Abstract
Introduction: The aim of this study was to assess the incidence of Corynebacterium striatum prosthetic joint infections (PJI) to determine if an increase has occurred recently. Moreover, susceptibility testing was conducted on C. striatum preserved isolates to determine antibiotic options for these infections. Methods: Retrospective review of PJI cases was conducted from 1/2017 through 1/2021 compared to 1/2021 through 7/2022 to determine how many cases of C. striatum have occurred for each of these time points. From these cases, demographics, outcomes and risk factors for C. striatum PJI were recorded. The preserved clinical isolates from these cases were tested for susceptibility to different antibiotics. Results: A statistically significant increase in the proportion of C. striatum PJI cases (1.98 to 7.84, p = 0.0489) has occurred over the past 16 months at a single institution. Chronic wounds and exposure to daptomycin were associated with the majority of these cases. Susceptibility testing of the clinical isolates showed uniform susceptibility to vancomycin, linezolid and dalbavancin. Uniform resistance was seen with ciprofloxacin, tetracycline and doxycycline as well. Interestingly, 85.7% of the isolates displayed inducible daptomycin resistance after overnight exposure to daptomycin. Conclusions: C. striatum is an emerging PJI pathogen. It is important for clinicians to be cognizant that this pathogen can have inducible high level daptomycin resistance and that daptomycin is likely not a reliable antibiotic for these infections. While vancomycin and linezolid are the traditional antibiotics to use in these infections, other antibiotics such as dalbavancin, may also have utility, but more research is needed to determine the effectiveness of this antibiotic in C. striatum infections.
Introduction
Staphylococcus aureus and bacteria that are part of the skin microbiome, most notably coagulase negative Staphylococcus, are the predominate causes of prosthetic joint infections (PJI) [1]. These bacteria typically cause PJI by infecting prosthetics during surgical interventions, from direct inoculation of wounds or hematogenous seeding of the prosthetic [1]. Bacterial biofilms make PJIs difficult to treat given conventional antibiotics have limited ability to eradicate bacteria in these sessile states. Consequently, to definitively cure chronic PJI, removal of the infected prosthetic is required and, if this is not feasible, patients often require chronic antibiotic suppression therapy to prevent recurrence [1].
A rare cause of PJI is Corynebacterium striatum, which is a commensal Gram-positive rod that is part of the skin microbiome [2]. This bacterium is associated with severe infections in immunocompromised hosts, especially in terminally ill oncological patients [3]. Prolonged use of intravenous antibiotics, central venous catheters and extended hospitalizations are known risk factors for acquiring C. striatum infections [3,4,5]. Recent studies have shown that the incidence of C. striatum infections is increasing among immunocompetent hosts [4,5,6]. Given the fact that this bacterium has been associated with numerous multidrug resistance mechanisms, treatment of C. striatum infections can be complex [7,8]. Consequently, the goals of this study were to: 1) determine if there has been an increased incidence of C. striatum PJI cases at a single academic center and 2) to evaluate the susceptibility pattern of preserved C. striatum PJI isolates to elucidate intravenous and oral antibiotic options for PJI patients.
Methods
Review and categorization of study population
Approval to conduct this retrospective cohort study was granted by the University of Maryland internal review board (HP-00103285). The only inclusion criterion was that patients had to have had PJI caused by C. striatum between 1 January 2017 and 1 July 2022 at the University of Maryland Medical Center. All other cases caused by other bacterial causes were excluded. Patients were retrospectively identified with the use of Current Procedural Terminology (CPT) codes which included: 27486, 27134, 27488, and 27091. For this study, PJI included only patients that met the standardized definition created by the Musculoskeletal Infection Society [9]. C. striatum was diagnosed with the use of Vitek 2 (BioMérieux, France) and the culture method used was the same during the two time periods. This culture method included streaking PJI samples on blood agar plates and incubating at 37°C for five days.
Patients that had C. striatum PJI over this time period had their demographics recorded, and treatments and outcomes were measured as were exposure to daptomycin and presence of chronic wounds. Recent exposure to daptomycin refers to patients being given doses of daptomycin within 3 months of the diagnosis of C. striatum PJI.
