Abstract
Monoclonal antibodies (mAbs), the new revolutionary class of medications, are fast becoming tools against various diseases thanks to a unique structure and function that allow them to bind highly specific targets or receptors. These specialized proteins can be produced in large quantities via the hybridoma technique introduced in 1975 or by means of modern technologies. Additional methods have been developed to generate mAbs with new biological properties such as humanized, chimeric, or murine. The inclusion of mAbs in therapeutic regimens is a major medical advance and will hopefully lead to significant improvements in infectious disease management. Since the first therapeutic mAb, muromonab-CD3, was approved by the U.S. Food and Drug Administration (FDA) in 1986, the list of approved mAbs and their clinical indications and applications have been proliferating. New technologies have been developed to modify the structure of mAbs, thereby increasing efficacy and improving delivery routes. Gene delivery technologies, such as non-viral synthetic plasmid DNA and messenger RNA vectors (DMabs or mRNA-encoded mAbs), built to express tailored mAb genes, might help overcome some of the challenges of mAb therapy, including production restrictions, cold-chain storage, transportation requirements, and expensive manufacturing and distribution processes. This paper reviews some of the recent developments in mAb discovery against viral infections and illustrates how mAbs can help to combat viral diseases and outbreaks.
1. Introduction
Viruses are microorganisms characterized by a wide range of features, such as shape, size and infectivity, and can cause mild to severe human and animal disease. Table S1 lists rare to pandemic viruses.
Our immune system has evolved strategies to neutralize viruses in various ways. The first contact between the virus and the cell host membrane represents the initial challenging step in the viral infectious life cycle. Antibodies, produced in response to virus detection, may act as a barrier, thereby preventing the virus from completing this crucial step. Antibodies may also clear viruses from the body before they have the possibility to enter a cell. They neutralize the pathogen by binding to free viruses (opsonization) and therefore blocking the interaction between the virus and the host cell. Exposure of portions of viral proteins (i.e., epitopes) on the cell surface through the major histocompatibility complex I (MHC I) allows T cells to kill infected cells. Other killing mechanisms are mediated by antibodies. These are antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), and complement-dependent cytotoxicity (CDC). These mechanisms help contain and clear the viral infection [1] (Figure 1). ADCC is the nonphagocytic killing of an antibody-coated target cell by a cytotoxic effector cell. The mechanism involves the release of cytotoxic granule content or the production of cell death-inducing molecules. The interaction of target-bound antibodies (IgG, IgA, or IgE classes) with specific Fc receptors (FcRs), glycoproteins on the effector cell surface that bind the Fc portion of immunoglobulins(Ig), triggers ADCC [2]. Monoclonal antibodies can enhance ADCC activity through their Fc portion, whose glycosylation pattern was shown to impact this effector function [3]. ADCP is a kind of cell-mediated immunity in which immune system cells phagocytose target cells or pathogens that have been bound by specific antibodies. In this case, the antibody Fc region engages with Fc receptors exposed on the surface of phagocytic cells and causes engulfment and killing [4]. Finally, CDC is a robust effector mechanism that consists in antibody binding to the complement component C1q to initiate the classical complement cascade, resulting in the assembly of the membrane attack complex (the complement cascade’s cytolytic end product) and lysis of the antibody-targeted infected cells [1].
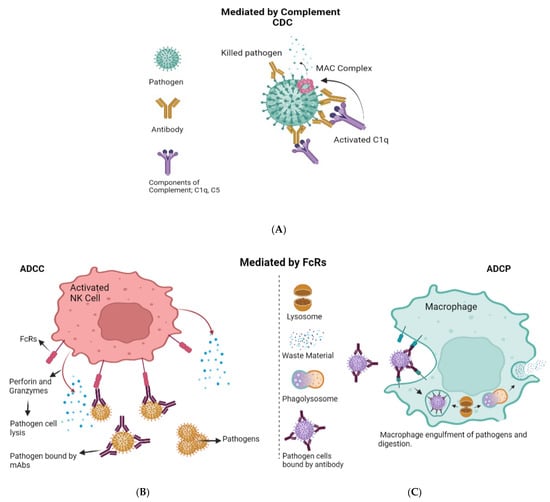
Figure 1.
Antibody effector functions. (A) Complement-dependent cytotoxicity (CDC). When an antibody binds to an antigen on the cell surface, the complement component C1q is activated and starts the cascade that leads to formation of the C5b-9 membrane attack complex (Mac), which causes cell lysis. (B) Antibody-dependent cellular cytotoxicity (ADCC) involves lysis of target cells that have been opsonized by antibodies. In the image shown here, the antibody Fc domain interacts with activated Fc receptors (FcR) on FcR-positive immune cells such as NK cells. This antibody–FcR interaction causes the production of cytokines such as IFN- and cytotoxic molecules such as perforin and granzymes, which induce pathogen cell death. (C) In antibody-dependent cell-mediated phagocytosis (ADCP), the interaction of the antibody Fc domain with the activated FcRs on phagocytes causes phagocytes to engulf the opsonized pathogens, resulting in clearance.
Antibodies are increasingly considered as an innovative and valuable class of therapeutic agents because of their unique target specificity, which promotes infection clearance [5]. Monoclonal antibodies (mAbs) derive their name from the clone of white blood cells which produced them and have gained interest in recent years as they can find application in diverse areas including medicine and biotechnology. Importantly, mAbs have been proposed as medications against viruses such as HIV and influenza and have recently been exploited in COVID-19 prophylaxis and therapy [5,6].
The mAb field has seen considerable progress since Emil von Behring and Shibasaburo Kitasato discovered antibodies in the 1890s. mAbs have the ability to target a wide variety of microbial organisms, including bacteria, viruses, parasites, fungi, and toxins [7]. Various mAbs are being used to conduct clinical trials against pathogenic viruses, including HIV and SARS-CoV-2 which, together, account for more than 40% of clinical trials (Figure 2). Other viral infections may be tackled with mAbs in the near future. For instance, influenza (10% of total clinical trials), Epstein−Barr virus (8% of total clinical trials), hepatitis C (8% of total clinical trials) and respiratory syncytial virus (7% of total clinical trials) (Figure 2). In this article, we will review the most recent advances, challenges and opportunities in mAb discovery and development against viruses.
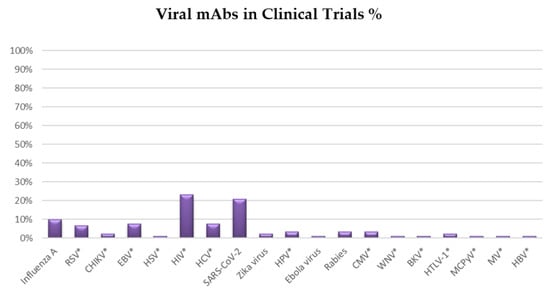
Figure 2.
Pie chart summarizing the distribution of clinical trials for mAbs against viral diseases. The graph displays the prevalence of monoclonal antibodies (mAbs) in clinical trials against various viruses. On 23 December 2021, the survey was concluded from https://clinicaltrials.gov/. *Abbreviations and acronyms used: Human immunodeficiency virus (HIV), Coronavirus Disease 2019 (COVID-19), Epstein-Barr Virus (EBV), Hepatitis C Virus (HCV), Hepatitis B virus (HBV), Respiratory Syncytial Virus (RSV), Human Papilloma Virus (HPV), Cytomegalovirus (CMV), Chikungunya virus (CHIKV), Human T-cell lymphotropic virus type 1 (HTLV-1), Herpes Simplex Virus (HSV), West Nile virus (WNV), BK virus (BKV), Merkel cell polyomavirus (MCPyV), Measles Virus (MV).
3. Standard and New Technologies for the Discovery and Development of Highly Effective mAbs
The recent technical progress in B cell sorting, sequencing, and cloning accelerated the mAb discovery and development process. In this section we will review the most recently developed methodologies which contribute to feeding the mAb discovery pipeline with increasingly safe and powerful candidates.
The hybridoma method is the most widely used technology for producing therapeutic antibodies from nonhuman sources. It consists in isolating B lymphocytes from mice that have been immunized with an antigen of interest and in fusing them with immortal myeloma cells to form hybrid cells, i.e., the hybridoma cells, which can then express mAbs against the specific antigen [153]. The advantage of this well-validated technology consists in the possibility to generate mAbs against virtually any antigen of interest. On the other hand, fusion efficiency can be low and mAbs obtained by the hybridoma method are usually nonhuman and this may be associated with downstream issues in the effector functions. Some examples of mAbs produced by means of the hybridoma methodology are presented below.
Eculizumab, a high-affinity humanized mAb that binds to the complement protein C5, prevents formation of the terminal complement complex C5b-9, which is involved in cell lysis, by inhibiting cleavage of C5 to C5a and C5b. By retaining early complement components, the C5 blockade has an indirect immunoprotective and immunoregulatory effect [154]. Leronlimab is a humanized IgG4 mAb directed against the C-C chemokine receptor type 5 (CCR5) which is being investigated as a therapy against HIV and various types of cancer. Other examples are represented by West Nile virus MGAWN1 (a neutralizing humanized mAb to West Nile virus E protein [155], MEDI-524 (Motavizumab), a humanized mAb with enhanced potency against respiratory syncytial virus (RSV) [140,156], KD-247, an anti-V3 humanized antibody that suppresses human immunodeficiency virus type 1 ex vivo and provides monkeys with sterile defense against a heterologous simian/human immunodeficiency virus infection [157]. KD-247 has the potential to be useful not only as a passive immunization antibody for HIV prevention but also as immunotherapy for HIV suppression in phenotype-matched HIV-infected people [157].
In addition to the hybridoma method, other B cell immortalization methods exist and have been used for mAb production. The Epstein−Barr virus (EBV) can transform and immortalize B cells, thus ensuring rapid screening of candidate mAbs [158,159]. Other immortalization tools include expression of BCL-6 and BCL-XL (anti-apoptotic Bcl-2 protein family). Introduction of these genes into peripheral blood memory B cells generates highly proliferating cells which secrete mAbs [160]. BCL-6 and BCL-XL transduced cells express the enzyme activation-induced cytidine deaminase (AID), which mediates somatic hypermutation and class-switch recombination and therefore increases diversity of the mAb repertoire. Immortalized cells can be maintained in culture for a long time.
