Association of Immune Semaphorins with COVID-19 Severity and Outcomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Patients
2.2. COVID-19 Disease Severity Classification
2.3. Data Collection
2.4. Measurement of Semaphorin Serum Concentrations
2.5. Statistical Analysis
3. Results
3.1. Baseline Patients’ Characteristics
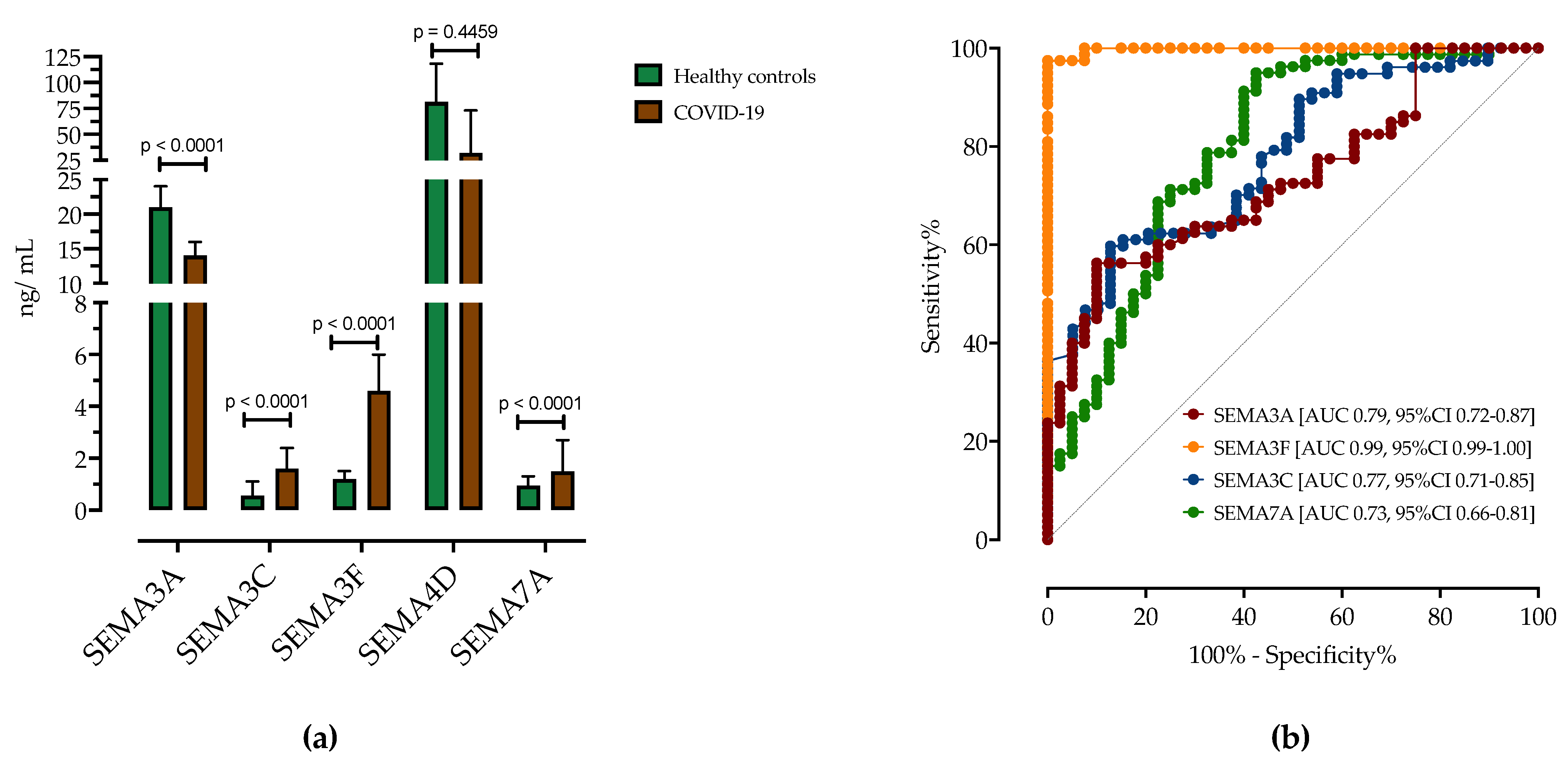
3.2. Analysis of Serum Semaphorin Concentrations in Patients with COVID-19 and Healthy Controls
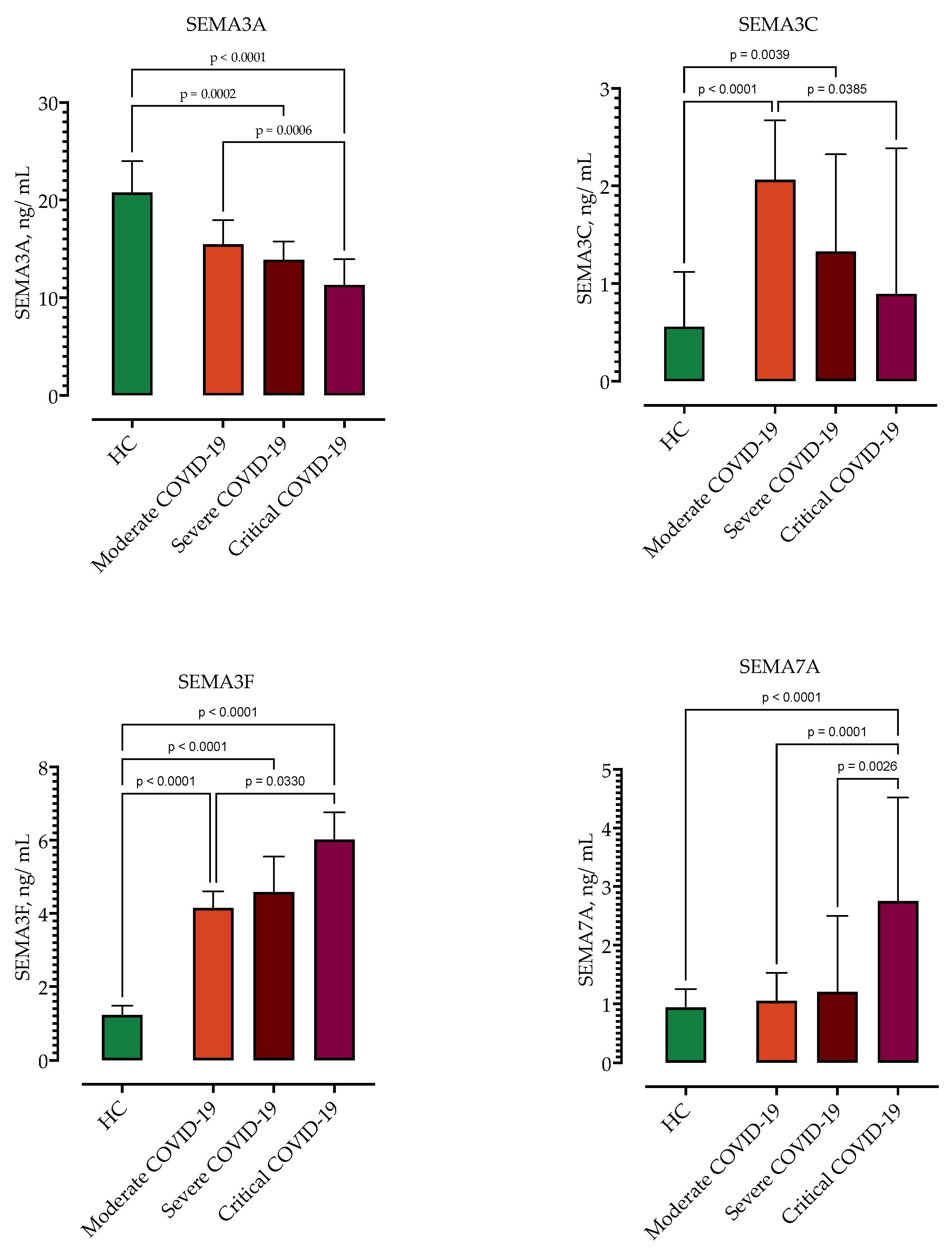
3.3. Correlation of Semaphorin Serum Concentrations with COVID-19 Severity
3.4. ROC Analysis of Serum Semaphorins Concentrations in Predicting COVID-19 Severity
3.5. Association of Serum Semaphorin Concentrations with COVID-19 Clinical Outcomes and Complications
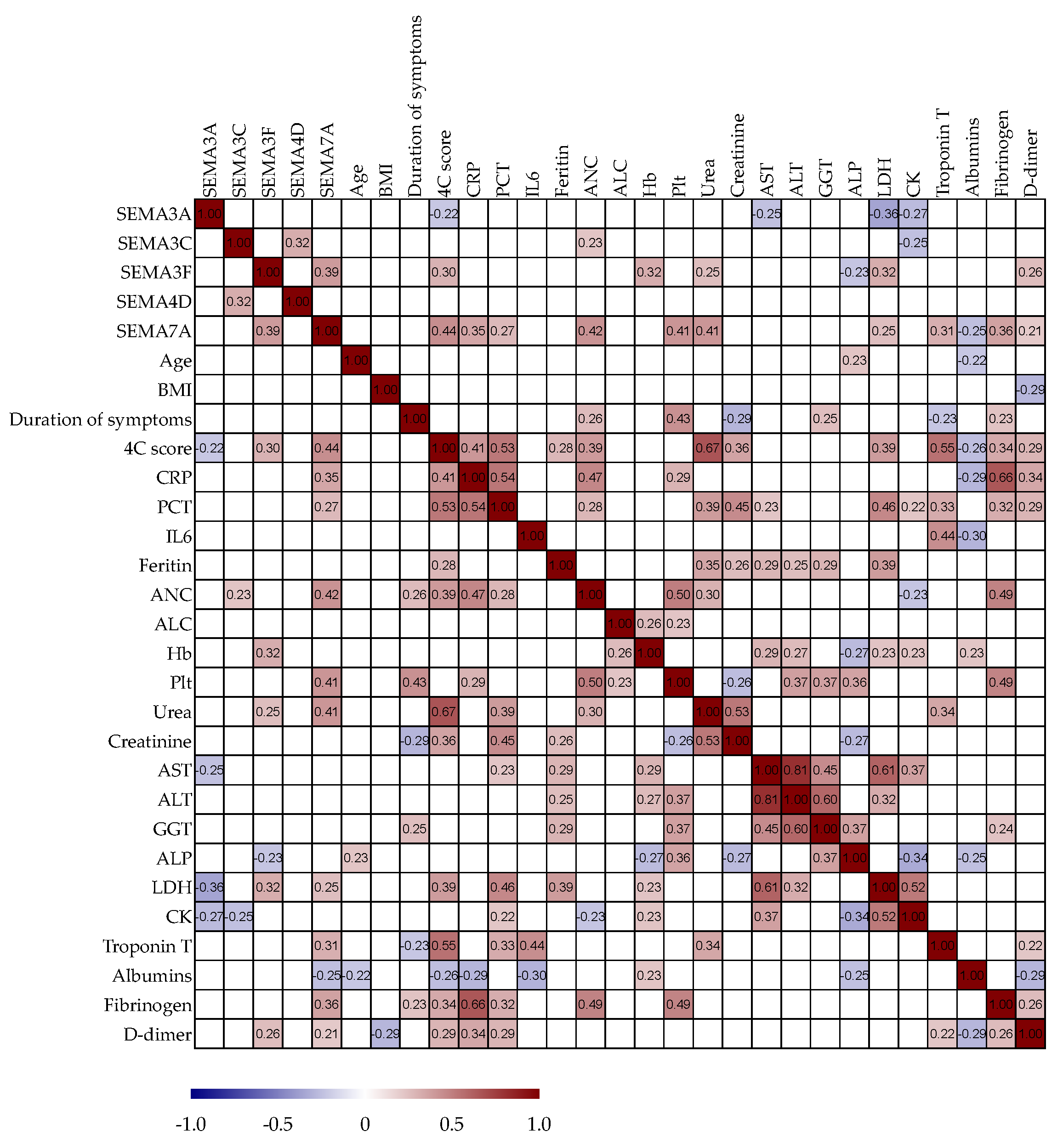
3.6. Correlation Analysis of Serum Semaphorin Concentrations with Routine Clinical and Laboratory Parameters
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berlin, D.A.; Gulick, R.M.; Martinez, F.J. Severe COVID-19. N. Engl. J. Med. 2020, 383, 2451–2460. [Google Scholar] [CrossRef]
- Schultze, J.L.; Aschenbrenner, A.C. COVID-19 and the human innate immune system. Cell 2021, 184, 1671–1692. [Google Scholar] [CrossRef]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef]
- Garg, S.S.; Sharma, A.; Gupta, J. Immunomodulation and immunotherapeutics of COVID-19. Clin. Immunol. 2021, 231, 108842. [Google Scholar] [CrossRef]
- Yazdani, U.; Terman, J.R. The semaphorins. Genome Biol. 2006, 7, 211. [Google Scholar] [CrossRef]
- Alto, L.T.; Terman, J.R. Semaphorins and their Signaling Mechanisms. Methods Mol. Biol. 2017, 1493, 1–25. [Google Scholar] [CrossRef]
- Fard, D.; Tamagnone, L. Semaphorins in health and disease. Cytokine Growth Factor. Rev. 2021, 57, 55–63. [Google Scholar] [CrossRef]
- Chong, V.; Nguyen, Q.D.; Sepah, Y.; Giani, A.; Pearce, E. HORNBILL: A phase I/IIa trial examining the safety, tolerability and early response of BI 764524 in patients with diabetic retinopathy and diabetic macular ischaemia-rationale, study design and protocol. Trials 2022, 23, 669. [Google Scholar] [CrossRef]
- Gurrapu, S.; Tamagnone, L. Semaphorins as Regulators of Phenotypic Plasticity and Functional Reprogramming of Cancer Cells. Trends Mol. Med. 2019, 25, 303–314. [Google Scholar] [CrossRef]
- Kalmarzi, R.N.; Rajabinejad, M.; Lotfi, R. Immune semaphorins: Crucial regulatory signals and novel therapeutic targets in asthma and allergic diseases. Eur. J. Pharmacol. 2020, 881, 173209. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, J.; Xu, S.; Liu, J.; Xu, Y.; Yu, J.; Zhao, M.; Pan, W.; Wang, M.; Wan, J. The role of semaphorins in cardiovascular diseases: Potential therapeutic targets and novel biomarkers. FASEB J. 2022, 36, e22509. [Google Scholar] [CrossRef]
