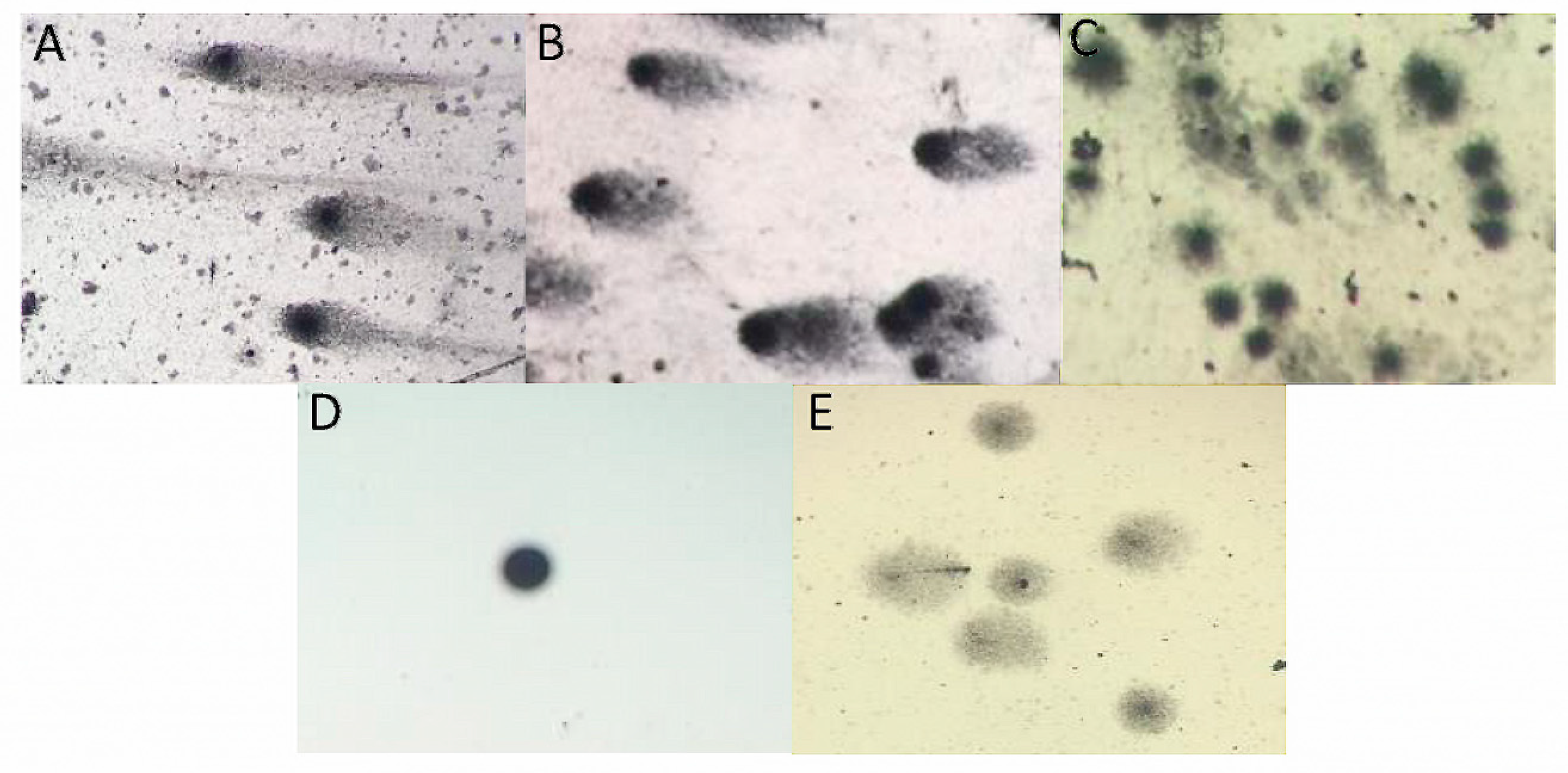
2.5.2. Single-Cell Gel Electrophoresis (SCGE) Assay (Comet Assay)
2.6. Safety Assessment (Hemolytic Activity)
2.7. Statistical Analyses
3. Results and Discussion
3.1. Stress Tolerance
3.2. Adhesion Ability
3.3. Anti-Pathogenic Assay
3.4. Functional Characteristics
3.5. Safety Assessment
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guarner, F.; Schaafsma, G. Probiotics. Int. J. Food Microbiol. 1998, 39, 237–238. [Google Scholar] [CrossRef]
- Buntin, N.; Suphitchaya, C.; Tipparat, H. Screening of lactic acid bacteria from gastrointestinal tracts of marine fish for their potential use as probiotics. Songklanakarin J. Sci. Technol. 2008, 30, 141–148. [Google Scholar]
- Castro-Rodríguez, D.; Hernández-Sánchez, H.; Yáñez, F.I. Probiotic properties of Leuconostoc mesenteroides isolated from aguamiel of Agave salmiana. Probiotics Antimicrob. 2015, 7, 107–117. [Google Scholar] [CrossRef]
- Shori, B.A. The potential applications of probiotics on dairy and non-dairy foods focusing on viability during storage. Biocatal. Agric. Biotechnol. 2015, 4, 423–431. [Google Scholar] [CrossRef]
- Liu, S.Q. Lactic Acid Bacteria: Leuconostoc spp.; Elsevier Inc.: Philadelphia, PA, USA; National University of Singapore: Singapore, 2016; pp. 1–6. [Google Scholar]
- Puerari, C.; Magalhães, T.K.; Schwan, F.R. New cocoa pulp-based kefir beverages: Microbiological, chemical composition and sensory analysis. Food Res. Int. 2012, 48, 634–640. [Google Scholar] [CrossRef]
- Diosma, G.; Romanin, E.D.; Rey-Burusco, F.M.; Londero, A.; Garrote, L.G. Yeasts from kefir grains: Isolation, identification, and probiotic characterization. World J. Microbiol. Biotechnol. 2014, 30, 43–53. [Google Scholar] [CrossRef]
- Zanirati, F.D.; Abatemarco, M.; Sandes, C.H.S.; Nicoli, R.J.; Nunes, C.A.; Neumann, E. Selection of lactic acid bacteria from Brazilian kefir grains for potential use as starter or probiotic cultures. Anaerobe 2015, 32, 70–76. [Google Scholar] [CrossRef]
- FAO/WHO. FAO/WHO Joint Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food (30 April 2002 and 1 May 2002); Scientific Research Publishing: London, ON, Canada, 2002. [Google Scholar]
- De Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães, A.I., Jr.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef]
- Mantzourani, I.; Kazakos, S.; Terpou, A.; Alexopoulos, A.; Bezirtzoglou, E.; Bekatorou, A.; Plessas, S. Potential of the probiotic Lactobacillus plantarum ATCC 14917 strain to produce functional fermented pomegranate juice. Foods 2018, 8, 4. [Google Scholar] [CrossRef]