The phage display technique relies on using bacteriophages (i.e., viruses that infect bacteria) to express a unique protein variant (such as antibody fragments). A gene encoding the protein of interest is implanted into a phage, allowing the phage to display the protein on the surface. The so-called phagemid plasmid, a recombinant phage display plasmid, improves the possibility of expression of both chains of target antibodies. Phage display benefits from the relatively easy manipulation of phagemid plasmids and this partially compensates for limitations imposed by phage size and proteins that can be accommodated on its surface. This system has been used to isolate antibodies which can neutralize a spectrum of viruses such as severe acute respiratory syndrome coronavirus (SARS-CoV), Ebola virus, yellow fever virus, hepatitis C virus, measles virus, rabies virus, and influenza virus. Moreover, using yeast phage display technology has led to producing novel mAbs against HIV-1 [161]. For example, 4Dm2m is a broadly neutralizing CD4-antibody fusion protein that is remarkably successful against HIV-1 [162]. Chen and co-workers enhanced flexibility, stability and half-life of 4Dm2m by introducing numerous modifications into the original molecule by taking advantage of phage-display library technologies and structure-guided design [162].
The HexaBody technology by Genmab is based on the observation that IgG antibodies may form ordered hexamers on cell surfaces after binding to their antigen. These hexamers engage the first component of the complement cascade, C1, thereby causing complement-dependent responses. When the antigen binds, conformational changes govern the exposure of the C1 binding site and complement activation [163,164]. de Jong and colleagues discovered a mutation in the Fc region of IgG that favors hexamer formation significantly more quickly upon mAb interaction with the antigen. Consequently, the complement system exerts higher activity levels which result in improved mAb effector functions. This platform has been acknowledged as a safe and effective method for the improvement of mAb activity [165] and is being exploited for optimizing mAb candidates. For instance, Genmab has been developing an antibody against multiple myeloma, named HexaBody-CD38 (GEN3014), which is undergoing Phase 1/2 clinical studies (NCT04824794) in patients affected by hematological malignancies. HexaBody-CD38 demonstrated significant increase in CDC and significant anti-tumor action [9,165]. Another example is represented by the hexabody isoform of mAb 2C7, which binds to a Neisseria gonorrhoeae lipooligosaccharide epitope expressed by >95% clinical isolates and hastens gonococcal vaginal clearance in mice [166].
The DuoBody® platform by Genmab is another interesting comprehensive technology for the discovery and production of bispecific antibodies that might help with cancer, autoimmune, infectious, and central nervous system disease antibody treatment. As reported above, bispecific antibodies bind to two epitopes on the same or separate targets. This might increase the specificity and effectiveness of the antibodies in inactivating target cells or pathogens [167,168,169]. The FDA has approved the first therapy created using DuoBody® technology platform [170].
Finally, the DuoHexaBody platform by Genmab combines the two technologies described above (Hexabody and DuoBody) and was used to develop DuoHexaBody-CD37 (GEN3009), a bispecific antibody that targets two non-overlapping CD37 epitopes. A Phase 1/2 clinical trial in patients suffering from hematologic malignancies is now under way [171]. Overall, HexaBody, DuoBody, and DuoHexaBody represent extremely promising and innovative methodologies which need robust approval and implementation into clinical trials.
IgM antibodies, unlike IgG molecules, already exist as pentameric oligomers held together by covalent bonds. The covalent linkage of IgG monomers via disulfide bonds in an IgM-derived 18 amino acid carboxyterminal extension, and furthermore between cysteine residues inserted at position 309, has been used to activate complement [172,173].
DNA and messenger RNA (mRNA) technologies have become increasingly popular since the most recent achievements in COVID-19 vaccine design and development. Administration of DNA-encoded mAbs and vaccines is a new and promising avenue that deserves exploration and exploitation, although some concerns must be addressed, such as the need for adjuvants or optimized delivery devices to obtain high immune response efficiency. In the use of DNA vaccines in humans, advances have been achieved using two approaches. The first one includes methods for physical delivery, such as a gene gun [174] or electroporation [175]. These have boosted the immunogenicity of DNA vaccines in human volunteers dramatically [176,177]. The second is the creation of a heterologous prime-boost algorithm [178] whereby the donors are primarily inoculated with a DNA vaccine, then boosted with either recombinant protein antigens or standard dead or live attenuated vaccines [179,180,181]. It is well accepted that DNA immunization can result in high-quality B-cell responses, which can be used to make highly functional mAbs [182,183]. When it comes to developing mAbs against more complex targets, such as membrane proteins, DNA immunization is more effective than traditional protein-based immunization methods [184].
For example, Elliott and colleagues developed synthetic plasmid DNA to encode two new influenza A and B mAbs that are broadly cross-protective [185]. They showed that this method generates strong quantities of functional antibodies directed against influenza A and B viruses in mouse serum via accelerated in vivo delivery of these plasmid DNA-encoded mAb (DMAb) constructs. For the first time, the authors demonstrated that FluA and FluB DMAbs are functionally comparable to recombinant mAbs produced in vitro by standard cell lines. The advantages of the DMAb technology make it a potential delivery method while offering benefits at every step of the supply chain. Indeed, DNA delivery is considerably less expensive than the traditional way of generating mAbs [185].
mRNA-based vaccines and therapies represent a new class of revolutionary medications. mRNA-1944, encoding a mAb against Chikungunya virus, has been the first mAb encoded by mRNA to be tested in a human trial [186] and will provide critical information on how mRNA may be employed to produce mAbs systemically in a dose-dependent and accessible way. mRNA-1944 encodes a completely human IgG antibody that was first isolated from B cells of a patient with a history of significant Chikungunya immunity. Within Moderna’s patented lipid nanoparticle (LNP) technology, it comprises two mRNAs that encode the heavy and light chains of this anti-Chikungunya antibody. Clinical investigations using mRNA-1944 demonstrated a linear dose-dependent relationship [186,187]. An injectable formulation containing a lipid nanoparticle–encapsulated mRNA molecule encoding this antibody protected mice against viral infection and triggered protective serum antibody responses in macaques [186]. The in vivo findings of this research opened the way to clinical trials of mRNA-based passive immunotherapy for human Chikungunya infection.
Modern medicine is being transformed by antibody immunotherapy. However, mAbs have certain drawbacks, such as production restrictions, cold-chain storage and transportation requirements, and expensive manufacturing and distribution processes. Transient in vivo gene delivery technologies, such as non-viral synthetic plasmid DNA and messenger RNA vectors built to express tailored mAb genes, might help overcome some of these obstacles. In the case of DMabs or mRNA-encoded mAbs, the body itself operates as a factory for antibody production, thus reducing both costs and the number of procedures necessary in bioprocesses [179].
Both DNA- and mRNA-mAbs have the potential to be used quickly as tools for the control of new infectious diseases. However, one of the advantages of DNA-mAbs over mRNA-mAbs is the speed with which the final formulation for distribution may be achieved. Overall, in silico approaches can improve mAb sequence, which can then be delivered either by a viral vector or as synthetic DNA. While synthetic DNA does not trigger an anti-vector backbone immunological response, viral vector integration within the host genome is possible [180]. On the other hand, mRNA must go through additional processes before being packed and inoculated or delivered. Inside cells, mRNA allows for fast protein expression by skipping the transcription step required when DNA is provided and by directly interacting with the cytoplasmic ribosomes to translate the desired protein. Conversely, DNA-mAbs should first enter the nucleus, be transcribed and then translated into the desired product. In this case, the amount of plasmid that might go into the nucleus is the relevant aspect to be considered. Several investigations have demonstrated that DNA-mAbs have protective effects in mice against various viral pathogens: Dengue virus [188], influenza A and B viruses [185,189,190], Ebolavirus [31,189], Zika virus [190], CHIKV [191], rabies [192], and HIV [193].
By integrating in vitro somatic hypermutation (SHM) with mammalian cell display, Peter M. Bowers and colleagues devised a unique approach for the collection and maturation of human antibodies that replicates fundamental characteristics of the adaptive immune system. SHM is dependent on the action of the B cell-specific enzyme activation-induced cytidine deaminase (AID) and can be replicated in non-B cells through expression of recombinant AID [194]. This method addresses many of the earlier constraints of mammalian cell display, allowing for direct antibody selection and maturation as full-length, glycosylated IgGs. Starting with a small number of variable region genes, the immune system has evolved to produce a high frequency of functional antibodies. By directly deaminating cytidine residues in Ig genes, AID is required for the beginning of SHM in B cells [195,196]. To do this, AID is directed towards V-region DNA sequences known as hot spots (e.g., WRCH), which cause mutations and amino acid changes in sites that are biased to alter antigen binding [197]. This method allows for de novo antibody maturation from a naive antibody library, as well as the maturation of preexisting antibodies. SHM affinity maturation in human B cell lymphoma lines has been reported in vitro [198].
Strategies to enhance antibody effector functions through Fc engineering represent an extremely promising avenue that deserves attention in the field of antiviral mAb discovery and development [1]. Mutations in the constant part of the mAb can extend antibody half-life: examples are represented by the anti-SARS-CoV-2 VIR-7831 and VIR-7832 mAbs, which contain the M428L/N434S mutations [199], and by the anti-RSV antibody Motavizumab, which is characterized by the M252Y/S254T/T256E substitutions [200]. Additional amino acid replacements, such as L234A, L235A, and P329G, have been found to completely abolish detectable binding to FcRIIa, FcRI, IIB, and IIC for IgG1 and IgG4, thereby preventing antibody-dependent enhancement of disease (ADE) [201]. The GAALIE modification (G236A/A330L/I332E) was shown to favor maturation of dendritic cells and induce protective CD8+ T cell responses by an anti-influenza antibody [202]. Taken together, these reports underline the capacity for IgG antibodies to promote functions which go beyond virus neutralization and encompass the so-called “vaccine-like effects” [1].
Computational techniques can predict antibody/antigen structures, engineer antibody function, and build antibody−antigen complexes with superior attributes based on high-throughput sequencing and a growing number of experimental structures of antibodies/antibody−antigen complexes. Numerous in silico approaches can be used to generate effective antibodies. For instance, prediction of (i) antibody−antigen binding, epitope mapping, and affinity maturation, (ii) aggregation, stability, and immunogenicity of antibodies, (iii) antibodies’ allosteric effects, (iv) modulation of the effector functions, and (v) structure prediction of variable domain, complementarity-determining regions, and (vi) vaccine design. Antibody−antigen binding affinities can be improved via in silico modifications of antibody residues. An example is represented by the modification of an anti-lysozyme antibody [203]. Lippow et al. were able to obtain a tenfold increase in affinity by docking the antibody onto its epitope on the antigen’s surface [204]. SnugDock [205] was proposed as a new method for predicting high-resolution antibody-antigen complex structures by physically optimizing the antibody−antigen rigid-body locations concurrently. When the crystal structure of an antibody is not available, this technique is especially advantageous since it allows for flaws in an antibody homology model that would otherwise impede rigid backbone docking predictions. Models of the West Nile virus envelope protein DIII in combination with the neutralizing E16 antibody Fab have been successfully reached by Aroop Sircar and colleagues by applying SnugDock [205]. Sefid and co-workers engineered a VHH nanobody against the Bap antigen in Acinetobacter baumannii utilizing in silico modeling [206].