- Lu, Q.; Zhu, L. The Role of Semaphorins in Metabolic Disorders. Int. J. Mol. Sci. 2020, 21, 5641. [Google Scholar] [CrossRef]
- Papic, N.; Zidovec Lepej, S.; Gorenec, L.; Grgic, I.; Gasparov, S.; Filipec Kanizaj, T.; Vince, A. The association of semaphorins 3C, 5A and 6D with liver fibrosis stage in chronic hepatitis C. PLoS ONE 2018, 13, e0209481. [Google Scholar] [CrossRef]
- Samadan, L.; Papic, N.; Mijic, M.; Knezevic Stromar, I.; Gasparov, S.; Vince, A. Do Semaphorins Play a Role in Development of Fibrosis in Patients with Nonalcoholic Fatty Liver Disease? Biomedicines 2022, 10, 3014. [Google Scholar] [CrossRef]
- Nishide, M.; Kumanogoh, A. The role of semaphorins in immune responses and autoimmune rheumatic diseases. Nat. Rev. Rheumatol. 2018, 14, 19–31. [Google Scholar] [CrossRef]
- Matloubi, M.; Koussih, L.; Shan, L.; Lukawy, C.; Gounni, A.S. Targeting the Semaphorin3E-plexinD1 complex in allergic asthma. Pharmacol. Ther. 2023, 242, 108351. [Google Scholar] [CrossRef]
- Ryu, S.; Broussard, L.; Youn, C.; Song, B.; Norris, D.; Armstrong, C.A.; Kim, B.; Song, P.I. Therapeutic Effects of Synthetic Antimicrobial Peptides, TRAIL and NRP1 Blocking Peptides in Psoriatic Keratinocytes. Chonnam Med. J. 2019, 55, 75–85. [Google Scholar] [CrossRef]
- Chen, X.; Wang, H.; Jia, K.; Wang, H.; Ren, T. Anti-Semaphorin-7A single chain antibody demonstrates beneficial effects on pulmonary inflammation during acute lung injury. Exp. Ther. Med. 2018, 15, 2356–2364. [Google Scholar] [CrossRef]
- Kanth, S.M.; Gairhe, S.; Torabi-Parizi, P. The Role of Semaphorins and Their Receptors in Innate Immune Responses and Clinical Diseases of Acute Inflammation. Front. Immunol. 2021, 12, 672441. [Google Scholar] [CrossRef]
- Qiu, Q.; Yu, X.; Chen, Q.; He, X. Sema3A inactivates the ERK/JNK signalling pathways to alleviate inflammation and oxidative stress in lipopolysaccharide-stimulated rat endothelial cells and lung tissues. Autoimmunity 2023, 56, 2200908. [Google Scholar] [CrossRef]
- ECDC. Data on SARS-CoV-2 Variants in the EU/EEA. Available online: https://www.ecdc.europa.eu/en/publications-data/data-virus-variants-covid-19-eueea (accessed on 1 October 2023).