- Gao, Y.; Hamid, N.; Gutierrez-Maddox, N.; Kantono, K.; Kitundu, E. Development of a Probiotic Beverage Using Breadfruit Flour as a Substrate. Foods 2019, 8, 214. [Google Scholar] [CrossRef]
- Fiorda, F.A.; Pereira, G.V.M.; Thomaz-Soccol, V.; Medeiros, A.P.; Rakshit, S.K.; Soccol, C.R. Development of kefir-based probiotic beverages with DNA protection and antioxidant activities using soybean hydrolyzed extract, colostrum and honey. LWT-Food Sci. Technol. 2016, 86, 690–697. [Google Scholar] [CrossRef]
- Barszczewski, W.; Robak, M. Differentiation of contaminating yeasts in brewery by PCR-based techniques. Food Microbiol. 2004, 21, 227–231. [Google Scholar] [CrossRef]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef]
- Perelmuter, K.; Fraga, M.; Zunino, P. In vitro activity of potential probiotic Lactobacillus murinus isolated from the dog. J. Appl. Microbiol. 2008, 104, 1718–1725. [Google Scholar] [CrossRef] [PubMed]
- Chelliah, R.; Ramakrishnan, R.S.; Prabhu, R.P.; Antony, U. Evaluation of antimicrobial activity and probiotic properties of wild-strain Pichia kudriavzevii isolated from frozen idli batter. Yeast 2016, 33, 385–401. [Google Scholar] [CrossRef]
- Ogunremi, R.O.; Sanni, I.A.; Agrawal, R. Probiotic potentials of yeasts isolated from some cereal-based Nigerian traditional fermented food products. J. Appl. Microbiol. 2015, 119, 797–808. [Google Scholar] [CrossRef]
- Brand, W.W.; Cuvelier, E.M.; Berse, C. Use of a free radical method to evaluate antioxidant activity. Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Singh, P.N.; McCoy, T.M.; Tice, R.R.; Schneider, L.E. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef]
- Collins, R.A. The comet assay for DNA damage and repair. Mol. Biotechnol. 2004, 26, 249–261. [Google Scholar] [CrossRef]
- Foulquié-Moreno, M.R.; Callewaert, R.; Devreese, B.; Van Beeumen, J.; De Vuyst, L. Isolation and biochemical characterization of enterocins produced by enterococci from different sources. J. Appl. Microbiol. 2003, 94, 214–229. [Google Scholar] [CrossRef]
- Usman, H.A. Viability of Lactobacillus gasseri and its cholesterol-binding and antimutagenic activities during subsequent refrigerated storage in non-fermented milk. J. Dairy Sci. 1999, 82, 2536–2542. [Google Scholar] [CrossRef]
