Exploring the Regulatory Role of XIST-microRNAs/mRNA Network in Circulating CD4+ T Cells of Hepatocellular Carcinoma Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Patient Population
2.2. Isolation of Circulating CD4+ Cells and RNA Extraction
2.3. RNA Quality Determination and NGS
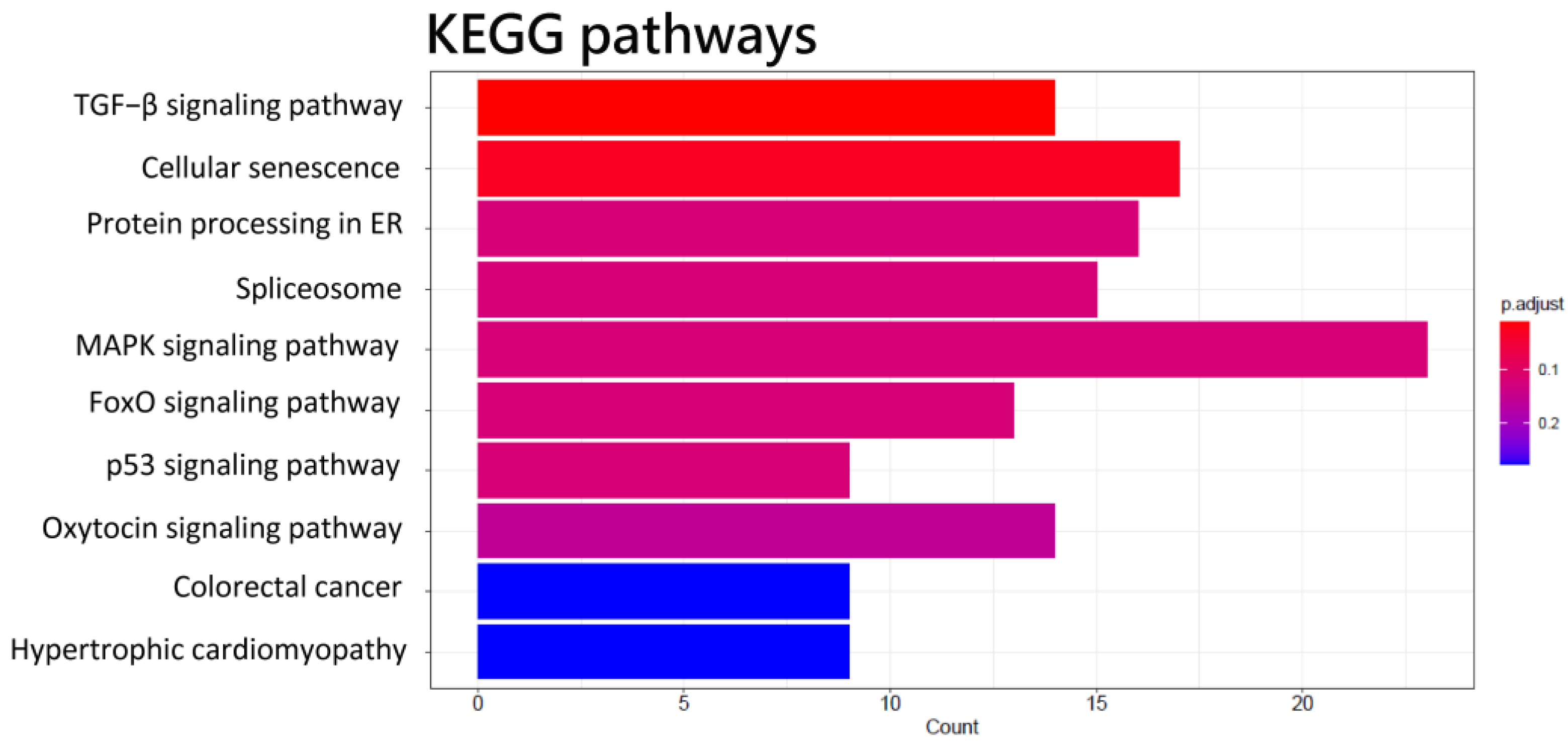
2.4. GO and KEGG Enrichment Analysis
2.5. Real-Time Quantitative Polymerase Chain Reaction
2.6. The Interaction of miRNA-lncRNA and miRNA–mRNA
2.7. CD4+ T Cells Infiltration Analysis
2.8. Statistical Analysis
3. Results
3.1. NGS Analysis
3.2. The miRNA-lncRNA or miRNA–mRNA Interaction by ENCORI
3.3. Validation of the Expression of Transcripts
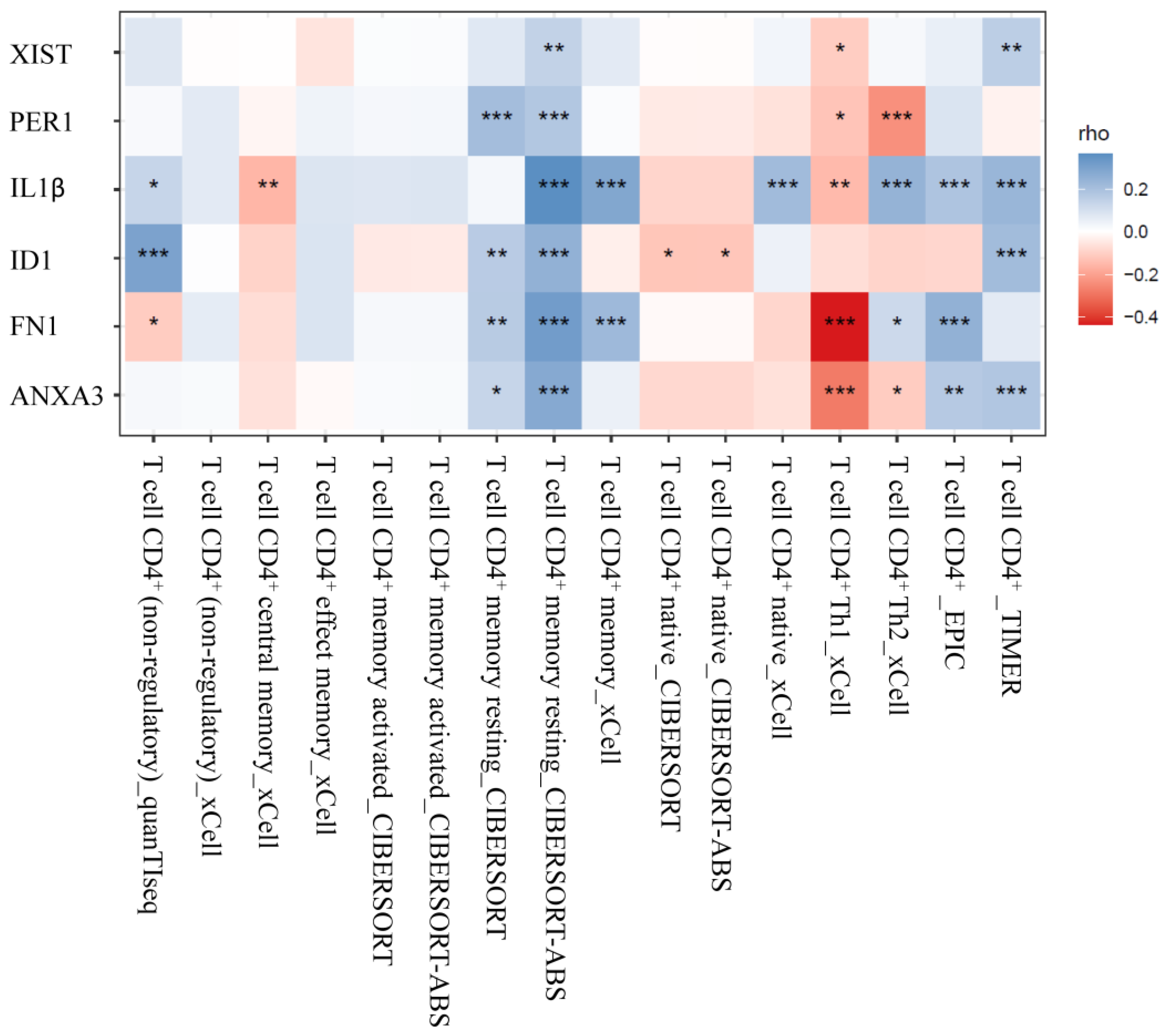
3.4. Analysis of CD4+ T Cells Infiltrates Correlation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sia, D.; Villanueva, A.; Friedman, S.L.; Llovet, J.M. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology 2017, 152, 745–761. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Gea, V.; Toffanin, S.; Friedman, S.L.; Llovet, J.M. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology 2013, 144, 512–527. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Yapali, S.; Tozun, N. Epidemiology and viral risk factors for hepatocellular carcinoma in the Eastern Mediterranean countries. Hepatoma Res. 2018, 4, 24. [Google Scholar] [CrossRef]
- Lawal, G.; Xiao, Y.; Rahnemai-Azar, A.A.; Tsilimigras, D.I.; Kuang, M.; Bakopoulos, A.; Pawlik, T.M. The Immunology of Hepatocellular Carcinoma. Vaccines 2021, 9, 1184. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.Q.; Cai, D.; Gong, J.P.; Lai, X. Innate immune cells and their interaction with T cells in hepatocellular carcinoma. Oncol. Lett. 2021, 21, 57. [Google Scholar] [CrossRef]
- Schneider, S.L.; Ross, A.L.; Grichnik, J.M. Do inflammatory pathways drive melanomagenesis? Exp. Dermatol. 2015, 24, 86–90. [Google Scholar] [CrossRef]
- Singh, B.; Kumar Rai, A. Loss of immune regulation in aged T-cells: A metabolic review to show lack of ability to control responses within the self. Hum. Immunol. 2022, 83, 808–817. [Google Scholar] [CrossRef]
- Huang, L.H.; Hsieh, T.M.; Huang, C.Y.; Liu, Y.W.; Wu, S.C.; Chien, P.C.; Hsieh, C.H. Disparity of Hepatocellular Carcinoma in Tumor Microenvironment-Related Genes and Infiltrating Immune Cells between Asian and Non-Asian Populations. Genes 2021, 12, 1274. [Google Scholar] [CrossRef]