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: A cohort study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Jones, R.P.; Ponomarenko, A. COVID-19-Related Age Profiles for SARS-CoV-2 Variants in England and Wales and States of the USA (2020 to 2022): Impact on All-Cause Mortality. Infect. Dis. Rep. 2023, 15, 600–634. [Google Scholar] [CrossRef]
- COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. National Institutes of Health. Available online: https://www.covid19treatmentguidelines.nih.gov/ (accessed on 19 May 2023).
- Subbe, C.P.; Kruger, M.; Rutherford, P.; Gemmel, L. Validation of a modified Early Warning Score in medical admissions. QJM 2001, 94, 521–526. [Google Scholar] [CrossRef]
- Vincent, J.L.; Moreno, R.; Takala, J.; Willatts, S.; De Mendonca, A.; Bruining, H.; Reinhart, C.K.; Suter, P.M.; Thijs, L.G. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996, 22, 707–710. [Google Scholar] [CrossRef]
- Fine, M.J.; Auble, T.E.; Yealy, D.M.; Hanusa, B.H.; Weissfeld, L.A.; Singer, D.E.; Coley, C.M.; Marrie, T.J.; Kapoor, W.N. A prediction rule to identify low-risk patients with community-acquired pneumonia. N. Engl. J. Med. 1997, 336, 243–250. [Google Scholar] [CrossRef]
- Knight, S.R.; Ho, A.; Pius, R.; Buchan, I.; Carson, G.; Drake, T.M.; Dunning, J.; Fairfield, C.J.; Gamble, C.; Green, C.A.; et al. Risk stratification of patients admitted to hospital with COVID-19 using the ISARIC WHO Clinical Characterisation Protocol: Development and validation of the 4C Mortality Score. BMJ 2020, 370, m3339. [Google Scholar] [CrossRef]
- Kiseleva, E.P.; Rutto, K.V. Semaphorin 3A in the Immune System: Twenty Years of Study. Biochemistry 2022, 87, 640–657. [Google Scholar] [CrossRef]
- Wen, H.; Lei, Y.; Eun, S.Y.; Ting, J.P. Plexin-A4-semaphorin 3A signaling is required for Toll-like receptor- and sepsis-induced cytokine storm. J. Exp. Med. 2010, 207, 2943–2957. [Google Scholar] [CrossRef]
- Dai, X.; Okon, I.; Liu, Z.; Wu, Y.; Zhu, H.; Song, P.; Zou, M.H. A novel role for myeloid cell-specific neuropilin 1 in mitigating sepsis. FASEB J. 2017, 31, 2881–2892. [Google Scholar] [CrossRef]
- Dhawan, M.; Rabaan, A.A.; Alwarthan, S.; Alhajri, M.; Halwani, M.A.; Alshengeti, A.; Najim, M.A.; Alwashmi, A.S.S.; Alshehri, A.A.; Alshamrani, S.A.; et al. Regulatory T Cells (Tregs) and COVID-19: Unveiling the Mechanisms, and Therapeutic Potentialities with a Special Focus on Long COVID. Vaccines 2023, 11, 699. [Google Scholar] [CrossRef]
- Ma, C.; Li, F.; Wang, Z.; Luo, H. A Novel Immune-Related Gene Signature Predicts Prognosis of Lung Adenocarcinoma. Biomed. Res. Int. 2022, 2022, 4995874. [Google Scholar] [CrossRef]