Since the SARS-CoV-2 pandemic began, in silico approaches have played a major role, as represented by antibodies against COVID-19 that, despite the emergence of a number of mutations, can still bind to the virus [207]. For example, docking studies revealed that tixagevimab, bamlanivimab, and sotrovimab can form a stable complex with the Delta variant, while neutralizing the majority of the SARS-CoV-2 Alpha strains. According to the simulations, tixagevimab, regdanvimab, and cilgavimab can successfully neutralize most B.1.1.7 strains, whereas bamlanivimab, tixagevimab, and sotrovimab can effectively suppress Delta. The same study showed that while presently available mAbs might be utilized to treat COVID-19 caused by variants of SARS-CoV-2, chimeric antibodies could provide superior outcomes [207].
Effector functions such as ADCC, CDC, and ADCP can also be engineered using high-throughput computational techniques and the available structures [208]. Structure prediction of variable domain and complementarity-determining regions is one aspect of in silico antibody research that has received a lot of attention, especially from the industry. Chemical Computer Group (CCG), Schrödinger Inc. (New York, NY, USA) [209], and Accelrys Inc. (San Diego, CA, USA) are just a few examples of companies that have created techniques to achieve these goals (e.g., PIGS server [210]). Another way to predict the structure of variable domain and complementarity-determining regions is to use homology modeling [211]. RosettaAntibody uses homology to anticipate and optimize the heavy chain variable domain (VH)/light chain variable domain (VL) [212,213].
4. Conclusions
Despite tremendous progress in developing new mAbs, there are still many obstacles to overcome in every step of the developing process, as well as clinical and market challenges such as viral antigenic escape, viral variability, and short duration of viral diseases that makes them commercially less attractive than chronic diseases. Many efforts are required to overcome these challenges, including advancing more potent mAbs, development of new formulation (liquid vs. lyophilized) and delivery (injection vs. aerosol) methods, efficient clinical trials which include mAb combinations, and engagement with organizations operating in low- and middle-income countries in order to favor technology transfer and access to these new bioproducts.
Since the approval of the first murine mAb in 1986, mAb-based therapy has revealed that antiviral mAbs may be used to recruit the endogenous immune systems of infected organisms to induce long-lasting vaccine-like effects and reduce the clinical and economic impact of these infections. The ability to engineer these molecules in order to improve their properties as well as to target intracellular compartments, bind two different antigens simultaneously, deliver drug conjugates, and generate Fc fusions revolutionized the treatment of human diseases, especially viral infections. Out of 104 currently approved mAbs, 6 (5.76%) were approved in the 1990s, 16 (15.38%) from 2000 to 2010, 70 (67.3%) from 2011 to 2020, and 12 (11.5%) in the past two years (2021–2022), thus showing an upward growth in the production and marketing authorization of therapeutic mAbs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10081861/s1, Table S1: Baltimore classification of viruses based on rare to pandemic diseases caused by viruses. Table S2: mAbs related to specific antigen in viral infections followed by clinical phase.
Author Contributions
P.M., Z.P. and C.S. wrote the article, which was revised by A.A.M. and R.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
R.R. is an employee of the GSK group of companies. The other authors have no conflict of interest to declare.
References
- Pelegrin, M.; Naranjo-Gomez, M.; Piechaczyk, M. Antiviral monoclonal antibodies: Can they be more than simple neutralizing agents? Trends Microbiol. 2015, 23, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Román, V.R.G.; Murray, J.C.; Weiner, L.M. Antibody-dependent cellular cytotoxicity (ADCC). In Antibody Fc; Elsevier: Amsterdam, The Netherlands, 2014; pp. 1–27. [Google Scholar]
- Iannello, A.; Ahmad, A. Role of antibody-dependent cell-mediated cytotoxicity in the efficacy of therapeutic anti-cancer monoclonal antibodies. Cancer Metastasis Rev. 2005, 24, 487–499. [Google Scholar] [CrossRef]
- Tay, M.Z.; Wiehe, K.; Pollara, J. Antibody-dependent cellular phagocytosis in antiviral immune responses. Front. Immunol. 2019, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Sanna, P.P.; Burton, D.R. Role of antibodies in controlling viral disease: Lessons from experiments of nature and gene knockouts. J. Virol. 2000, 74, 9813–9817. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.C.; Adams, A.C.; Hufford, M.M.; De La Torre, I.; Winthrop, K.; Gottlieb, R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021, 21, 382–393. [Google Scholar] [CrossRef]
- Kantha, S.S. A centennial review; the 1890 tetanus antitoxin paper of von Behring and Kitasato and the related developments. Keio J. Med. 1991, 40, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Krupovic, M.; Agol, V.I. The Baltimore classification of viruses 50 years later: How does it stand in the light of virus evolution? Microbiol. Mol. Biol. Rev. 2021, 85, e00053-e21. [Google Scholar] [CrossRef]
- White, M.K.; Wollebo, H.S.; David Beckham, J.; Tyler, K.L.; Khalili, K. Zika virus: An emergent neuropathological agent. Ann. Neurol. 2016, 80, 479–489. [Google Scholar] [CrossRef]
- Petersen, L.R.; Jamieson, D.J.; Powers, A.M.; Honein, M.A. Zika virus. N. Engl. J. Med. 2016, 374, 1552–1563. [Google Scholar] [CrossRef]
- Richman, R.; Diallo, D.; Diallo, M.; Sall, A.A.; Faye, O.; Diagne, C.T.; Dia, I.; Weaver, S.C.; Hanley, K.A.; Buenemann, M. Ecological niche modeling of Aedes mosquito vectors of chikungunya virus in southeastern Senegal. Parasit. Vectors 2018, 11, 255. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Kraemer, M.U.G.; Brady, O.J.; Pigott, D.M.; Shearer, F.M.; Weiss, D.J.; Golding, N.; Ruktanonchai, C.W.; Gething, P.W.; Cohn, E.; et al. Mapping global environmental suitability for Zika virus. eLife 2016, 5, e15272. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zou, J.; Shan, C.; Shi, P.-Y. Small molecules and antibodies for Zika therapy. J. Infect. Dis. 2017, 216, S945–S950. [Google Scholar] [CrossRef] [PubMed]
- Van Rompay, K.K.A.; Coffey, L.L.; Kapoor, T.; Gazumyan, A.; Keesler, R.I.; Jurado, A.; Peace, A.; Agudelo, M.; Watanabe, J.; Usachenko, J.; et al. A combination of two human monoclonal antibodies limits fetal damage by Zika virus in macaques. Proc. Natl. Acad. Sci. USA 2020, 117, 7981–7989. [Google Scholar] [CrossRef] [PubMed]
- Magnani, D.M.; Rogers, T.F.; Beutler, N.; Ricciardi, M.J.; Bailey, V.K.; Gonzalez-Nieto, L.; Briney, B.; Sok, D.; Le, K.; Strubel, A.; et al. Neutralizing human monoclonal antibodies prevent Zika virus infection in macaques. Sci. Transl. Med. 2017, 9, eaan8184. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, H.; Geisbert, T.W. Ebola haemorrhagic fever. Lancet 2011, 377, 849–862. [Google Scholar] [CrossRef]
- Feldmann, H.; Jones, S.; Klenk, H.-D.; Schnittler, H.-J. Ebola virus: From discovery to vaccine. Nat. Rev. Immunol. 2003, 3, 677–685. [Google Scholar] [CrossRef] [PubMed]
- FDA Approves First Treatment for Ebola Virus 2020; US Food and Drug Administration: Silver Spring, MD, USA, 2020.
- Fan, P.; Chi, X.; Liu, G.; Zhang, G.; Chen, Z.; Liu, Y.; Fang, T.; Li, J.; Banadyga, L.; He, S.; et al. Potent neutralizing monoclonal antibodies against Ebola virus isolated from vaccinated donors. MAbs 2020, 12, 1742457. [Google Scholar] [CrossRef]
- Baize, S.; Pannetier, D.; Oestereich, L.; Rieger, T.; Koivogui, L.; Magassouba, N.; Soropogui, B.; Sow, M.S.; Keïta, S.; De Clerck, H.; et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 2014, 371, 1418–1425. [Google Scholar] [CrossRef]
- Qiu, X.; Wong, G.; Audet, J.; Bello, A.; Fernando, L.; Alimonti, J.B.; Fausther-Bovendo, H.; Wei, H.; Aviles, J.; Hiatt, E.; et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 2014, 514, 47–53. [Google Scholar] [CrossRef]
- Qiu, X.; Audet, J.; Lv, M.; He, S.; Wong, G.; Wei, H.; Luo, L.; Fernando, L.; Kroeker, A.; Bovendo, H.F.; et al. Two-mAb cocktail protects macaques against the Makona variant of Ebola virus. Sci. Transl. Med. 2016, 8, 329ra33. [Google Scholar] [CrossRef]
- Corti, D.; Misasi, J.; Mulangu, S.; Stanley, D.A.; Kanekiyo, M.; Wollen, S.; Ploquin, A.; Doria-Rose, N.A.; Staupe, R.P.; Bailey, M.; et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 2016, 351, 1339–1342. [Google Scholar] [CrossRef] [PubMed]
- Pascal, K.E.; Dudgeon, D.; Trefry, J.C.; Anantpadma, M.; Sakurai, Y.; Murin, C.D.; Turner, H.L.; Fairhurst, J.; Torres, M.; Rafique, A.; et al. Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in nonhuman primates. J. Infect. Dis. 2018, 218, S612–S626. [Google Scholar] [CrossRef] [PubMed]
- Gaudinski, M.R.; Coates, E.E.; Novik, L.; Widge, A.; Houser, K.V.; Burch, E.; Holman, L.A.; Gordon, I.J.; Chen, G.L.; Carter, C.; et al. Safety, tolerability, pharmacokinetics, and immunogenicity of the therapeutic monoclonal antibody mAb114 targeting Ebola virus glycoprotein (VRC 608): An open-label phase 1 study. Lancet 2019, 393, 889–898. [Google Scholar] [CrossRef]
- Lee, A. Ansuvimab: First Approval. Drugs 2021, 81, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Olinger, G.G.; Pettitt, J.; Kim, D.; Working, C.; Bohorov, O.; Bratcher, B.; Hiatt, E.; Hume, S.D.; Johnson, A.K.; Morton, J.; et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc. Natl. Acad. Sci. USA 2012, 109, 18030–18035. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.L.; Marzi, A. Immunotherapeutics for Ebola Virus Disease: Hope on the Horizon. Biol. Targets Ther. 2021, 15, 79. [Google Scholar] [CrossRef]
- Qiu, X.; Audet, J.; Wong, G.; Pillet, S.; Bello, A.; Cabral, T.; Strong, J.E.; Plummer, F.; Corbett, C.R.; Alimonti, J.B.; et al. Successful treatment of Ebola virus—Infected cynomolgus macaques with monoclonal antibodies. Sci. Transl. Med. 2012, 4, 138ra81. [Google Scholar] [CrossRef] [PubMed]
- Pallesen, J.; Murin, C.D.; De Val, N.; Cottrell, C.A.; Hastie, K.M.; Turner, H.L.; Fusco, M.L.; Flyak, A.I.; Zeitlin, L.; Crowe, J.E.; et al. Structures of Ebola virus GP and sGP in complex with therapeutic antibodies. Nat. Microbiol. 2016, 1, 16128. [Google Scholar] [CrossRef]
- Patel, A.; Park, D.H.; Davis, C.W.; Smith, T.R.F.; Leung, A.; Tierney, K.; Bryan, A.; Davidson, E.; Yu, X.; Racine, T.; et al. In vivo delivery of synthetic human DNA-encoded monoclonal antibodies protect against ebolavirus infection in a mouse model. Cell Rep. 2018, 25, 1982–1993. [Google Scholar] [CrossRef]
- Palache, A. Influenza Prevention Can Help Meet Wider Public Health Objectives. Health (Irvine. Calif). Available online: https://www.scirp.org/journal/paperinformation.aspx?paperid=24256 (accessed on 24 May 2022).