Evaluating susceptibility with Kirby-Bauer disc diffusion
Seven C. striatum PJI isolates from the time period mentioned above that had been preserved at -80°C were thawed and streaked onto blood agar plates (Hardy Diagnostics, USA) and then incubated at 37°C for 24 hours. After a 0.5 McFarland standard was achieved, each isolate was placed onto individual 150 mm Mueller-Hinton agar plates with 5% sheep blood plates (Thermo Scientific, USA). The agar was let to dry for approximately 15 minutes before antibiotic discs were placed. Antibiotic discs used in this study were vancomycin, linezolid, ciprofloxacin, (Hardy Diagnostics, USA) given standardized breakpoints were available to define if the isolates were were susceptible or non-susceptible to these antibiotics (reference M100). Additional antibiotic discs of sulfamethoxazole-trimethoprim, cefoxitin, cefuroxime, ceftriaxone, ampicillin, amoxicillin-clavulanate and ceftaroline were tested without official breakpoints. Antibiotic discs were placed at least 20 mm apart from the other discs and more than 15 mm from the edge of the agar plate. After all the discs were placed each plate was incubated at 36°C and read 20 hours later. The experiment was conducted in triplicates and the diameter of the zone of inhibition for each isolate was determined by recording the smallest diameter.
Susceptibility to dalbavancin
Microbroth dilution was used to determine dalbavancin MIC for the seven clinical isolates. This was conducted by dissolving dalbavancin (MedChemExpress, USA) into D5W and then diluting to concentrations desired. Microbroth dilution was conducted by following the International standard protocol (ISO 20776-1). A 96 microwell plate (Costar) was used to conduct the experiment. Optical density was measured at time zero and 24 hours with a SpectraMax ID3 microplate reader. The experiment was conducted in triplicate and the highest MIC for each isolate was recorded.
Evaluation of inducible daptomycin resistance
Each C. striatum strain was tested for baseline daptomycin susceptibility by taking individual colonies and following the same procedure discussed above that was used for the Kirby-Bauer disc diffusion. Instead of discs being placed, three daptomycin gradient diffusion test strips (BioMérieux, France) were placed onto each 150 mm Mueller-Hinton agar with 5% sheep blood plate (Thermo Scientific, USA). Each plate was incubated at 36 °C and daptomycin minimal inhibitory concentrations (MIC) were read 20 h later to determine the MIC to daptomycin for each isolate. The isolates were then exposed to daptomycin by cutting a daptomycin gradient diffusion test strip in half and incubating each clinical isolate independently overnight.10 Daptomycin susceptibility was then repeated again by using daptomycin gradient diffusion test strips. The isolates that showed daptomycin resistance were then serially grown on blood agar plates (Hardy Diagnostics, USA) on daily basis. Evaluation for retained inducible daptomycin resistance was evaluated on day 3 and day 5 with three daptomycin gradient diffusion test strips. In correlation, vancomycin susceptibility with vancomycin disc diffusion was evaluated in tandem on days 1, 3 and 5.
Results
Table 1 shows the baseline characteristics of C. striatum PJI from 1/2017. Notably, there had been three cases (3/151) of C. striatum from 1/2017-1/2021 but there have been four cases (4/51) of C. striatum from 12021-7/2022 (p=0.0489). Of the seven patients only one patient was immunocompromised. Six of the seven patients had had recent exposure to daptomycin and had chronic wounds. Most patients were treated with two-stage revision surgery and were treated with vancomycin or dalbavancin. One patient did not undergo revision surgery and has been on amoxicillin-clavulanate oral suppression therapy with no evidence of recurrence.

Table 1.
Baseline characteristics of C. striatum PJI.