- Malik, M.F.; Satherley, L.K.; Davies, E.L.; Ye, L.; Jiang, W.G. Expression of Semaphorin 3C in Breast Cancer and its Impact on Adhesion and Invasion of Breast Cancer Cells. Anticancer. Res. 2016, 36, 1281–1286. [Google Scholar]
- De Angelis Rigotti, F.; Wiedmann, L.; Hubert, M.O.; Vacca, M.; Hasan, S.S.; Moll, I.; Carvajal, S.; Jimenez, W.; Starostecka, M.; Billeter, A.T.; et al. Semaphorin 3C exacerbates liver fibrosis. Hepatology 2023, 78, 1092–1105. [Google Scholar] [CrossRef]
- Cai, A.; Ye, G.; Placier, S.; Frere, P.; Surin, B.; Vandermeersch, S.; Kormann, R.; Xu-Dubois, Y.C.; Genest, M.; Lannoy, M.; et al. Genetic inactivation of Semaphorin 3C protects mice from acute kidney injury. Kidney Int. 2022, 101, 720–732. [Google Scholar] [CrossRef]
- Lepore, J.J.; Mericko, P.A.; Cheng, L.; Lu, M.M.; Morrisey, E.E.; Parmacek, M.S. GATA-6 regulates semaphorin 3C and is required in cardiac neural crest for cardiovascular morphogenesis. J. Clin. Investig. 2006, 116, 929–939. [Google Scholar] [CrossRef]
- Plein, A.; Calmont, A.; Fantin, A.; Denti, L.; Anderson, N.A.; Scambler, P.J.; Ruhrberg, C. Neural crest-derived SEMA3C activates endothelial NRP1 for cardiac outflow tract septation. J. Clin. Investig. 2015, 125, 2661–2676. [Google Scholar] [CrossRef]
- Yang, W.J.; Hu, J.; Uemura, A.; Tetzlaff, F.; Augustin, H.G.; Fischer, A. Semaphorin-3C signals through Neuropilin-1 and PlexinD1 receptors to inhibit pathological angiogenesis. EMBO Mol. Med. 2015, 7, 1267–1284. [Google Scholar] [CrossRef]
- Madureira, G.; Soares, R. The misunderstood link between SARS-CoV-2 and angiogenesis. A narrative review. Pulmonology 2023, 29, 323–331. [Google Scholar] [CrossRef]
- Plant, T.; Eamsamarng, S.; Sanchez-Garcia, M.A.; Reyes, L.; Renshaw, S.A.; Coelho, P.; Mirchandani, A.S.; Morgan, J.M.; Ellett, F.E.; Morrison, T.; et al. Semaphorin 3F signaling actively retains neutrophils at sites of inflammation. J. Clin. Investig. 2020, 130, 3221–3237. [Google Scholar] [CrossRef]
- Kang, S.; Okuno, T.; Takegahara, N.; Takamatsu, H.; Nojima, S.; Kimura, T.; Yoshida, Y.; Ito, D.; Ohmae, S.; You, D.J.; et al. Intestinal epithelial cell-derived semaphorin 7A negatively regulates development of colitis via alphavbeta1 integrin. J. Immunol. 2012, 188, 1108–1116. [Google Scholar] [CrossRef]
- Morote-Garcia, J.C.; Napiwotzky, D.; Kohler, D.; Rosenberger, P. Endothelial Semaphorin 7A promotes neutrophil migration during hypoxia. Proc. Natl. Acad. Sci. USA 2012, 109, 14146–14151. [Google Scholar] [CrossRef]
- Roth, J.M.; Kohler, D.; Schneider, M.; Granja, T.F.; Rosenberger, P. Semaphorin 7A Aggravates Pulmonary Inflammation during Lung Injury. PLoS ONE 2016, 11, e0146930. [Google Scholar] [CrossRef]