- Traynor, K. First recombinant flu vaccine approved. Am. J. Health-Syst. Pharm. 2013, 70, 382. [Google Scholar] [CrossRef]
- Pebody, R.; McMenamin, J.; Nohynek, H. Live attenuated influenza vaccine (LAIV): Recent effectiveness results from the USA and implications for LAIV programmes elsewhere. Arch. Dis. Child. 2018, 103, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Gresset-Bourgeois, V.; Leventhal, P.S.; Pepin, S.; Hollingsworth, R.; Kazek-Duret, M.-P.; De Bruijn, I.; Samson, S.I. Quadrivalent inactivated influenza vaccine (VaxigripTetraTM). Expert Rev. Vaccines 2018, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liao, H.; Liu, Y.; Liu, L.; Wang, F.; Song, H.; Cheng, J.; Liu, X.; Xu, D. Drug-resistant and genetic evolutionary analysis of influenza virus from patients during the 2013 and 2014 influenza season in Beijing. Microb. Drug Resist. 2017, 23, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Shobugawa, Y.; Saito, R.; Dapat, C.; Dapat, I.C.; Kondo, H.; Saito, K.; Sato, I.; Kawashima, T.; Suzuki, Y.; Suzuki, H. Clinical effectiveness of neuraminidase inhibitors—Oseltamivir, zanamivir, laninamivir, and peramivir—For treatment of influenza A (H3N2) and A (H1N1) pdm09 infection: An observational study in the 2010–2011 influenza season in Japan. J. Infect. Chemother. 2012, 18, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Corti, D.; Cameroni, E.; Guarino, B.; Kallewaard, N.L.; Zhu, Q.; Lanzavecchia, A. Tackling influenza with broadly neutralizing antibodies. Curr. Opin. Virol. 2017, 24, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.J.; Nilsson, A.C.; Silverman, M.; Assy, N.; Kulkarni, P.; McBride, J.M.; Deng, R.; Li, C.; Yang, X.; Nguyen, A.; et al. A phase 2 randomized, double-blind, placebo-controlled trial of MHAA4549A, a monoclonal antibody, plus oseltamivir in patients hospitalized with severe influenza A virus infection. Antimicrob. Agents Chemother. 2020, 64, e00352-20. [Google Scholar] [CrossRef]
- Lim, J.J.; Deng, R.; Derby, M.A.; Larouche, R.; Horn, P.; Anderson, M.; Maia, M.; Carrier, S.; Pelletier, I.; Burgess, T.; et al. Two phase 1, randomized, double-blind, placebo-controlled, single-ascending-dose studies to investigate the safety, tolerability, and pharmacokinetics of an anti-influenza A virus monoclonal antibody, MHAA4549A, in healthy volunteers. Antimicrob. Agents Chemother. 2016, 60, 5437–5444. [Google Scholar] [CrossRef]
- Gupta, P.; Kamath, A.V.; Park, S.; Chiu, H.; Lutman, J.; Maia, M.; Tan, M.-W.; Xu, M.; Swem, L.; Deng, R. Preclinical pharmacokinetics of MHAA4549A, a human monoclonal antibody to influenza A virus, and the prediction of its efficacious clinical dose for the treatment of patients hospitalized with influenza A. MAbs 2016, 8, 991–997. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, P.; Wu, M.; Yang, K.; Guo, J.; Wang, X.; Li, J.; Fang, Z.; Wang, G.; Xing, M.; et al. Adenovirus delivery of encoded monoclonal antibody protects against different types of influenza virus infection. NPJ Vaccines 2020, 5, 57. [Google Scholar] [CrossRef]
- Sutton, T.C.; Lamirande, E.W.; Bock, K.W.; Moore, I.N.; Koudstaal, W.; Rehman, M.; Weverling, G.J.; Goudsmit, J.; Subbarao, K. In vitro neutralization is not predictive of prophylactic efficacy of broadly neutralizing monoclonal antibodies CR6261 and CR9114 against lethal H2 influenza virus challenge in mice. J. Virol. 2017, 91, e01603-17. [Google Scholar] [CrossRef] [PubMed]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly conserved protective epitopes on influenza B viruses. Science 2012, 337, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Frace, A.M.; Klimov, A.I.; Rowe, T.; Black, R.A.; Katz, J.M. Modified M2 proteins produce heterotypic immunity against influenza A virus. Vaccine 1999, 17, 2237–2244. [Google Scholar] [CrossRef]
- Padilla-Quirarte, H.O.; Lopez-Guerrero, D.V.; Gutierrez-Xicotencatl, L.; Esquivel-Guadarrama, F. Protective antibodies against influenza proteins. Front. Immunol. 2019, 10, 1677. [Google Scholar] [CrossRef]
- Hershberger, E.; Sloan, S.; Narayan, K.; Hay, C.A.; Smith, P.; Engler, F.; Jeeninga, R.; Smits, S.; Trevejo, J.; Shriver, Z.; et al. Safety and efficacy of monoclonal antibody VIS410 in adults with uncomplicated influenza A infection: Results from a randomized, double-blind, phase-2, placebo-controlled study. EBioMedicine 2019, 40, 574–582. [Google Scholar] [CrossRef]
- Ramos, E.L.; Mitcham, J.L.; Koller, T.D.; Bonavia, A.; Usner, D.W.; Balaratnam, G.; Fredlund, P.; Swiderek, K.M. Efficacy and safety of treatment with an anti-m2e monoclonal antibody in experimental human influenza. J. Infect. Dis. 2015, 211, 1038–1044. [Google Scholar] [CrossRef]
- Wollacott, A.M.; Boni, M.F.; Szretter, K.J.; Sloan, S.E.; Yousofshahi, M.; Viswanathan, K.; Bedard, S.; Hay, C.A.; Smith, P.F.; Shriver, Z.; et al. Safety and upper respiratory pharmacokinetics of the hemagglutinin stalk-binding antibody VIS410 support treatment and prophylaxis based on population modeling of seasonal influenza A outbreaks. EBioMedicine 2016, 5, 147–155. [Google Scholar] [CrossRef]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Renn, A.; Fu, Y.; Hu, X.; Hall, M.D.; Simeonov, A. Fruitful neutralizing antibody pipeline brings hope to defeat SARS-CoV2. Trends Pharmacol. Sci. 2020, 41, 815–829. [Google Scholar] [CrossRef]
- Bioworld Biopharma Products in Development for COVID-19. Available online: https://www.bioworld.com/COVID19products (accessed on 22 April 2020).