- Chen, Z.; Zheng, X.; Zhao, J.; Tang, J.; Hu, L.; Wang, S. Glucose oxidase-loaded colloidal stable WS(2) nanobowls for combined starvation/photothermal therapy of colorectal tumors. Int. J. Pharm. 2023, 636, 122848. [Google Scholar] [CrossRef]
- Zheng, X.; Jin, W.; Wang, S.; Ding, H. Progression on the Roles and Mechanisms of Tumor-Infiltrating T Lymphocytes in Patients with Hepatocellular Carcinoma. Front. Immunol. 2021, 12, 729705. [Google Scholar] [CrossRef]
- Tay, R.E.; Richardson, E.K.; Toh, H.C. Revisiting the role of CD4+ T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther. 2021, 28, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Luckheeram, R.V.; Zhou, R.; Verma, A.D.; Xia, B. CD4+ T cells: Differentiation and functions. Clin. Dev. Immunol. 2012, 2012, 925135. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zhang, Z.; Zhou, L.; Qi, Z.; Xing, S.; Lv, J.; Shi, J.; Fu, B.; Liu, Z.; Zhang, J.Y.; et al. Impairment of CD4+ cytotoxic T cells predicts poor survival and high recurrence rates in patients with hepatocellular carcinoma. Hepatology 2013, 58, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, J.K.; Peng, Y.; He, W.; Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 2020, 19, 77. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Gholipour, M.; Hussen, B.M.; Taheri, M. The Impact of Long Non-Coding RNAs in the Pathogenesis of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 649107. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, L.; Gu, J.; Zhang, H.; Yuan, J.; Lian, Q.; Lv, G.; Wang, S.; Wu, Y.; Yang, Y.T.; et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat. Commun. 2017, 8, 14421. [Google Scholar] [CrossRef] [PubMed]
- West, K.A.; Lagos, D. Long Non-Coding RNA Function in CD4+ T Cells: What We Know and What Next? Non-Coding RNA 2019, 5, 43. [Google Scholar] [CrossRef]
- Hirsova, P.; Bamidele, A.O.; Wang, H.; Povero, D.; Revelo, X.S. Emerging Roles of T Cells in the Pathogenesis of Nonalcoholic Steatohepatitis and Hepatocellular Carcinoma. Front. Endocrinol. 2021, 12, 760860. [Google Scholar] [CrossRef]
- Plasek, L.M.; Valadkhan, S. lncRNAs in T lymphocytes: RNA regulation at the heart of the immune response. Am. J. Physiol. Cell Physiol. 2021, 320, C415–C427. [Google Scholar] [CrossRef]