- Kuklina, E. Semaphorin 4D as a guidance molecule in the immune system. Int. Rev. Immunol. 2021, 40, 268–273. [Google Scholar] [CrossRef]
- Smith, E.S.; Jonason, A.; Reilly, C.; Veeraraghavan, J.; Fisher, T.; Doherty, M.; Klimatcheva, E.; Mallow, C.; Cornelius, C.; Leonard, J.E.; et al. SEMA4D compromises blood-brain barrier, activates microglia, and inhibits remyelination in neurodegenerative disease. Neurobiol. Dis. 2015, 73, 254–268. [Google Scholar] [CrossRef]
- Yoshida, Y.; Ogata, A.; Kang, S.; Ebina, K.; Shi, K.; Nojima, S.; Kimura, T.; Ito, D.; Morimoto, K.; Nishide, M.; et al. Semaphorin 4D Contributes to Rheumatoid Arthritis by Inducing Inflammatory Cytokine Production: Pathogenic and Therapeutic Implications. Arthritis Rheumatol. 2015, 67, 1481–1490. [Google Scholar] [CrossRef]
- Rajabinejad, M.; Asadi, G.; Ranjbar, S.; Afshar Hezarkhani, L.; Salari, F.; Gorgin Karaji, A.; Rezaiemanesh, A. Semaphorin 4A, 4C, and 4D: Function comparison in the autoimmunity, allergy, and cancer. Gene 2020, 746, 144637. [Google Scholar] [CrossRef]
- Baudet, M.L.; Bellon, A.; Holt, C.E. Role of microRNAs in Semaphorin function and neural circuit formation. Semin. Cell Dev. Biol. 2013, 24, 146–155. [Google Scholar] [CrossRef]
- Fayyad-Kazan, M.; Makki, R.; Skafi, N.; El Homsi, M.; Hamade, A.; El Majzoub, R.; Hamade, E.; Fayyad-Kazan, H.; Badran, B. Circulating miRNAs: Potential diagnostic role for coronavirus disease 2019 (COVID-19). Infect. Genet. Evol. 2021, 94, 105020. [Google Scholar] [CrossRef]
- Altounian, M.; Bellon, A.; Mann, F. Neuronal miR-17-5p contributes to interhemispheric cortical connectivity defects induced by prenatal alcohol exposure. Cell Rep. 2023, 42, 113020. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, H.; Zhang, W.; Duan, D.; Zhu, G.; Cao, W.; Liu, B. Increased Expression of Sema3C Indicates a Poor Prognosis and Is Regulated by miR-142-5p in Glioma. Biol. Pharm. Bull. 2020, 43, 639–648. [Google Scholar] [CrossRef]
- Xiang, G.; Cheng, Y. MiR-126-3p inhibits ovarian cancer proliferation and invasion via targeting PLXNB2. Reprod. Biol. 2018, 18, 218–224. [Google Scholar] [CrossRef]
- Wei, Y.; Guo, S.; Tang, J.; Wen, J.; Wang, H.; Hu, X.; Gu, Q. MicroRNA-19b-3p suppresses gastric cancer development by negatively regulating neuropilin-1. Cancer Cell Int. 2020, 20, 193. [Google Scholar] [CrossRef]
- Yang, T.; Huang, H.; Shao, Q.; Yee, S.; Majumder, T.; Liu, G. miR-92 Suppresses Robo1 Translation to Modulate Slit Sensitivity in Commissural Axon Guidance. Cell Rep. 2018, 24, 2694–2708. [Google Scholar] [CrossRef]