- Piccaluga, P.P.; Di Guardo, A.; Lagni, A.; Lotti, V.; Diani, E.; Navari, M.; Gibellini, D. COVID-19 Vaccine: Between Myth and Truth. Vaccines 2022, 10, 349. [Google Scholar] [CrossRef]
- Catalan-Dibene, J. Human antibodies can neutralize SARS-CoV-2. Nat. Rev. Immunol. 2020, 20, 350. [Google Scholar] [CrossRef] [PubMed]
- Copin, R.; Baum, A.; Wloga, E.; Pascal, K.E.; Giordano, S.; Fulton, B.O.; Zhou, A.; Negron, N.; Lanza, K.; Chan, N.; et al. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell 2021, 184, 3949–3961. [Google Scholar] [CrossRef]
- Torres, J.L.; Ozorowski, G.; Andreano, E.; Liu, H.; Copps, J.; Piccini, G.; Donnici, L.; Conti, M.; Planchais, C.; Planas, D.; et al. Structural Insights of a Highly Potent Pan-Neutralizing SARS-CoV-2 Human Monoclonal Antibody. bioRxiv 2021, 119. [Google Scholar] [CrossRef] [PubMed]
- Andreano, E.; Paciello, I.; Piccini, G.; Manganaro, N.; Pileri, P.; Hyseni, I.; Leonardi, M.; Pantano, E.; Abbiento, V.; Benincasa, L.; et al. Hybrid immunity improves B cells and antibodies against SARS-CoV-2 variants. Nature 2021, 600, 530–535. [Google Scholar] [CrossRef]
- Cho, H.; Gonzales-Wartz, K.K.; Huang, D.; Yuan, M.; Peterson, M.; Liang, J.; Beutler, N.; Torres, J.L.; Cong, Y.; Postnikova, E.; et al. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci. Transl. Med. 2021, 13, eabj5413. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, S.; Zhang, G.; Peng, W.; Chang, Z.; Zhang, X.; Fan, Z.; Chai, Y.; Wang, F.; Zhao, X.; et al. An engineered bispecific human monoclonal antibody against SARS-CoV-2. Nat. Immunol. 2022, 23, 423–430. [Google Scholar] [CrossRef] [PubMed]
- De Gasparo, R.; Pedotti, M.; Simonelli, L.; Nickl, P.; Muecksch, F.; Cassaniti, I.; Percivalle, E.; Lorenzi, J.C.C.; Mazzola, F.; Magrì, D.; et al. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature 2021, 593, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Cathcart, A.L.; Havenar-Daughton, C.; Lempp, F.A.; Ma, D.; Schmid, M.; Agostini, M.L.; Guarino, B.; Rosen, L.; Tucker, H.; Dillen, J.; et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef]
- Van Vo, G.; Bagyinszky, E.; An, S.S.A. COVID-19 Genetic Variants and Their Potential Impact in Vaccine Development. Microorganisms 2022, 10, 598. [Google Scholar]
- Moore, J.P. Approaches for Optimal Use of Different COVID-19 Vaccines: Issues of Viral Variants and Vaccine Efficacy. JAMA 2021, 325, 1251–1252. [Google Scholar] [CrossRef]
- Fernandes, Q.; Inchakalody, V.P.; Merhi, M.; Mestiri, S.; Taib, N.; Moustafa Abo El-Ella, D.; Bedhiafi, T.; Raza, A.; Al-Zaidan, L.; Mohsen, M.O.; et al. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann. Med. 2022, 54, 524–540. [Google Scholar] [CrossRef] [PubMed]
- Bannas, P.; Hambach, J.; Koch-Nolte, F. Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics. Front. Immunol. 2017, 8, 1603. [Google Scholar] [CrossRef] [PubMed]
- Schoof, M.; Faust, B.; Saunders, R.A.; Sangwan, S.; Rezelj, V.; Hoppe, N.; Boone, M.; Billesbølle, C.B.; Puchades, C.; Azumaya, C.M.; et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 2020, 370, 1473–1479. [Google Scholar] [CrossRef] [PubMed]
- Arbabi-Ghahroudi, M. Camelid Single-Domain Antibodies: Promises and Challenges as Lifesaving Treatments. Int. J. Mol. Sci. 2022, 23, 5009. [Google Scholar] [CrossRef]
- Deb, P.; Molla, M.M.A.; Saif-Ur-Rahman, K.M. An update to monoclonal antibody as therapeutic option against COVID-19. Biosaf. Health 2021, 3, 87–91. [Google Scholar] [CrossRef]
- COVID-19 Biologics Tracker. Available online: https://www.antibodysociety.org/covid-19-biologics-tracker/ (accessed on 24 May 2022).
- Du, L.; Yang, Y.; Zhou, Y.; Lu, L.; Li, F.; Jiang, S. MERS-CoV spike protein. Expert Opin. Ther. Targets 2017, 21, 131–143. [Google Scholar] [CrossRef]
- Corti, D.; Zhao, J.; Pedotti, M.; Simonelli, L.; Agnihothram, S.; Fett, C.; Fernandez-Rodriguez, B.; Foglierini, M.; Agatic, G.; Vanzetta, F.; et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl. Acad. Sci. USA 2015, 112, 10473–10478. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, N.; Zuo, T.; Shi, X.; Poon, K.-M.V.; Wu, Y.; Gao, F.; Li, D.; Wang, R.; Guo, J.; et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014, 6, 234ra59. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, S.; Jiang, L.; Cui, Y.; Li, D.; Wang, D.; Wang, N.; Fu, L.; Shi, X.; Li, Z.; et al. Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27. Sci. Rep. 2015, 5, 13133. [Google Scholar] [CrossRef] [PubMed]
- Ying, T.; Prabakaran, P.; Du, L.; Shi, W.; Feng, Y.; Wang, Y.; Wang, L.; Li, W.; Jiang, S.; Dimitrov, D.S.; et al. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat. Commun. 2015, 6, 8223. [Google Scholar] [CrossRef]
- Agrawal, A.S.; Ying, T.; Tao, X.; Garron, T.; Algaissi, A.; Wang, Y.; Wang, L.; Peng, B.-H.; Jiang, S.; Dimitrov, D.S.; et al. Passive transfer of a germline-like neutralizing human monoclonal antibody protects transgenic mice against lethal Middle East respiratory syndrome coronavirus infection. Sci. Rep. 2016, 6, 31629. [Google Scholar] [CrossRef]
- Ying, T.; Du, L.; Ju, T.W.; Prabakaran, P.; Lau, C.C.Y.; Lu, L.; Liu, Q.; Wang, L.; Feng, Y.; Wang, Y.; et al. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 2014, 88, 7796–7805. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wan, Y.; Liu, P.; Zhao, J.; Lu, G.; Qi, J.; Wang, Q.; Lu, X.; Wu, Y.; Liu, W.; et al. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015, 25, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.; Subbarao, N. A comparative study of human betacoronavirus spike proteins: Structure, function and therapeutics. Arch. Virol. 2021, 166, 697–714. [Google Scholar] [CrossRef] [PubMed]
- Pascal, K.E.; Coleman, C.M.; Mujica, A.O.; Kamat, V.; Badithe, A.; Fairhurst, J.; Hunt, C.; Strein, J.; Berrebi, A.; Sisk, J.M.; et al. Pre-and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA 2015, 112, 8738–8743. [Google Scholar] [CrossRef]
- Luciw, P.A. Human immunodeficiency viruses and their replication. Fields Virol. 1996, 2, 1881–1952. [Google Scholar]
- Jaworski, J.P.; Cahn, P. Preventive and therapeutic features of broadly neutralising monoclonal antibodies against HIV-1. Lancet HIV 2018, 5, e723–e731. [Google Scholar] [CrossRef]
- Schoofs, T.; Klein, F.; Braunschweig, M.; Kreider, E.F.; Feldmann, A.; Nogueira, L.; Oliveira, T.; Lorenzi, J.C.C.; Parrish, E.H.; Learn, G.H.; et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science 2016, 352, 997–1001. [Google Scholar] [CrossRef]
- Herbeuval, J.-P.; Grivel, J.-C.; Boasso, A.; Hardy, A.W.; Chougnet, C.; Dolan, M.J.; Yagita, H.; Lifson, J.D.; Shearer, G.M. CD4+ T-cell death induced by infectious and noninfectious HIV-1: Role of type 1 interferon—Dependent, TRAIL/DR5-mediated apoptosis. Blood 2005, 106, 3524–3531. [Google Scholar] [CrossRef]
- Spencer, D.A.; Shapiro, M.B.; Haigwood, N.L.; Hessell, A.J. Advancing HIV broadly neutralizing antibodies: From discovery to the clinic. Front. Public Health 2021, 9, 610. [Google Scholar] [CrossRef]
- Benjelloun, F.; Lawrence, P.; Verrier, B.; Genin, C.; Paul, S. Role of human immunodeficiency virus type 1 envelope structure in the induction of broadly neutralizing antibodies. J. Virol. 2012, 86, 13152–13163. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pegu, A.; Hessell, A.J.; Mascola, J.R.; Haigwood, N.L. Use of broadly neutralizing antibodies for HIV-1 prevention. Immunol. Rev. 2017, 275, 296–312. [Google Scholar] [CrossRef] [PubMed]
- Falkowska, E.; Ramos, A.; Feng, Y.; Zhou, T.; Moquin, S.; Walker, L.M.; Wu, X.; Seaman, M.S.; Wrin, T.; Kwong, P.D.; et al. PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J. Virol. 2012, 86, 4394–4403. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, S.S.; McGreevey, W.; Whiteside, A.; Shah, M.; Cohen, J.; Hecht, R.; Bollinger, L.A.; Kinghorn, A. Twenty years of antiretroviral therapy for people living with HIV: Global costs, health achievements, economic benefits. Health Aff. 2019, 38, 1163–1172. [Google Scholar] [CrossRef]
- Jaworski, J.P.; Vendrell, A.; Chiavenna, S.M. Neutralizing monoclonal antibodies to fight HIV-1: On the threshold of success. Front. Immunol. 2017, 7, 661. [Google Scholar] [CrossRef]
- Johnson, L.F.; May, M.T.; Dorrington, R.E.; Cornell, M.; Boulle, A.; Egger, M.; Davies, M.-A. Estimating the impact of antiretroviral treatment on adult mortality trends in South Africa: A mathematical modelling study. PLoS Med. 2017, 14, e1002468. [Google Scholar] [CrossRef] [PubMed]
- Scheid, J.F.; Horwitz, J.A.; Bar-On, Y.; Kreider, E.F.; Lu, C.-L.; Lorenzi, J.C.C.; Feldmann, A.; Braunschweig, M.; Nogueira, L.; Oliveira, T.; et al. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 2016, 535, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Bar, K.J.; Sneller, M.C.; Harrison, L.J.; Justement, J.S.; Overton, E.T.; Petrone, M.E.; Salantes, D.B.; Seamon, C.A.; Scheinfeld, B.; Kwan, R.W.; et al. Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N. Engl. J. Med. 2016, 375, 2037–2050. [Google Scholar] [CrossRef] [PubMed]
- Caskey, M.; Klein, F.; Lorenzi, J.C.C.; Seaman, M.S.; West, A.P.; Buckley, N.; Kremer, G.; Nogueira, L.; Braunschweig, M.; Scheid, J.F.; et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 2015, 522, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Tobin, N.H.; Aldrovandi, G.M. Immunology of pediatric HIV infection. Immunol. Rev. 2013, 254, 143–169. [Google Scholar] [CrossRef]
- Wen, M.; Arora, R.; Wang, H.; Liu, L.; Kimata, J.T.; Zhou, P. GPI-anchored single chain Fv-an effective way to capture transiently-exposed neutralization epitopes on HIV-1 envelope spike. Retrovirology 2010, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Cimakasky, L.M.; Hampton, R.; Nguyen, D.H.; Hildreth, J.E.K. Lipid rafts and HIV pathogenesis: Host membrane cholesterol is required for infection by HIV type 1. AIDS Res. Hum. Retrovir. 2001, 17, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Carter, G.C.; Bernstone, L.; Sangani, D.; Bee, J.W.; Harder, T.; James, W. HIV entry in macrophages is dependent on intact lipid rafts. Virology 2009, 386, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Kozak, S.L.; Heard, J.M.; Kabat, D. Segregation of CD4 and CXCR4 into distinct lipid microdomains in T lymphocytes suggests a mechanism for membrane destabilization by human immunodeficiency virus. J. Virol. 2002, 76, 1802–1815. [Google Scholar] [CrossRef]