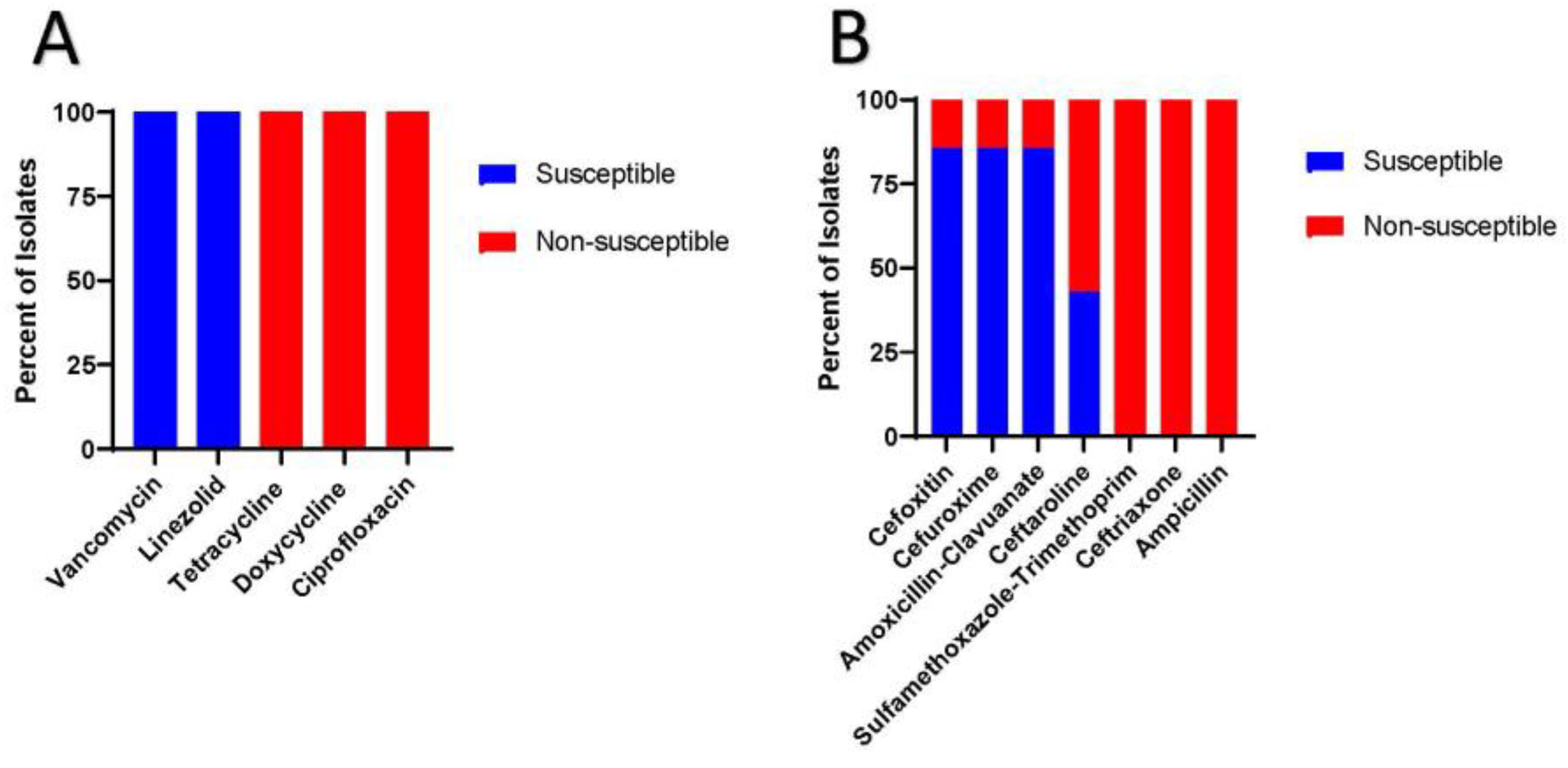
Figure 1 shows the results of Kirby-Bauer testing. Notably all the PJI isolates were susceptible to vancomycin and linezolid with zones of inhibited growth greater than 24 mm and 26 mm, respectively. Conversely all the isolates were non-susceptible to ciprofloxacin, tetracycline and doxycycline, with zones of bacterial growth inhibition all less than 15 mm. Supplemental Figure 1 shows the Kirby-Bauer results for antibiotics with no official breakpoints.