- Likotrafiti, E.; Tuohy, K.M.; Gibson, G.R.; Rastall, R.A. Development of antimicrobial synbiotics using potentially-probiotic faecal isolates of Lactobacillus fermentum and Bifidobacterium longum. Anaerobe 2013, 20, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Lian, W.C.; Hsiao, H.C.; Chou, C.C. Viability of microencapsulated bifidobacteria in simulated gastric juice and bile solution. Int. J. Food Microbiol. 2003, 86, 293–301. [Google Scholar] [CrossRef]
- Saulnier, A.M.D.; Spinler, K.J.; Gibson, R.R.G.; Versalovic, J. Mechanisms of probiosis and prebiosis: Considerations for enhanced functional foods. Curr. Opin. Biotech. 2009, 20, 135–141. [Google Scholar] [CrossRef]
- Valdéz, J.C.; Peral, M.C.; Rachid, M.; Santana, M.; Perdigón, G. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: The potential use of probiotics in wound treatment. Clin. Microbiol. Infect. 2005, 11, 472–479. [Google Scholar] [CrossRef]
- Tsai, C.C.; Lin, P.P.P.; Hsieh, Y.M. Three Lactobacillus strains from healthy infant stool inhibit enterotoxigenic Escherichia coli grown in vitro. Anaerobe 2008, 14, 61–67. [Google Scholar] [CrossRef]
- Ramos, A.N.; Sesto Cabral, M.E.; Noseda, D.; Bosch, A.; Yantorno, O.M. Antipathogenic properties of Lactobacillus plantarum on Pseudomonas aeruginosa: The potential use of its supernatants in the treatment of infected chronic wounds. Wound Repair Regen. 2012, 20, 552–562. [Google Scholar] [CrossRef]
- Yamazakia, M.; Ohtsua, H.; Yakabea, Y.; Kishimab, M.; Abea, H. In vitro screening of Lactobacilli isolated from chicken excreta to control Salmonella enteritidis and Typhimurium. Brit. Poult. Sci. 2012, 53, 183–189. [Google Scholar] [CrossRef]
- Ramirez-Chavarin, M.L.; Wacher, C.; Eslava-Campos, C.A.; Perez-Chabela, M.L. Probiotic potential of thermotolerant lactic acid bacteria strains isolated from cooked meat products. Int. Food Res. 2013, 20, 991–1000. [Google Scholar]
- Keller, M.K.; Hasslöf, P.; Stecksén-Blicks, C.; Twetman, S. Co-aggregation and growth inhibition of probiotic lactobacilli and clinical isolates of mutans streptococci: An in vitro study. Acta Odontol. Scand. 2011, 69, 263–268. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, M.; Dai, X. Biological characteristics and probiotic effect of Leuconostoc lactis strain isolated from the intestine of black porgy fish. Braz. J. Microbiol. 2013, 44, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Maragkoudakis, P.A.; Zoumpopoulou, G.; Miaris, C.; Kalantzopoulos, G.; Pot, B.; Tsakalidou, E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int. Dairy J. 2006, 3, 189–199. [Google Scholar] [CrossRef]