- Chazal, N.; Gerlier, D. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 2003, 67, 226–237. [Google Scholar] [CrossRef]
- Rossey, I.; Gilman, M.S.A.; Kabeche, S.C.; Sedeyn, K.; Wrapp, D.; Kanekiyo, M.; Chen, M.; Mas, V.; Spitaels, J.; Melero, J.A.; et al. Potent single-domain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state. Nat. Commun. 2017, 8, 14158. [Google Scholar] [CrossRef] [PubMed]
- Ledwith, B.J.; Manam, S.; Troilo, P.J.; Barnum, A.B.; Pauley, C.J.; Griffiths II, T.G.; Harper, L.B.; Beare, C.M.; Bagdon, W.J.; Nichols, W.W. Plasmid DNA vaccines: Investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology 2000, 43, 258–272. [Google Scholar] [CrossRef]
- Tiwari, P.M.; Vanover, D.; Lindsay, K.E.; Bawage, S.S.; Kirschman, J.L.; Bhosle, S.; Lifland, A.W.; Zurla, C.; Santangelo, P.J. Engineered mRNA-expressed antibodies prevent respiratory syncytial virus infection. Nat. Commun. 2018, 9, 3999. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.K.; Pazgier, M.; DeVico, A.L. Survivors remorse: Antibody-mediated protection against HIV-1. Immunol. Rev. 2017, 275, 271–284. [Google Scholar] [CrossRef]
- Poignard, P.; Sabbe, R.; Picchio, G.R.; Wang, M.; Gulizia, R.J.; Katinger, H.; Parren, P.W.H.I.; Mosier, D.E.; Burton, D.R. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity 1999, 10, 431–438. [Google Scholar] [CrossRef]
- Armbruster, C.; Stiegler, G.M.; Vcelar, B.A.; Jäger, W.; Köller, U.; Jilch, R.; Ammann, C.G.; Pruenster, M.; Stoiber, H.; Katinger, H.W.D. Passive immunization with the anti-HIV-1 human monoclonal antibody (hMAb) 4E10 and the hMAb combination 4E10/2F5/2G12. J. Antimicrob. Chemother. 2004, 54, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Ferrantelli, F.; Hofmann-Lehmann, R.; Rasmussen, R.A.; Wang, T.; Xu, W.; Li, P.-L.; Montefiori, D.C.; Cavacini, L.A.; Katinger, H.; Stiegler, G.; et al. Post-exposure prophylaxis with human monoclonal antibodies prevented SHIV89. 6P infection or disease in neonatal macaques. Aids 2003, 17, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Termini, J.M.; Martinez-Navio, J.M.; Gao, G.; Fuchs, S.P.; Desrosiers, R.C. Glycoengineering of AAV-delivered monoclonal antibodies yields increased ADCC activity. Mol. Ther. Methods Clin. Dev. 2021, 20, 204–217. [Google Scholar] [CrossRef]
- Arvin, G.; Campadelli-Fiume, G.; Mocarski, E.; Moore, P.S.; Roizman, B.; Whitley, R.; Yamanishi, K. (Eds.) Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Plotkin, S.A.; Boppana, S.B. Vaccination against the human cytomegalovirus. Vaccine 2019, 37, 7437–7442. [Google Scholar] [CrossRef]
- Patel, S.J.; Kuten, S.A.; Knight, R.J.; Hong, D.M.; Gaber, A.O. Resolution of mild ganciclovir-resistant cytomegalovirus disease with reduced-dose cidofovir and CMV-hyperimmune globulin. J. Transplant. 2014, 2014, 342319. [Google Scholar] [CrossRef][Green Version]
- Germer, M.; Herbener, P.; Schüttrumpf, J. Functional properties of human cytomegalovirus hyperimmunoglobulin and standard immunoglobulin preparations. Ann. Transplant. 2016, 21, 558–564. [Google Scholar] [CrossRef]
- Ha, S.; Li, F.; Troutman, M.C.; Freed, D.C.; Tang, A.; Loughney, J.W.; Wang, D.; Wang, I.-M.; Vlasak, J.; Nickle, D.C.; et al. Neutralization of diverse human cytomegalovirus strains conferred by antibodies targeting viral gH/gL/pUL128-131 pentameric complex. J. Virol. 2017, 91, e02033-16. [Google Scholar] [CrossRef] [PubMed]
- Bonaros, N.; Mayer, B.; Schachner, T.; Laufer, G.; Kocher, A. CMV-hyperimmune globulin for preventing cytomegalovirus infection and disease in solid organ transplant recipients: A meta-analysis. Clin. Transplant. 2008, 22, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Krishna, B.A.; Wills, M.R.; Sinclair, J.H. Advances in the treatment of cytomegalovirus. Br. Med. Bull. 2019, 131, 5–17. [Google Scholar] [CrossRef]
- Grossi, P.; Mohacsi, P.; Szabolcs, Z.; Potena, L. Cytomegalovirus immunoglobulin after thoracic transplantation: An overview. Transplantation 2016, 100, S1–S4. [Google Scholar] [CrossRef]
- Ye, X.; Ku, Z.; Zhang, N.; Fu, T.-M.; An, Z. Recent progress in development of monoclonal antibodies against human cytomegalovirus. Curr. Opin. Virol. 2022, 52, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Heim, K.P.; Che, Y.; Chi, X.; Qiu, X.; Han, S.; Dormitzer, P.R.; Yang, X. Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion. Sci. Adv. 2021, 7, eabf3178. [Google Scholar] [CrossRef]
- Baraniak, I.; Kropff, B.; McLean, G.R.; Pichon, S.; Piras-Douce, F.; Milne, R.S.B.; Smith, C.; Mach, M.; Griffiths, P.D.; Reeves, M.B. Epitope-specific humoral responses to human cytomegalovirus glycoprotein-B vaccine with MF59: Anti-AD2 levels correlate with protection from viremia. J. Infect. Dis. 2018, 217, 1907–1917. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Tokumoto, T.; Shirakawa, H.; Hashimoto, K.; Ikuta, K.; Kushida, N.; Yanagida, T.; Shishido, K.; Aikawa, K.; Toma, H.; et al. Lack of antibodies against the antigen domain 2 epitope of cytomegalovirus (CMV) glycoprotein B is associated with CMV disease after renal transplantation in recipients having the same glycoprotein H serotypes as their donors. Transpl. Infect. Dis. 2011, 13, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Bialas, K.M.; Westreich, D.; de la Rosa, E.; Nelson, C.S.; Kauvar, L.M.; Fu, T.-M.; Permar, S.R. Maternal Antibody Responses and Nonprimary Congenital Cytomegalovirus Infection of HIV-1—Exposed Infants. J. Infect. Dis. 2016, 214, 1916–1923. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.D.; Nikitin, P.; Gesner, T.; Lin, J.J.; Barkan, D.T.; Ciferri, C.; Carfi, A.; Akbarnejad Yazdi, T.; Skewes-Cox, P.; Wiedmann, B.; et al. In vitro characterization of human cytomegalovirus-targeting therapeutic monoclonal antibodies LJP538 and LJP539. Antimicrob. Agents Chemother. 2016, 60, 4961–4971. [Google Scholar] [CrossRef]
- Boeckh, M.; Bowden, R.A.; Storer, B.; Chao, N.J.; Spielberger, R.; Tierney, D.K.; Gallez-Hawkins, G.; Cunningham, T.; Blume, K.G.; Levitt, D.; et al. Randomized, placebo-controlled, double-blind study of a cytomegalovirus-specific monoclonal antibody (MSL-109) for prevention of cytomegalovirus infection after allogeneic hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2001, 7, 343–351. [Google Scholar] [CrossRef]
- Borucki, M.J.; Spritzler, J.; Asmuth, D.M.; Gnann, J.; Hirsch, M.S.; Nokta, M.; Aweeka, F.; Nadler, P.I.; Sattler, F.; Alston, B.; et al. A phase II, double-masked, randomized, placebo-controlled evaluation of a human monoclonal anti-Cytomegalovirus antibody (MSL-109) in combination with standard therapy versus standard therapy alone in the treatment of AIDS patients with Cytomegalovirus re. Antiviral Res. 2004, 64, 103–111. [Google Scholar] [CrossRef]
- Ishida, J.H.; Burgess, T.; Derby, M.A.; Brown, P.A.; Maia, M.; Deng, R.; Emu, B.; Feierbach, B.; Fouts, A.E.; Liao, X.C.; et al. Phase 1 randomized, double-blind, placebo-controlled study of RG7667, an anticytomegalovirus combination monoclonal antibody therapy, in healthy adults. Antimicrob. Agents Chemother. 2015, 59, 4919–4929. [Google Scholar] [CrossRef]
- Ishida, J.H.; Patel, A.; Mehta, A.K.; Gatault, P.; McBride, J.M.; Burgess, T.; Derby, M.A.; Snydman, D.R.; Emu, B.; Feierbach, B.; et al. Phase 2 randomized, double-blind, placebo-controlled trial of RG7667, a combination monoclonal antibody, for prevention of cytomegalovirus infection in high-risk kidney transplant recipients. Antimicrob. Agents Chemother. 2017, 61, e01794-16. [Google Scholar] [CrossRef]
- Kschonsak, M.; Rougé, L.; Arthur, C.P.; Hoangdung, H.; Patel, N.; Kim, I.; Johnson, M.C.; Kraft, E.; Rohou, A.L.; Gill, A.; et al. Structures of HCMV trimer reveal the basis for receptor recognition and cell entry. Cell 2021, 184, 1232–1244. [Google Scholar] [CrossRef] [PubMed]
- McVoy, M.M.; Tenorio, E.; Kauvar, L.M. A native human monoclonal antibody targeting HCMV gB (AD-2 site I). Int. J. Mol. Sci. 2018, 19, 3982. [Google Scholar] [CrossRef] [PubMed]
- Brey, C.U.; Proff, J.; Teufert, N.; Salzer, B.; Brozy, J.; Münz, M.; Pendzialek, J.; Ensser, A.; Holter, W.; Lehner, M. A gB/CD3 bispecific BiTE antibody construct for targeting Human Cytomegalovirus-infected cells. Sci. Rep. 2018, 8, 17453. [Google Scholar] [CrossRef]
- Meng, W.; Tang, A.; Ye, X.; Gui, X.; Li, L.; Fan, X.; Schultz, R.D.; Freed, D.C.; Ha, S.; Wang, D.; et al. Targeting human-cytomegalovirus-infected cells by redirecting T cells using an anti-CD3/anti-glycoprotein B bispecific antibody. Antimicrob. Agents Chemother. 2018, 62, e01719-17. [Google Scholar] [CrossRef]
- Li, F.; Freed, D.C.; Tang, A.; Rustandi, R.R.; Troutman, M.C.; Espeseth, A.S.; Zhang, N.; An, Z.; McVoy, M.; Zhu, H.; et al. Complement enhances in vitro neutralizing potency of antibodies to human cytomegalovirus glycoprotein B (gB) and immune sera induced by gB/MF59 vaccination. NPJ Vaccines 2017, 2. [Google Scholar] [CrossRef]
- Su, H.; Ye, X.; Freed, D.C.; Li, L.; Ku, Z.; Xiong, W.; Gao, P.; Liu, X.; Montgomery, D.; Xu, W.; et al. Potent Bispecific Neutralizing Antibody Targeting Glycoprotein B and the gH/gL/pUL128/130/131 Complex of Human Cytomegalovirus. Antimicrob. Agents Chemother. 2020, 65, e02422-20. [Google Scholar] [CrossRef] [PubMed]
- Battles, M.B.; McLellan, J.S. Respiratory syncytial virus entry and how to block it. Nat. Rev. Microbiol. 2019, 17, 233–245. [Google Scholar] [CrossRef]
- Collins, P.L.; Fearns, R.; Graham, B.S. Respiratory syncytial virus: Virology, reverse genetics, and pathogenesis of disease. In Challenges and Opportunities for Respiratory Syncytial Virus Vaccines; Springer: Berlin, Germany, 2013; pp. 3–38. [Google Scholar]
- Mejias, A.; Rodrìguez-Fernández, R.; Oliva, S.; Peeples, M.E.; Ramilo, O. The journey to a respiratory syncytial virus vaccine. Ann. Allergy Asthma Immunol. 2020, 125, 36–46. [Google Scholar] [CrossRef]
- Globulin, R.-R.S.V.I. Intravenous (Human) (RSV-IGIV), Drug Information; Medlmmune Inc.: Gaithersburg, MD, USA.