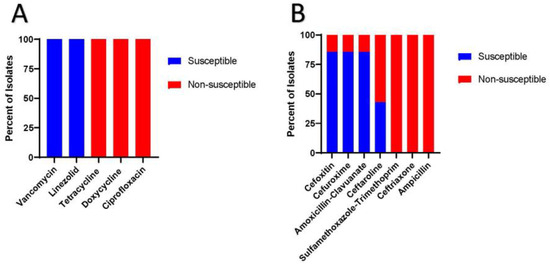
Figure 1.
Kirby-Bauer results for the seven C. striatum PJI isolates. 1a) Susceptibilities of antibiotics with known breakpoints. 1b) Susceptibilities of antibiotics with unknown breakpoints.
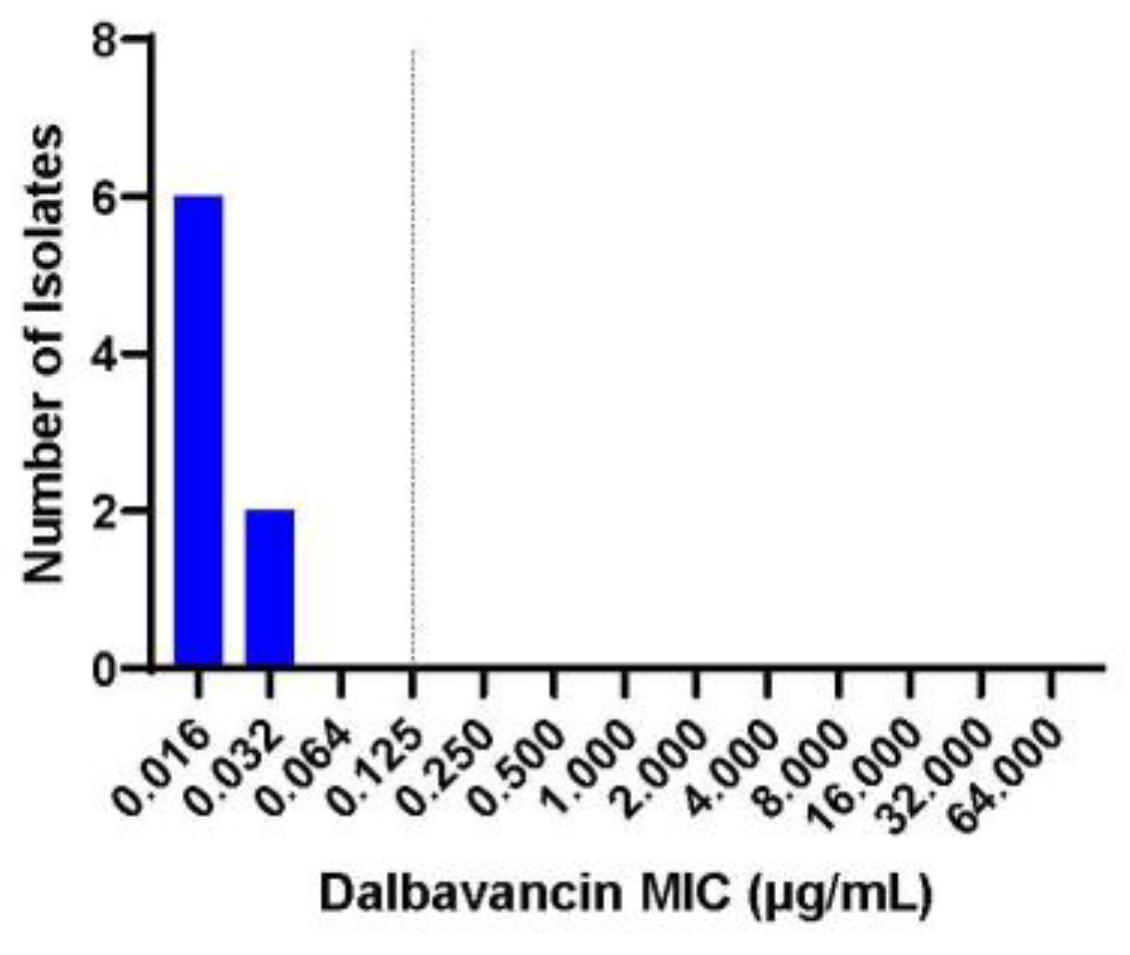
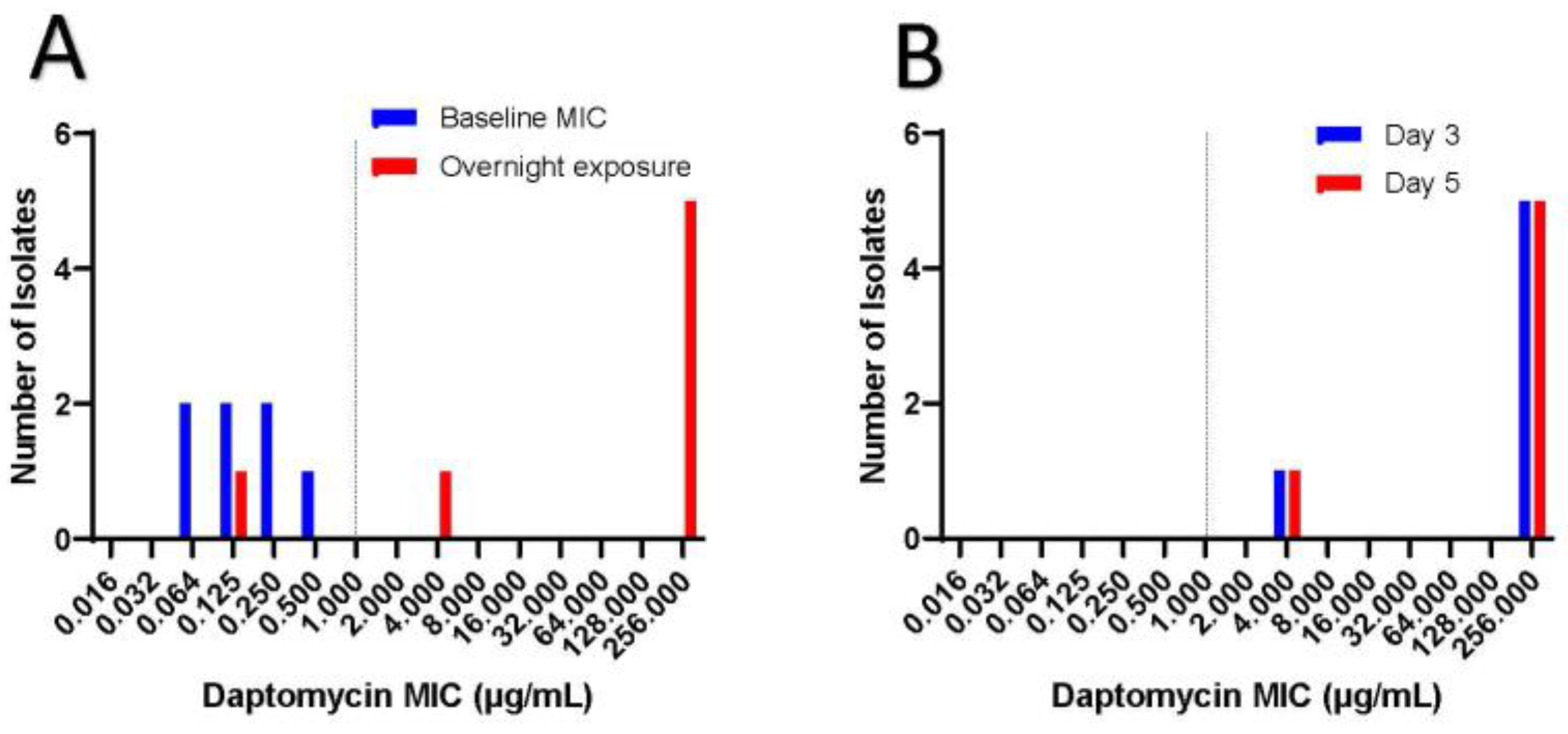
Figure 2 shows the MIC to dalbavancin for the seven clinical isolates in which all showed susceptibility to dalbavancin with MIC less or equal to 0.032 µg/mL. Interestingly, all clinical isolates were susceptible to daptomycin at baseline (Figure 3). However, after overnight exposure to daptomycin, six (85.7%) isolates showed MIC over 1 and five (71.4%) had MIC over 256. This daptomycin resistance persisted despite growing each isolate daily on blood agar plates and then testing for daptomycin non-susceptibility on days 3 and 5 (Figure 3). All isolates retained vancomycin susceptibility on day 1, 3 and 5.