- White, R.E.; Giffard, R.G. MicroRNA-320 induces neurite outgrowth by targeting ARPP-19. Neuroreport 2012, 23, 590–595. [Google Scholar] [CrossRef]
- Liang, Y.; Fang, D.; Gao, X.; Deng, X.; Chen, N.; Wu, J.; Zeng, M.; Luo, M. Circulating microRNAs as emerging regulators of COVID-19. Theranostics 2023, 13, 125–147. [Google Scholar] [CrossRef]

| Moderate n = 24 | Severe n = 32 | Critical n = 24 | p-Value | |
|---|---|---|---|---|
| Male sex | 13 (54.17%) | 19 (59.38%) | 12 (50%) | 0.7801 |
| Age, years | 64 (48–69) | 62 (44–71) | 63 (48–65) | 0.8829 |
| BMI, kg/m2 | 26 (24–29) | 29 (27–34) | 30 (25–31) | 0.0517 |
| Waist–hip ratio (WHR) | 0.97 (0.91–1) | 1 (0.97–1.1) | 0.98 (0.92–1.1) | 0.0744 |
| Comorbidities | ||||
| Charlson comorbidity index | 1.5 (0–3.8) | 2 (0.25–3) | 2 (1.3–3.8) | 0.5018 |
| Diabetes mellitus | 8 (33.33%) | 4 (12.50%) | 6 (25.00%) | 0.1706 |
| Arterial hypertension | 13 (54.17%) | 17 (53.13%) | 11 (45.83%) | 0.8152 |
| COPD | 2 (8.33%) | 3 (9.38%) | 2 (8.33%) | 0.9870 |
| Dyslipidemia | 3 (12.50%) | 7 (21.88%) | 5 (20.83%) | 0.6411 |
| Gastritis/GERD | 4 (16.67% | 1 (3.13%) | 2 (8.33%) | 0.2062 |
| Cardiovascular diseases | 5 (20.83%) | 4 (12.50%) | 5 (20.83%) | 0.6301 |
| No comorbidities | 7 (29.17%) | 10 (31.25%) | 6 (25.00%) | 0.8761 |
| Chronic medications | ||||
| ACE inhibitors | 5 (20.83%) | 15 (46.88%) | 13 (54.17%) | 0.0563 |
| Acetylsalicylic acid | 3 (12.50%) | 4 (12.50%) | 1 (4.17%) | 0.5230 |
| Beta blockers | 4 (16.67%) | 12 (37.50%) | 9 (37.50%) | 0.1832 |
| Proton pump inhibitors | 5 (20.83%) | 4 (12.50%) | 2 (8.33%) | 0.4379 |
| Hypolipidemic | 3 (12.50%) | 6 (18.75%) | 5 (20.83%) | 0.7279 |
| Peroral hypoglycemics | 5 (20.83%) | 7 (21.88%) | 6 (25.00%) | 0.9364 |
| No medications | 7 (29.17%) | 10 (31.25%) | 6 (25.00%) | 0.8761 |
| Disease severity at admission | ||||
| Duration of illness, days | 9 (7–9) | 10 (7–11) | 9 (7.3–9.8) | 0.8483 |
| Body temperature, °C | 38 (37–38) | 38 (37–38) | 37 (37–38) | 0.2829 |
| Heart rate/min | 93 (84–100) | 96 (80–105) | 98 (86–107) | 0.5774 |
| Mean arterial pressure, mmHg | 94 (87–103) | 93 (83–100) | 100 (89–108) | 0.1196 |
| Respiratory rate/min | 21 (18–24) | 25 (21–30) | 27 (22–31) | 0.0006 |
| SpO2 on room air, % | 95 (93–96) | 88 (82–89) | 82 (78–87) | <0.0001 |
| Pao2/FiO2 ratio | 347 (323–428) | 183 (137–250) | 126 (76–174) | <0.0001 |
| MEWS | 2 (1–3.5) | 3 (2–4) | 3 (2–3.8) | 0.0422 |
| SOFA | 1.5 (1–2) | 2 (2–3) | 2 (2–3) | 0.0008 |
| PSI | 54 (41–73) | 64 (52–77) | 84 (58–99) | 0.0078 |