- Shi, T.; Morishita, A.; Kobara, H.; Masaki, T. The Role of Long Non-Coding RNA and microRNA Networks in Hepatocellular Carcinoma and Its Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 10630. [Google Scholar] [CrossRef]
- El-Araby, R.E.G.; Saad, A.; Elamrosy, H.; Adel, R.; Elshafie, S.; Helal, H.; Adel, M.; Roshdy, F. LncRNA-miRNA crosstalk in HCC cases on top of HCV infection. Eur. PMC 2022, 1–16. [Google Scholar]
- Fernandes, J.C.R.; Acuna, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Syrett, C.M.; Paneru, B.; Sandoval-Heglund, D.; Wang, J.; Banerjee, S.; Sindhava, V.; Behrens, E.M.; Atchison, M.; Anguera, M.C. Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight 2019, 4, e126751. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Gibson, A.; Edberg, J.; Kimberly, R.P.; Absher, D.M. Skewed allelic expression on X chromosome associated with aberrant expression of XIST on systemic lupus erythematosus lymphocytes. Hum. Mol. Genet. 2020, 29, 2523–2534. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Syrett, C.M.; Kramer, M.C.; Basu, A.; Atchison, M.L.; Anguera, M.C. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl. Acad. Sci. USA 2016, 113, E2029–E2038. [Google Scholar] [CrossRef]
- Hart, M.; Walch-Rückheim, B.; Friedmann, K.S.; Rheinheimer, S.; Tänzer, T.; Glombitza, B.; Sester, M.; Lenhof, H.P.; Hoth, M.; Schwarz, E.C.; et al. miR-34a: A new player in the regulation of T cell function by modulation of NF-κB signaling. Cell Death Dis. 2019, 10, 46. [Google Scholar] [CrossRef]
- Hart, M.; Nickl, L.; Walch-Rueckheim, B.; Krammes, L.; Rheinheimer, S.; Diener, C.; Taenzer, T.; Kehl, T.; Sester, M.; Lenhof, H.P.; et al. Wrinkle in the plan: miR-34a-5p impacts chemokine signaling by modulating CXCL10/CXCL11/CXCR3-axis in CD4+, CD8+ T cells, and M1 macrophages. J. Immunother. Cancer 2020, 8, e001617. [Google Scholar] [CrossRef]
- Scalavino, V.; Liso, M.; Cavalcanti, E.; Gigante, I.; Lippolis, A.; Mastronardi, M.; Chieppa, M.; Serino, G. miR-369-3p modulates inducible nitric oxide synthase and is involved in regulation of chronic inflammatory response. Sci. Rep. 2020, 10, 15942. [Google Scholar] [CrossRef] [PubMed]
- Lenart, M.; Dzialo, E.; Kluczewska, A.; Weglarczyk, K.; Szaflarska, A.; Rutkowska-Zapala, M.; Surmiak, M.; Sanak, M.; Pituch-Noworolska, A.; Siedlar, M. miRNA Regulation of NK Cells Antiviral Response in Children with Severe and/or Recurrent Herpes Simplex Virus Infections. Front. Immunol. 2020, 11, 589866. [Google Scholar] [CrossRef] [PubMed]