- Simpson, S.; Burls, A. A Systematic Review of the Effectiveness and Cost-Effectiveness of Palivizumab (Synagis) in the Prevention of Respiratory Syncytial Virus (RSV) Infection in Infants at High Risk of Infection. The University of York. Centre for Reviews and Dissemination. Database of Abstracts of Reviews of Effects (DARE): Quality-assessed Reviews. 2001. Available online: https://www.ncbi.nlm.nih.gov/books/NBK68498/ (accessed on 24 May 2022).
- Cingoz, O. Motavizumab. MAbs 2009, 1, 439–442. [Google Scholar] [CrossRef]
- Shilovskiy, I.P.; Andreev, S.M.; Kozhikhova, K.V.; Nikolskii, A.A.; Khaitov, M.R. Prospects for the use of peptides against respiratory syncytial virus. Mol. Biol. 2019, 53, 484–500. [Google Scholar] [CrossRef]
- Huang, J.; Diaz, D.; Mousa, J.J. Antibody epitopes of pneumovirus fusion proteins. Front. Immunol. 2019, 10, 2778. [Google Scholar] [CrossRef] [PubMed]
- Committee on Infectious Diseases Committee on Fetus and Newborn. Respiratory syncytial virus immune globulin intravenous: Indications for use. Pediatrics 1997, 99, 645–650. [Google Scholar] [CrossRef]
- Palivizumab, A. Humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 1998, 102, 531–537. [Google Scholar]
- Jartti, T.; Bønnelykke, K.; Elenius, V.; Feleszko, W. Role of viruses in asthma. Semin. Immunopathol. 2020, 42, 61–74. [Google Scholar] [CrossRef]
- McLellan, J.S.; Chen, M.; Leung, S.; Graepel, K.W.; Du, X.; Yang, Y.; Zhou, T.; Baxa, U.; Yasuda, E.; Beaumont, T.; et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 2013, 340, 1113–1117. [Google Scholar] [CrossRef]
- Tang, A.; Chen, Z.; Cox, K.S.; Su, H.-P.; Callahan, C.; Fridman, A.; Zhang, L.; Patel, S.B.; Cejas, P.J.; Swoyer, R.; et al. A Potent Broadly Neutralizing Human RSV Antibody Targets Conserved Site IV of the Fusion Glycoprotein. Nat. Commun. 2019, 10, 4153. [Google Scholar] [CrossRef]
- Zhu, Q.; McLellan, J.S.; Kallewaard, N.L.; Ulbrandt, N.D.; Palaszynski, S.; Zhang, J.; Moldt, B.; Khan, A.; Svabek, C.; McAuliffe, J.M.; et al. A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci. Transl. Med. 2017, 9, eaaj1928. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.G.; Ritschel, T.; Pascual, G.; Brakenhoff, J.P.; Keogh, E.; Furmanova-Hollenstein, P.; Lanckacker, E.; Wadia, J.S.; Gilman, M.S.; Williamson, R.A.; et al. Structural basis for recognition of the central conserved region of RSV G by neutralizing human antibodies. PLoS Pathog. 2018, 14, e1006935. [Google Scholar] [CrossRef] [PubMed]
- Fedechkin, S.O.; George, N.L.; Wolff, J.T.; Kauvar, L.M.; DuBois, R.M. Structures of respiratory syncytial virus G antigen bound to broadly neutralizing antibodies. Sci. Immunol. 2018, 3, eaar3534. [Google Scholar] [CrossRef] [PubMed]
- Acero-Bedoya, S.; Wozniak, P.S.; Sánchez, P.J.; Ramilo, O.; Mejias, A. Recent trends in RSV immunoprophylaxis: Clinical implications for the infant. Am. J. Perinatol. 2019, 36, S63–S67. [Google Scholar] [CrossRef]
- Domachowske, J.B.; Anderson, E.J.; Goldstein, M. The future of respiratory syncytial virus disease prevention and treatment. Infect. Dis. Ther. 2021, 10, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Rocca, A.; Biagi, C.; Scarpini, S.; Dondi, A.; Vandini, S.; Pierantoni, L.; Lanari, M. Passive immunoprophylaxis against respiratory syncytial virus in children: Where are we now? Int. J. Mol. Sci. 2021, 22, 3703. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Tomar, P.C. Hybridoma technology; advancements, clinical significance, and future aspects. J. Genet. Eng. Biotechnol. 2021, 19, 159. [Google Scholar] [CrossRef]
- Diurno, F.; Numis, F.G.; Porta, G.; Cirillo, F.; Maddaluno, S.; Ragozzino, A.; De Negri, P.; Di Gennaro, C.; Pagano, A.; Allegorico, E.; et al. Eculizumab treatment in patients with COVID-19: Preliminary results from real life ASL Napoli 2 Nord experience. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4040–4047. [Google Scholar] [PubMed]
- Beigel, J.H.; Nordstrom, J.L.; Pillemer, S.R.; Roncal, C.; Goldwater, D.R.; Li, H.; Holland, P.C.; Johnson, S.; Stein, K.; Koenig, S. Safety and pharmacokinetics of single intravenous dose of MGAWN1, a novel monoclonal antibody to West Nile virus. Antimicrob. Agents Chemother. 2010, 54, 2431–2436. [Google Scholar] [CrossRef]
- Carbonell-Estrany, X.; Simões, E.A.; Dagan, R.; Hall, C.B.; Harris, B.; Hultquist, M.; Connor, E.M.; Losonsky, G.A.; Motavizumab Study Group. Motavizumab for prophylaxis of respiratory syncytial virus in high-risk children: A noninferiority trial. Pediatrics 2010, 125, e35–e51. [Google Scholar] [CrossRef] [PubMed]
- Eda, Y.; Murakami, T.; Ami, Y.; Nakasone, T.; Takizawa, M.; Someya, K.; Kaizu, M.; Izumi, Y.; Yoshino, N.; Matsushita, S.; et al. Anti-V3 humanized antibody KD-247 effectively suppresses ex vivo generation of human immunodeficiency virus type 1 and affords sterile protection of monkeys against a heterologous simian/human immunodeficiency virus infection. J. Virol. 2006, 80, 5563–5570. [Google Scholar] [CrossRef]
- Hui-Yuen, J.; Koganti, S.; Bhaduri-McIntosh, S. Human B cell immortalization for monoclonal antibody production. In Monoclonal Antibodies; Springer: Berlin, Germany, 2014; pp. 183–189. [Google Scholar]
- Steinitz, M.; Klein, G.; Koskimies, S.; Makel, O. EB virus-induced B lymphocyte cell lines producing specific antibody. Nature 1977, 269, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Kwakkenbos, M.J.; Diehl, S.A.; Yasuda, E.; Bakker, A.Q.; Van Geelen, C.M.M.; Lukens, M.V.; Van Bleek, G.M.; Widjojoatmodjo, M.N.; Bogers, W.M.J.M.; Mei, H.; et al. Generation of stable monoclonal antibody—Producing B cell receptor—Positive human memory B cells by genetic programming. Nat. Med. 2010, 16, 123–128. [Google Scholar] [CrossRef]
- Bowley, D.R.; Labrijn, A.F.; Zwick, M.B.; Burton, D.R. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng. Des. Sel. 2007, 20, 81–90. [Google Scholar] [CrossRef]
- Chen, W.; Bardhi, A.; Feng, Y.; Wang, Y.; Qi, Q.; Li, W.; Zhu, Z.; Dyba, M.A.; Ying, T.; Jiang, S.; et al. Improving the CH1-CK heterodimerization and pharmacokinetics of 4Dm2m, a novel potent CD4-antibody fusion protein against HIV-1. MAbs 2016, 8, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Burton, D.R. Is IgM-like dislocation a common feature of antibody function? Immunol. Today 1986, 7, 165–167. [Google Scholar] [CrossRef]
- Collins, C.; Tsui, F.W.L.; Shulman, M.J. Differential activation of human and guinea pig complement by pentameric and hexameric IgM. Eur. J. Immunol. 2002, 32, 1802–1810. [Google Scholar] [CrossRef]
- de Jong, R.N.; Beurskens, F.J.; Verploegen, S.; Strumane, K.; van Kampen, M.D.; Voorhorst, M.; Horstman, W.; Engelberts, P.J.; Oostindie, S.C.; Wang, G.; et al. A novel platform for the potentiation of therapeutic antibodies based on antigen-dependent formation of IgG hexamers at the cell surface. PLoS Biol. 2016, 14, e1002344. [Google Scholar] [CrossRef]
- Gulati, S.; McQuillen, D.P.; Mandrell, R.E.; Jani, D.B.; Rice, P.A. Immunogenicity of Neisseria gonorrhoeae lipooligosaccharide epitope 2C7, widely expressed in vivo with no immunochemical similarity to human glycosphingolipids. J. Infect. Dis. 1996, 174, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Campi, G.; Charlish, P. The Bispecific Approach. MedNous 2021, 15. Available online: https://www.mednous.com/system/files/2021-03/MedNous-WEB%20MAR2021.pdf (accessed on 24 May 2022).