Figure 2.
Dalbavancin MIC for the seven C. striatum clinical isolates. The breakpoint of 0.125 µg/mL is extrapolated from staphylococcal infections.
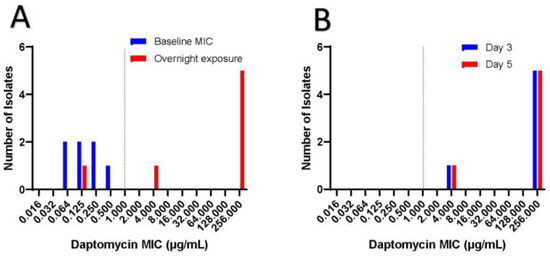
Figure 3.
Inducible daptomycin resistance.
Discussion
C. striatum is a rare PJI pathogen but the statistically significant increase in C. striatum cases over the past eighteen months at a single institution raises concern that this might be an emerging PJI pathogen even in immunocompetent hosts. This increase in cases does not seem to be correlated with COVID-19 given the annual number of PJI cases conducted at this medical center has not significantly changed compared to before the pandemic. Rather a potential explanation for the increase is cases is because since 1/2021 at the University of Maryland, there has been an effort to reduce acute kidney injury with less use of vancomycin and more use of daptomycin for Gram-positive bacterial infections that are not pulmonic in etiology. This is important, because as seen here and reinforced by others, C. striatum has the ability to induce high levels of daptomycin resistance after brief exposures (Figure 2) [10]. Clinicians should be cognizant about this because C. striatum is part of the skin microbiome, and patients with chronic wounds and recent exposure to daptomycin may be at risk for infections with this pathogen (Table 1).
Figure 3a shows baseline daptomycin MIC in which all isolates were susceptible to daptomycin with MIC less than 1. Dotted line represents MIC of 1, which is considered susceptible. When exposed to daptomycin for 16 hours, 71.4 % of the isolates displayed high levels of daptomycin resistance with MIC over 256 µg/mL. Figure 3b shows that high grade daptomycin resistance continued despite isolation of individual colonies from each clinical isolate and plating them daily on blood agar plates. Daptomycin MIC was tested on day 3 and day 5, in which all isolates retained the same daptomycin resistance.
The mechanism of inducible daptomycin resistance is different than that seen with Enterococcus spp. and Staphylococcus spp., where changes in cell wall charge, composition and/or fluidity cause daptomycin resistance [11,12]. Rather, C. striatum resistance is thought to be from mutations in the pgsA2 gene, which encodes an enzyme that helps form peptidoglycan from diphosphate diglyceride [10]. This daptomycin resistance is persistent and does not revert back to daptomycin susceptibility even when isolates are subcultured without daptomycin (Figure 3). This is different than the daptomycin resistance seen with Enterococcus spp. and Staphylococcus spp., which often reverts to daptomycin susceptibility on serial passage of bacteria without daptomycin pressures [10]. Moreover, as seen here, vancomycin resistance is often not associated with the development of daptomycin resistance in C. striatum. This has important clinical ramification because daptomycin is often considered an effective antibiotic for Gram-positive bacteria, but the findings here support the need to check for inducible daptomycin resistance before using this antibiotic to treat these infections.
Consequently, daptomycin is likely not an antibiotic that should be used for most C. striatum PJI. Obstinately, there is also a paucity of data on alternative antibiotics to treat this pathogen beyond vancomycin or linezolid [5,7]. Vancomycin and linezolid can be associated with adverse drug reactions which make the use of other agents desirable to mitigate these, especially when treating PJI patients as outpatients. As a result, this study evaluated the potential of dalbavancin to treat these infections. Dalbavancin is similar to vancomycin, but has been associated with less drug reactions and is typically dosed on weekly basis given its prolonged pK data [13,14]. This antibiotic has also been shown to stay in cortical bones at concentrations above 0.125 µg/mL for over 4-6 weeks [15,16]. These properties allow this antibiotic to be an advantageous therapeutic in the musculoskeletal infections, but limited data is available assessing the activity of dalbavancin for C. striatum infections. Here dalbavancin had robust activity to all the clinical isolates with MIC less than or equal to 0.032 µg/mL. This suggests that dalbavancin is a potential alternative treatment of C. striatum PJI, while also circumventing the need for peripheral inserted central catheters.