| 4C mortality score | 5 (3–8) | 9 (6–11) | 11 (8–12) | <0.0001 |
| Moderate n = 24 | Severe n = 32 | Critical n = 24 | p-Value | |
|---|---|---|---|---|
| CRP, mg/L | 83 (30–120) | 101 (67–171) | 133 (75–213) | 0.0214 |
| Procalcitonin, µg/L | 0.11 (0.058–0.19) | 0.13 (0.073–0.41) | 0.2 (0.094–0.69) | 0.0611 |
| Interleukin-6, pg/mL | 15 (6.5–54) | 61 (18–107) | 68 (18–142) | 0.0102 |
| Ferritin, µg/L | 711 (443–1222) | 841 (623–1468) | 945 (656–2063) | 0.2179 |
| WBC, ×109/L | 5.9 (4.8–8.7) | 6.3 (4.1–8.2) | 6.4 (5.1–11) | 0.3815 |
| Lymphocyte count, 109/L | 0.83 (0.55–1.1) | 0.6 (0.44–0.88) | 0.68 (0.49–0.83) | 0.3235 |
| Neutrophil count, 109/L | 4.4 (3.5–6.4) | 5.1 (3.2–6.7) | 5.4 (4.2–9) | 0.2769 |
| Hemoglobin, g/L | 135 (117–147) | 134 (127–145) | 140 (126–148) | 0.5138 |
| Platelets, ×109/L | 166 (120–277) | 190 (137–247) | 197 (141–246) | 0.6331 |
| Glucose, mmol/L | 6.5 (5.8–7.9) | 7.2 (6.6–8.1) | 8.2 (6.9–9.8) | 0.0367 |
| Urea, mmol/L | 4.4 (3.2–5.1) | 6.2 (4.5–9.2) | 6.1(4.7–9.3) | 0.0003 |
| Creatinine, µmol/L | 72 (64–83) | 80 (67–97) | 69 (60–99) | 0.2905 |
| eGFR, ml/min/1.73 m2 | 99 (80–107) | 89 (71–106) | 91 (57–100) | 0.1748 |
| Bilirubin, µmol/L | 10 (8–12) | 13 (10–16) | 11 (8.3–14) | 0.1045 |
| AST, IU/L | 39 (26–51) | 59 (34–86) | 49 (37–90) | 0.0104 |
| ALT, IU/L | 29 (21–57) | 51 (28–75) | 31 (24–65) | 0.1682 |
| GGT, IU/L | 50 (30–55) | 47 (26–124) | 43 (2293) | 0.5901 |
| LDH, IU/L | 262 (199–319) | 382 (251–490) | 483 (370–598) | <0.0001 |
| CK, IU/L | 104 (56–272) | 173 (66–319) | 269 (120–524) | 0.0352 |
| Albumins, g/L | 38 (36–41) | 37 (35–41) | 37 (35–39) | 0.2015 |
| Fibrinogen, g/L | 5.6 (5–6.4) | 6.2 (5.4–6.9) | 5.8 (5.4–6.9) | 0.3437 |
| D-dimer, mg/L | 0.95 (0.63–2) | 0.86 (0.5–1.6) | 1 (0.52–1.3) | 0.7481 |
| Healthy Controls (n = 40) | COVID-19 Patients (n = 80) | Difference (95% CI) | p-Value | |
| SEMA3A, ng/mL | 21 (15–24) | 14 (11–16) | −7.2 (−8.6–−4.2) | <0.0001 |
| SEMA3C, ng/mL | 0.56 (0.25–1.1) | 1.6 (0.84–2.4) | 1.1 (0.4–1.2) | <0.0001 |
| SEMA3F, ng/mL | 1.2 (0.95–1.5) | 4.6 (4.1–6) | 3.4 (3.2–3.9) | <0.0001 |
| SEMA4D, ng/mL | 47 (21–81) | 32 (31–73) | −14 (−17–11) | 0.4459 |
| SEMA7A, ng/mL | 0.9 (0.2–1.3) | 1.5 (0.78–2.7) | 0.5 (0.3–1.1) | <0.0001 |
| COVID-19 vs. HC | Sensitivity (95% CI) | Specificity (95% CI) | AUC (95% CI) | p-Value |
| SEMA3A < 15.50 ng/mL | 71.25% (62.34%–78.77%) | 72.50% (59.75%–82.40%) | 0.7953 (0.7188–0.8718) | <0.0001 |
| SEMA3C > 1.00 ng/mL | 63.64% (54.30%–72.05%) | 66.67% (53.53%–77.64%) | 0.7792 (0.7086–0.8498) | <0.0001 |