- van Ingen, E.; Foks, A.C.; Woudenberg, T.; van der Bent, M.L.; de Jong, A.; Hohensinner, P.J.; Wojta, J.; Bot, I.; Quax, P.H.A.; Nossent, A.Y. Inhibition of microRNA-494-3p activates Wnt signaling and reduces proinflammatory macrophage polarization in atherosclerosis. Mol. Ther. Nucleic Acids 2021, 26, 1228–1239. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; He, J.C.; Yang, Y.; Wang, J.M.; Qian, Y.W.; Yang, T.; Ji, L. The Prognostic Value of Tumor-infiltrating Lymphocytes in Hepatocellular Carcinoma: A Systematic Review and Meta-analysis. Sci. Rep. 2017, 7, 7525. [Google Scholar] [CrossRef]
- Capelle, C.M.; Chen, A.; Zeng, N.; Baron, A.; Grzyb, K.; Arns, T.; Skupin, A.; Ollert, M.; Hefeng, F.Q. Stress hormone signalling inhibits Th1 polarization in a CD4 T-cell-intrinsic manner via mTORC1 and the circadian gene PER1. Immunology 2022, 165, 428–444. [Google Scholar] [CrossRef]
- Li, N.; Yamamoto, G.; Fuji, H.; Kisseleva, T. Interleukin-17 in Liver Disease Pathogenesis. Semin. Liver Dis. 2021, 41, 507–515. [Google Scholar] [CrossRef]
- Granito, A.; Muratori, L.; Lalanne, C.; Quarneti, C.; Ferri, S.; Guidi, M.; Lenzi, M.; Muratori, P. Hepatocellular carcinoma in viral and autoimmune liver diseases: Role of CD4+ CD25+ Foxp3+ regulatory T cells in the immune microenvironment. World J. Gastroenterol. 2021, 27, 2994–3009. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Liu, S.; Yang, M. Regulatory T cells and their associated factors in hepatocellular carcinoma development and therapy. World J. Gastroenterol. 2022, 28, 3346–3358. [Google Scholar] [CrossRef]
- Chen, D.; Li, X.; Li, H.; Wang, K.; Tian, X. Identification of Immune-Related Prognostic mRNA and lncRNA in Patients with Hepatocellular Carcinoma. J. Oncol. 2022, 2022, 5313149. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, M.; Zhang, X.; Zhang, J.; Guo, H.; Wu, C. An extracellular matrix-based signature associated with immune microenvironment predicts the prognosis of patients with hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2022, 46, 101877. [Google Scholar] [CrossRef]
- Liu, C.; Jin, R.; Wang, H.C.; Tang, H.; Liu, Y.F.; Qian, X.P.; Sun, X.Y.; Ge, Q.; Sun, X.H.; Zhang, Y. Id1 expression promotes peripheral CD4+ T cell proliferation and survival upon TCR activation without co-stimulation. Biochem. Biophys. Res. Commun. 2013, 436, 47–52. [Google Scholar] [CrossRef]