Beyond dalbavancin, ceftaroline has been shown to have sporadic susceptibility in C. striatum isolates and thus should be used with caution especially given the lack of official breakpoints [17]. Furthermore, the tetracyclines, ciprofloxacin and ceftriaxone, ampicillin and sulfamethoxazole-trimethoprim (Figure 1a,b) have been shown to display limited susceptibilities in C. striatum isolates [7,8,11]. Interestingly, a sizable portion of C. striatum isolates have been shown to retain susceptibilities to cefoxitin, cefuroxime and amoxicillin-clavulanate (Figure 1b) [7,8]. This occurs because this bacterium typically has a bla gene encoding class A beta-lactamase [7]. Class A extended spectrum beta-lactamase confers resistance to penicillins and 1st and 3rd generation cephalosporins but retains susceptibility to specific 2nd generation cephalosporins most notably cefoxitin and beta-lactamase inhibitor, such as clavulanic acid [7,8]. This is important for clinicians to be cognizant about because amoxicillin-clavulanate and cefuroxime can be potential oral suppression antibiotic options in C. striatum PJI. However, given the paucity of data on appropriate breakpoints for these antibiotics, further research is needed.
This study is associated with some limitations. The small number of clinical isolates was a result of the rarity of this infection, which limits generalizability of the findings seen here. The use of Vitek 2 (BioMérieux, France) was also used to identify C. striatum, as is commonly used in many hospitals and has been proven to have excellent reliability in diagnosing these infections [18]. However, genetic testing was not conducted on the clinical isolates to elucidate the exact mechanism of their phenotypic susceptibilities or if these were similar strains of C. striatum. Correlated with this, the lack of standardized dalbavancin breakpoints limits the ability to make definitive statements on susceptibility for this antibiotic. Rather the breakpoints for S. aureus were used to determine relative dalbavancin susceptibilities. Further research is therefore needed to clarify the appropriate breakpoints of antibiotics for C. striatum. Moreover, Gram-positive rods are often considered a contaminant in PJI, which may lead this to be an underreported pathogen in PJI and this study should raise awareness of this potential emerging PJI pathogen. Nonetheless, the findings here support the need to test for phenotypic antibiotic susceptibilities to various antibiotics to allow clinicians options in treating these patients.
Conclusions
In conclusion, C. striatum is a PJI pathogen that clinicians need to be aware of. This is in part because this bacterium can have unique high level daptomycin resistance that can develop with short exposures to this antibiotic. This is important to realize given the fact that daptomycin is a commonly used antibiotic for Gram-positive infections. Moreover, this study reinforces that vancomycin, linezolid and potentially dalbavancin are the preferred agents for C. striatum infections. However, other agents such as second generation cephalosporins and beta-lactam plus beta-lactamase inhibitors may also be effective, but the absence of standardized breakpoints for these antibiotics requires further research.
Author Contributions
JBD devised and conducted this study and experimentation. JBD wrote the manuscript and approved the final version for publication.
Funding
None to declare.
Institutional Review Board Statement
This is an observational study. The University of Maryland Research Ethics Committee has confirmed that no ethical approval is required.
Data Availability Statement
Data can be shared upon reasonable request.