| SEMA3F > 2.10 ng/mL | 97.47% (92.63%–99.33%) | 97.5% (89.54%–99.74%) | 0.9981 (0.9951–1.0) | <0.0001 |
| SEMA7A > 1.15 ng/mL | 63.75% (54.59%–72.01%) | 65% (52.01%–76.09%) | 0.7339 (0.6591–0.8087) | <0.0001 |
| Sensitivity (95% CI) | Specificity (95% CI) | AUC (95% CI) | p-Value | |
|---|---|---|---|---|
| Moderate vs. severe/critical | ||||
| SEMA3A > 14 ng/mL | 62.5% (51.5%–72.3%) | 66.6% (49.8%–80.0%) | 0.7586 (0.66–0.85) | 0.0003 |
| SEMA3C > 1.80 ng/mL | 60.3% (49.1%–70.6%) | 58.3% (41.7%–73.2%) | 0.6612 (0.56–0.76) | 0.0242 |
| SEMA3F < 4.35 ng/mL | 72.7% (61.9%–81.3%) | 70.8% (54.1%–83.3%) | 0.739 (0.54–0.84) | 0.0008 |
| SEMA7A < 1.23 ng/mL | 66.1% (55.1%–75.5%) | 58.3% (41.7%–73.2%) | 0.7154 (0.62–0.80) | 0.0024 |
| CRP < 95 mg/L | 60.7% (49.7%–70.7%) | 58.3% (41.7%–73.2%) | 0.7428 (0.63–0.85) | 0.0087 |
| IL-6 < 53 pg/mL | 60.5% (47.2%–72.4%) | 75.0% (56.7%–87.2%) | 0.7428 (0.63–0.85) | 0.0025 |
| LDH < 300 IU/L | 80.3% (70.3%–87.6%) | 70.8% (54.1%–83.3%) | 0.779 (0.68–0.86) | <0.0001 |
| Critical vs. moderate/severe | ||||
| SEMA3A < 13.0 ng/mL | 69.6% (58.8%–78.6%) | 70.8% (54.1%–83.3%) | 0.7865 (0.69–0.87) | <0.0001 |
| SEMA3F > 4.7 ng/mL | 63.6% (52.5%–73.4%) | 70.8% (54.7%–83.3%) | 0.7591 (0.66–0.85) | 0.0003 |
| SEMA7A > 2.0 ng/mL | 82.1% (72.3%–89.0%) | 70.8% (54.1%–83.3%) | 0.8177 (0.73–0.89) | <0.0001 |
| CRP > 100 mg/L | 50.0% (39.2%–60.7%) | 54.1% (37.8%–69.6%) | 0.5432 (0.42–0.65) | 0.5426 |
| IL-6 > 60 pg/ml | 68.4% (55.1%–79.2%) | 55.0% (37.2%–71.5%) | 0.6447 (0.51–0.77) | 0.0719 |
| LDH > 450 IU/L | 76.7% (66.4%–84.7%) | 58.3% (41.7%–73.2%) | 0.7563 (0.66–0.85) | 0.0003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargovic, M.; Papic, N.; Samadan, L.; Balen Topic, M.; Vince, A. Association of Immune Semaphorins with COVID-19 Severity and Outcomes. Biomedicines 2023, 11, 2786. https://doi.org/10.3390/biomedicines11102786
Vargovic M, Papic N, Samadan L, Balen Topic M, Vince A. Association of Immune Semaphorins with COVID-19 Severity and Outcomes. Biomedicines. 2023; 11(10):2786. https://doi.org/10.3390/biomedicines11102786
Chicago/Turabian StyleVargovic, Martina, Neven Papic, Lara Samadan, Mirjana Balen Topic, and Adriana Vince. 2023. "Association of Immune Semaphorins with COVID-19 Severity and Outcomes" Biomedicines 11, no. 10: 2786. https://doi.org/10.3390/biomedicines11102786
APA StyleVargovic, M., Papic, N., Samadan, L., Balen Topic, M., & Vince, A. (2023). Association of Immune Semaphorins with COVID-19 Severity and Outcomes. Biomedicines, 11(10), 2786. https://doi.org/10.3390/biomedicines11102786