- Adimpong, D.B.; Nielsen, D.S.; Sørensen, K.I.; Derkx, P.M.; Jespersen, L. Genotypic characterization and safety assessment of lactic acid bacteria from indigenous African fermented food products. BMC Microbiol. 2012, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]

| Microorganism | Inhibition Zone (mm) * | |
|---|---|---|
| Escherichia coli | Staphylococcus aureus | |
| Lactobacillus satsumensis (LBPF1) | 12.5 ± 0.50 Ca ** | 10.5 ± 0.50 Ba |
| Leuconostoc mesenteroides (LBPF2) | 10.5 ± 0.50 Ca | 12.0 ± 1.00 Ba |
| Saccharomyces cerevisiae (LBPF3) | 8.0 ± 0.10 Ca | 8.5 ± 0.50 Ba |
| Honey kefir beverage | 27.5 ± 1.50 Aa | 19.5 ± 1.50 Ab |
| Control (Ampicillin 50 mg/mL) | 42.5 ± 1.50 Ba | 23.5 ± 0.50 Aa |
| Microorganism | Damage Index * | |
|---|---|---|
| 1 h | 24 h | |
| Lactobacillus satsumensis | 3.05 ± 0.25 aA ** | 3.45 ± 0.13 aA |
| Leuconostoc mesenteroides | 3.26 ± 0.21 aA | 3.65 ± 0.24 aA |
| Saccharomyces cerevisiae | 2.44 ± 0.06 bA | 2.39 ± 0.16 bA |
| Positive control | 3.79 ± 0.19 cA | 3.86 ± 0.06 aA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira Coelho, B.; Fiorda-Mello, F.; de Melo Pereira, G.V.; Thomaz-Soccol, V.; Rakshit, S.K.; de Carvalho, J.C.; Soccol, C.R. In Vitro Probiotic Properties and DNA Protection Activity of Yeast and Lactic Acid Bacteria Isolated from A Honey-Based Kefir Beverage. Foods 2019, 8, 485. https://doi.org/10.3390/foods8100485
de Oliveira Coelho B, Fiorda-Mello F, de Melo Pereira GV, Thomaz-Soccol V, Rakshit SK, de Carvalho JC, Soccol CR. In Vitro Probiotic Properties and DNA Protection Activity of Yeast and Lactic Acid Bacteria Isolated from A Honey-Based Kefir Beverage. Foods. 2019; 8(10):485. https://doi.org/10.3390/foods8100485
Chicago/Turabian Stylede Oliveira Coelho, Bruna, Fernanda Fiorda-Mello, Gilberto V. de Melo Pereira, Vanete Thomaz-Soccol, Sudip K. Rakshit, Júlio Cesar de Carvalho, and Carlos Ricardo Soccol. 2019. "In Vitro Probiotic Properties and DNA Protection Activity of Yeast and Lactic Acid Bacteria Isolated from A Honey-Based Kefir Beverage" Foods 8, no. 10: 485. https://doi.org/10.3390/foods8100485
APA Stylede Oliveira Coelho, B., Fiorda-Mello, F., de Melo Pereira, G. V., Thomaz-Soccol, V., Rakshit, S. K., de Carvalho, J. C., & Soccol, C. R. (2019). In Vitro Probiotic Properties and DNA Protection Activity of Yeast and Lactic Acid Bacteria Isolated from A Honey-Based Kefir Beverage. Foods, 8(10), 485. https://doi.org/10.3390/foods8100485