References
- Tande, A.J.; Patel, R. Prosthetic joint infection. Clin Microbiol Rev. 2014, 27, 302–345. [Google Scholar] [CrossRef]
- Alibi, S.; Ferjani, A.; Boukadida, J.; et al. Occurrence of Corynebacterium striatum as an emerging antibiotic-resistant nosocomial pathogen in a Tunisian hospital. Sci Rep. 2017, 7, 9704. [Google Scholar] [CrossRef]
- Chen, F.L.; Hsueh, P.R.; Teng, S.O.; Ou, T.Y.; Lee, W.S. Corynebacterium striatum bacteremia associated with central venous catheter infection. J Microbiol Immunol Infect. 2012, 45, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.P.; Ferguson, D.A.; Jr Sarubbi, F.A. Corynebacterium striatum: An underappreciated community and nosocomial pathogen. J Infect. 2005, 50, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, L.; Suárez, A.I.; Rodríguez-Baño, J.; Bernard, K.; Muniáin, M.A. Clinical significance of Corynebacterium striatum isolated from human samples. Clin Microbiol Infect. 1997, 3, 634–639. [Google Scholar] [CrossRef]
- Lee, Y.W.; Huh, J.W.; Hong, S.B.; et al. Severe pneumonia caused by Corynebacterium striatum in adults, Seoul, South Korea, 2014-2019. Emerg Infect Dis. 2022, 28, 2147–2154. [Google Scholar] [CrossRef] [PubMed]
- Leyton, B.; Ramos, J.N.; Baio, P.V.P.; et al. Treat me well or will resist: Uptake of mobile genetic elements determine the resistome of Corynebacterium striatum. Int J Mol Sci. 2021, 22, 7499. [Google Scholar] [CrossRef] [PubMed]
- Milosavljevic, M.N.; Milosavljevic, J.Z.; Kocovic, A.G.; et al. Antimicrobial treatment of Corynebacterium striatum invasive infections: A systematic review. Rev Inst Med Trop Sao Paulo. 2021, 63, e49. [Google Scholar] [CrossRef] [PubMed]
- Parvizi, J.; Tan, T.L.; Goswami, K.; et al. The 2018 definition of periprosthetic hip and knee infection: An evidence-based and validated criteria. J Arthroplasty. 2018, 33, 1309–1314.e2. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.F.; McElvania, E.; Wallace, M.A.; et al. Evaluating the rapid emergence of daptomycin resistance in Corynebacterium: A multicenter study. J Clin Microbiol. 2021, 59, e02052–20. [Google Scholar] [CrossRef] [PubMed]
- Gómez Casanova, N.; Siller Ruiz, M.; Muñoz Bellido, J.L. Mechanisms of resistance to daptomycin in Staphylococcus aureus. Rev Esp Quimioter. 2017, 30, 391–396. [Google Scholar] [PubMed]
- Ernst, C.M.; Peschel, A. MprF-mediated daptomycin resistance. Int J Med Microbiol. 2019, 309, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Rappo, U.; Puttagunta, S.; Shevchenko, V.; et al. Dalbavancin for the treatment of osteomyelitis in adult patients: A randomized clinical trial of efficacy and safety. Open Forum Infect Dis. 2018, 6, ofy331. [Google Scholar] [CrossRef] [PubMed]
- Almangour, T.A.; Alhifany, A.A. Dalbavancin for the management of osteomyelitis: A major step forward? J Antimicrob Chemother. 2020, 75, 2717–2722. [Google Scholar] [CrossRef] [PubMed]
- Bouza, E.; Valerio, M.; Soriano, A.; et al. Dalbavancin in the treatment of different gram-positive infections: A real-life experience. Int J Antimicrob Agents. 2018, 51, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.W.; Puttagunta, S.; Sprenger, C.R.; Rubino, C.; Van Wart, S.; Baldassarre, J. Extended-duration dosing and distribution of dalbavancin into bone and articular tissue. Antimicrob Agents Chemother. 2015, 59, 1849–1855. [Google Scholar] [CrossRef] [PubMed]
- Sader, H.S.; Jones, R.N.; Stilwell, M.G.; Flamm, R.K. Ceftaroline activity tested against uncommonly isolated Gram-positive pathogens: Report from the SENTRY Antimicrobial Surveillance Program (2008-2011). Int J Antimicrob Agents. 2014, 43, 284–286. [Google Scholar] [CrossRef] [PubMed]
- Rennie, R.P.; Brosnikoff, C.; Turnbull, L.; et al. Multicenter evaluation of the Vitek 2 anaerobe and Corynebacterium identification card. J Clin Microbiol. 2008, 46, 2646–2651. [Google Scholar] [CrossRef] [PubMed]
© GERMS 2023.