- Ben-Sasson, S.Z.; Hu-Li, J.; Quiel, J.; Cauchetaux, S.; Ratner, M.; Shapira, I.; Dinarello, C.A.; Paul, W.E. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 7119–7124. [Google Scholar] [CrossRef]
- Hebel, K.; Rudolph, M.; Kosak, B.; Chang, H.D.; Butzmann, J.; Brunner-Weinzierl, M.C. IL-1β and TGF-β act antagonistically in induction and differentially in propagation of human proinflammatory precursor CD4+ T cells. J. Immunol. 2011, 187, 5627–5635. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, N.R.J.; Reilly, N.S.; Schrock, D.C.; Hocking, D.C.; Oakes, P.W.; Fowell, D.J. CD4+ T Cell Interstitial Migration Controlled by Fibronectin in the Inflamed Skin. Front. Immunol. 2020, 11, 1501. [Google Scholar] [CrossRef] [PubMed]
- Cano-Gamez, E.; Soskic, B.; Roumeliotis, T.I.; So, E.; Smyth, D.J.; Baldrighi, M.; Willé, D.; Nakic, N.; Esparza-Gordillo, J.; Larminie, C.G.C.; et al. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines. Nat. Commun. 2020, 11, 1801. [Google Scholar] [CrossRef] [PubMed]




| Genes | Fold Change (log2) | P-adj | Chromosome | Type |
|---|---|---|---|---|
| RPS9 | −7.13 | 0.0090 | chr19 | protein_coding |
| TAP2 | −7.09 | 0.0104 | chr6 | protein_coding |
| LILRB1 | −5.71 | 0.0020 | chr19 | protein_coding |
| XIST | −3.08 | 0.0001 | chrX | lncRNA |
| SNORD3A | −2.95 | 0.0000 | chr17 | snoRNA |
| PCDH9 | −2.76 | 0.0163 | chr13 | protein_coding |
| FCRL5 | −2.15 | 0.0001 | chr1 | protein_coding |
| IGHM | −2.04 | 0.0006 | chr14 | IG_C_gene |
| GATA2 | −1.98 | 0.0000 | chr3 | protein_coding |
| MS4A2 | −1.89 | 0.0194 | chr11 | protein_coding |
| FCRL2 | −1.73 | 0.0125 | chr1 | protein_coding |
| TCL1A | −1.70 | 0.0000 | chr14 | protein_coding |
| MS4A1 | −1.45 | 0.0000 | chr11 | protein_coding |
| CD79A | −1.40 | 0.0442 | chr19 | protein_coding |
| LRRN3 | −1.28 | 0.0079 | chr7 | protein_coding |
| LGALS12 | −1.27 | 0.0471 | chr11 | protein_coding |
| FCRL1 | −1.20 | 0.0389 | chr1 | protein_coding |
| PTPRS | −1.16 | 0.0156 | chr19 | protein_coding |
| RETN | −1.13 | 0.0021 | chr19 | protein_coding |
| AEBP1 | −1.12 | 0.0184 | chr7 | protein_coding |
| U2AF1 | 1.03 | 0.0064 | chr21 | protein_coding |
| PER1 | 1.03 | 0.0000 | chr17 | protein_coding |
| IL1B | 1.32 | 0.0000 | chr2 | protein_coding |
| G0S2 | 1.36 | 0.0119 | chr1 | protein_coding |
| RN7SL1 | 1.46 | 0.0061 | chr14 | misc_RNA |
| MIR222HG | 1.64 | 0.0473 | chrX | lncRNA |
| MAFF | 2.15 | 0.0000 | chr22 | protein_coding |
| CEACAM8 | 2.54 | 0.0021 | chr19 | protein_coding |
| CTSG | 2.67 | 0.0079 | chr14 | protein_coding |
| ID1 | 2.86 | 0.0000 | chr20 | protein_coding |
| ANXA3 | 2.89 | 0.0104 | chr4 | protein_coding |
| MMP8 | 3.03 | 0.0021 | chr11 | protein_coding |
| CAMP | 3.15 | 0.0015 | chr3 | protein_coding |
| FN1 | 3.76 | 0.0471 | chr2 | protein_coding |
| miRNAs | Fold Change (log2) | P-adj |
|---|---|---|
| hsa-miR-34a-5p | 1.8277 | 2.96 × 10−7 |
| hsa-miR-11400 | −2.3323 | 5.06 × 10−3 |
| hsa-miR-495-3p | −1.4377 | 8.24 × 10−5 |
| hsa-miR-369-3p | −1.3807 | 4.40 × 10−4 |
| hsa-miR-493-5p | −1.3633 | 1.75 × 10−3 |
| hsa-miR-889-3p | −1.2994 | 3.01 × 10−2 |
| hsa-miR-493-3p | −1.2379 | 6.44 × 10−3 |
| hsa-miR-494-3p | −1.2308 | 7.77 × 10−3 |
| hsa-miR-370-3p | −1.2273 | 1.10 × 10−5 |
| hsa-miR-411-5p | −1.1649 | 3.01 × 10−2 |
| hsa-miR-379-5p | −1.1397 | 2.53 × 10−3 |
| hsa-miR-654-3p | −1.1235 | 6.53 × 10−3 |
| hsa-miR-543 | −1.0776 | 1.43 × 10−3 |
| miRNA | Predicted Target Genes | Expression Data in the NGS Analysis |
|---|---|---|
| hsa-miR-34a-5p | PER1 | Up-regulated |
| hsa-miR-34a-5p | IL1B | Up-regulated |
| hsa-miR-369-3p | MS4A1 | Down-regulated |
| hsa-miR-369-3p | PCDH9 | Down-regulated |
| hsa-miR-369-3p | ANXA3 | Up-regulated |
| hsa-miR-370-3p | PER1 | Up-regulated |
| hsa-miR-494-3p | MS4A1 | Down-regulated |
| hsa-miR-494-3p | PCDH9 | Down-regulated |
| hsa-miR-494-3p | ID1 | Up-regulated |
| hsa-miR-495-3p | PCDH9 | Down-regulated |
| hsa-miR-495-3p | IL1B | Up-regulated |
| hsa-miR-495-3p | ID1 | Up-regulated |
| hsa-miR-543 | PCDH9 | Down-regulated |
| hsa-miR-543 | FN1 | Up-regulated |
| hsa-miR-543 | GATA2 | Down-regulated |
| hsa-miR-654-3p | MS4A1 | Down-regulated |
| hsa-miR-654-3p | ID1 | Up-regulated |
| hsa-miR-889-3p | LILRB1 | Down-regulated |
| hsa-miR-889-3p | ID1 | Up-regulated |
| hsa-miR-889-3p | GATA2 | Down-regulated |
| miRNAs | LncRNA | Target Sequence |
|---|---|---|
| hsa-miR-34a-5p | XIST | gcugaCACAUACAUACACUGCCu |
| hsa-miR-369-3p | XIST | acauuuuugaaAAGUAUUAUu |
| hsa-miR-369-3p | XIST | accacccccugAUGUAUUAUu |
| hsa-miR-369-3p | XIST | uuAGAUUGA-CA-GUAUUAUg |
| hsa-miR-370-3p | XIST | ugaccacugcugggCAGCAGGa |
| hsa-miR-370-3p | XIST | gagagcugaguCUUCAGCAGGu |
| hsa-miR-370-3p | XIST | cuuucuUUUCCUCCCAGCAGGg |
| hsa-miR-370-3p | XIST | ccccuuUUGCAGUACAGCAGGg |
| hsa-miR-370-3p | XIST | cccuuuUUGCAGUACAGCAGGg |
| hsa-miR-370-3p | XIST | ugcccuUUGCAGUACAGCAGGg |
| hsa-miR-370-3p | XIST | cuuccuUUCCCUUCCAGCAGGg |
| hsa-miR-370-3p | XIST | ccccuuUUGCAGUACAGCAGGg |
| hsa-miR-370-3p | XIST | ucaccuUUCCCUUCCAGCAGGg |
| hsa-miR-370-3p | XIST | ccacuuUUCCCUUCCAGCAGGa |
| hsa-miR-370-3p | XIST | ccuccuuggCAAAGCAGCAGGa |
| hsa-miR-370-3p | XIST | uauuccUUUUGUCUUACAGCAGGg |
| hsa-miR-370-3p | XIST | cucccuUUGUAUUCCAGCAGGg |
| hsa-miR-370-3p | XIST | ccccauUUGCA-UUCAGCAGGg |
| hsa-miR-494-3p | XIST | cucugcaagguacUAUGUUUCg |
| hsa-miR-494-3p | XIST | ugucUUACCCAUUUCCAUGUUUCu |
| hsa-miR-495-3p | XIST | uaaAAGGGCUCAGGGUUUGUUc |
| hsa-miR-543 | XIST | caaGACUGUCACUUGGAAUGUUc |
| hsa-miR-654-3p | XIST | --------------CAGACAUu |
| hsa-miR-654-3p | XIST | acuucccucaggAGCAGACAUu |
| hsa-miR-889-3p | XIST | cuucUGGUAGAGUGGGAUAUUAu |
| hsa-miR-889-3p | XIST | uuuuacagcaaggGAUAUUAa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.-H.; Rau, C.-S.; Liu, Y.-W.; Wu, C.-J.; Chien, P.-C.; Lin, H.-P.; Wu, Y.-C.; Huang, C.-Y.; Hsieh, T.-M.; Hsieh, C.-H. Exploring the Regulatory Role of XIST-microRNAs/mRNA Network in Circulating CD4+ T Cells of Hepatocellular Carcinoma Patients. Biomedicines 2023, 11, 1848. https://doi.org/10.3390/biomedicines11071848
Huang L-H, Rau C-S, Liu Y-W, Wu C-J, Chien P-C, Lin H-P, Wu Y-C, Huang C-Y, Hsieh T-M, Hsieh C-H. Exploring the Regulatory Role of XIST-microRNAs/mRNA Network in Circulating CD4+ T Cells of Hepatocellular Carcinoma Patients. Biomedicines. 2023; 11(7):1848. https://doi.org/10.3390/biomedicines11071848
Chicago/Turabian StyleHuang, Lien-Hung, Cheng-Shyuan Rau, Yueh-Wei Liu, Chia-Jung Wu, Peng-Chen Chien, Hui-Ping Lin, Yi-Chan Wu, Chun-Ying Huang, Ting-Min Hsieh, and Ching-Hua Hsieh. 2023. "Exploring the Regulatory Role of XIST-microRNAs/mRNA Network in Circulating CD4+ T Cells of Hepatocellular Carcinoma Patients" Biomedicines 11, no. 7: 1848. https://doi.org/10.3390/biomedicines11071848
APA StyleHuang, L.-H., Rau, C.-S., Liu, Y.-W., Wu, C.-J., Chien, P.-C., Lin, H.-P., Wu, Y.-C., Huang, C.-Y., Hsieh, T.-M., & Hsieh, C.-H. (2023). Exploring the Regulatory Role of XIST-microRNAs/mRNA Network in Circulating CD4+ T Cells of Hepatocellular Carcinoma Patients. Biomedicines, 11(7), 1848. https://doi.org/10.3390/biomedicines11071848