- Labrijn, A.F.; Meesters, J.I.; de Goeij, B.E.C.G.; van den Bremer, E.T.J.; Neijssen, J.; van Kampen, M.D.; Strumane, K.; Verploegen, S.; Kundu, A.; Gramer, M.J.; et al. Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Proc. Natl. Acad. Sci. USA 2013, 110, 5145–5150. [Google Scholar] [CrossRef] [PubMed]
- Oostindie, S.C.; van der Horst, H.J.; Kil, L.P.; Strumane, K.; Overdijk, M.B.; van den Brink, E.N.; van den Brakel, J.H.N.; Rademaker, H.J.; van Kessel, B.; van den Noort, J.; et al. DuoHexaBody-CD37®, a novel biparatopic CD37 antibody with enhanced Fc-mediated hexamerization as a potential therapy for B-cell malignancies. Blood Cancer J. 2020, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Genmab 2021 Annual Report. 2022. Available online: https://ir.genmab.com/news-releases/news-release-details/genmab-publishes-2021-annual-report (accessed on 24 May 2022).
- Hutchings, M.; Lugtenburg, P.; Mous, R.; Clausen, M.R.; Chamuleau, M.; Linton, K.; Rule, S.; Lopez, J.S.; Oliveri, R.S.; DeMarco, D.; et al. Epcoritamab (GEN3013; DuoBody-CD3 × CD20) to induce complete response in patients with relapsed/refractory B-cell non-Hodgkin lymphoma (B-NHL): Complete dose escalation data and efficacy results from a phase I/II trial. J. Clin. Oncol. 2020, 38, 8009. [Google Scholar] [CrossRef]
- Smith, R.I.; Coloma, M.J.; Morrison, S.L. Addition of a mu-tailpiece to IgG results in polymeric antibodies with enhanced effector functions including complement-mediated cytolysis by IgG4. J. Immunol. 1995, 154, 2226–2236. [Google Scholar]
- Sopp, J.M.; Peters, S.J.; Rowley, T.F.; Oldham, R.J.; James, S.; Mockridge, I.; French, R.R.; Turner, A.; Beers, S.A.; Humphreys, D.P.; et al. On-target IgG hexamerisation driven by a C-terminal IgM tail-piece fusion variant confers augmented complement activation. Commun. Biol. 2021, 4, 1031. [Google Scholar] [CrossRef] [PubMed]
- Raska, M.; Turanek, J. DNA vaccines for the induction of immune responses in mucosal tissues. In Mucosal Immunology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 1307–1335. [Google Scholar]
- Xu, Z.; Patel, A.; Tursi, N.J.; Zhu, X.; Muthumani, K.; Kulp, D.W.; Weiner, D.B. Harnessing recent advances in synthetic DNA and electroporation technologies for rapid vaccine development against COVID-19 and other emerging infectious diseases. Front. Med. Technol. 2020, 5, 571030. [Google Scholar] [CrossRef] [PubMed]
- Kalams, S.A.; Parker, S.D.; Elizaga, M.; Metch, B.; Edupuganti, S.; Hural, J.; De Rosa, S.; Carter, D.K.; Rybczyk, K.; Frank, I.; et al. Safety and comparative immunogenicity of an HIV-1 DNA vaccine in combination with plasmid interleukin 12 and impact of intramuscular electroporation for delivery. J. Infect. Dis. 2013, 208, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Evans, K.; McElwaine-Johnn, H.; Sharpe, M.; Oxford, J.; Lambkin-Williams, R.; Mant, T.; Nolan, A.; Zambon, M.; Ellis, J.; et al. DNA vaccination protects against an influenza challenge in a double-blind randomised placebo-controlled phase 1b clinical trial. Vaccine 2009, 27, 2506–2512. [Google Scholar] [CrossRef] [PubMed]
- Lu, S. Heterologous prime–boost vaccination. Curr. Opin. Immunol. 2009, 21, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kennedy, J.S.; West, K.; Montefiori, D.C.; Coley, S.; Lawrence, J.; Shen, S.; Green, S.; Rothman, A.L.; Ennis, F.A.; et al. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime–protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 2008, 26, 3947–3957. [Google Scholar] [CrossRef]
- Ledgerwood, J.E.; Wei, C.-J.; Hu, Z.; Gordon, I.J.; Enama, M.E.; Hendel, C.S.; McTamney, P.M.; Pearce, M.B.; Yassine, H.M.; Boyington, J.C.; et al. DNA priming and influenza vaccine immunogenicity: Two phase 1 open label randomised clinical trials. Lancet Infect. Dis. 2011, 11, 916–924. [Google Scholar] [CrossRef]
- Vaine, M.; Wang, S.; Liu, Q.; Arthos, J.; Montefiori, D.; Goepfert, P.; McElrath, M.J.; Lu, S. Profiles of human serum antibody responses elicited by three leading HIV vaccines focusing on the induction of Env-specific antibodies. PLoS ONE 2010, 5, e13916. [Google Scholar] [CrossRef]
- Boyer, J.D.; Chattergoon, M.A.; Ugen, K.E.; Shah, A.; Bennett, M.; Cohen, A.; Nyland, S.; Lacy, K.E.; Bagarazzi, M.L.; Higgins, T.J.; et al. Enhancement of cellular immune response in HIV-1 seropositive individuals: A DNA-based trial. Clin. Immunol. 1999, 90, 100–107. [Google Scholar] [CrossRef]
- Boyer, J.D.; Chattergoon, M.; Shah, A.; Ginsberg, R.; MacGregor, R.R.; Weiner, D.B. HIV-1 DNA based vaccine induces a CD8 mediated cross-clade CTL response. Dev. Biol. Stand. 1998, 95, 147–153. [Google Scholar]
- Liu, S.; Wang, S.; Lu, S. DNA immunization as a technology platform for monoclonal antibody induction. Emerg. Microbes Infect. 2016, 5, e33. [Google Scholar] [CrossRef]
- Elliott, S.T.C.; Kallewaard, N.L.; Benjamin, E.; Wachter-Rosati, L.; McAuliffe, J.M.; Patel, A.; Smith, T.R.F.; Schultheis, K.; Park, D.H.; Flingai, S.; et al. DMAb inoculation of synthetic cross reactive antibodies protects against lethal influenza A and B infections. NPJ Vaccines 2017, 2, 18. [Google Scholar] [CrossRef]
- August, A.; Attarwala, H.Z.; Himansu, S.; Kalidindi, S.; Lu, S.; Pajon, R.; Han, S.; Lecerf, J.-M.; Tomassini, J.E.; Hard, M.; et al. A phase 1 trial of lipid-encapsulated mRNA encoding a monoclonal antibody with neutralizing activity against Chikungunya virus. Nat. Med. 2021, 27, 2224–2233. [Google Scholar] [CrossRef]
- Moderna. Moderna Announces Positive Phase 1 Results for the First Systemic Messenger RNA Therapeutic Encoding a Secreted Protein (MRNA-1944). Bus. wire Cambridge. 2019. Available online: https://www.sec.gov/Archives/edgar/data/1682852/000119312519243385/d796420dex992.htm (accessed on 24 May 2022).
- Flingai, S.; Plummer, E.M.; Patel, A.; Shresta, S.; Mendoza, J.M.; Broderick, K.E.; Sardesai, N.Y.; Muthumani, K.; Weiner, D.B. Protection against dengue disease by synthetic nucleic acid antibody prophylaxis/immunotherapy. Sci. Rep. 2015, 5, 12616. [Google Scholar] [CrossRef]
- Yamazaki, T.; Nagashima, M.; Ninomiya, D.; Arai, Y.; Teshima, Y.; Fujimoto, A.; Ainai, A.; Hasegawa, H.; Chiba, J. Passive immune-prophylaxis against influenza virus infection by the expression of neutralizing anti-hemagglutinin monoclonal antibodies from plasmids. Jpn. J. Infect. Dis. 2011, 64, 40–49. [Google Scholar] [CrossRef]
- Andrews, C.D.; Luo, Y.; Sun, M.; Yu, J.; Goff, A.J.; Glass, P.J.; Padte, N.N.; Huang, Y.; Ho, D.D. In vivo production of monoclonal antibodies by gene transfer via electroporation protects against lethal influenza and Ebola infections. Mol. Ther. Clin. Dev. 2017, 7, 74–82. [Google Scholar] [CrossRef]
- Esquivel, R.N.; Patel, A.; Kudchodkar, S.B.; Park, D.H.; Stettler, K.; Beltramello, M.; Allen, J.W.; Mendoza, J.; Ramos, S.; Choi, H.; et al. In vivo delivery of a DNA-encoded monoclonal antibody protects non-human primates against Zika virus. Mol. Ther. 2019, 27, 974–985. [Google Scholar] [CrossRef]
- Muthumani, K.; Block, P.; Flingai, S.; Muruganantham, N.; Chaaithanya, I.K.; Tingey, C.; Wise, M.; Reuschel, E.L.; Chung, C.; Muthumani, A.; et al. Rapid and long-term immunity elicited by DNA-encoded antibody prophylaxis and DNA vaccination against chikungunya virus. J. Infect. Dis. 2016, 214, 369–378. [Google Scholar] [CrossRef]
- Thran, M.; Mukherjee, J.; Pönisch, M.; Fiedler, K.; Thess, A.; Mui, B.L.; Hope, M.J.; Tam, Y.K.; Horscroft, N.; Heidenreich, R.; et al. mRNA mediates passive vaccination against infectious agents, toxins, and tumors. EMBO Mol. Med. 2017, 9, 1434–1447. [Google Scholar] [CrossRef]
- Pardi, N.; Secreto, A.J.; Shan, X.; Debonera, F.; Glover, J.; Yi, Y.; Muramatsu, H.; Ni, H.; Mui, B.L.; Tam, Y.K.; et al. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017, 8, 14630. [Google Scholar] [CrossRef]
- Chaudhuri, J.; Khuong, C.; Alt, F.W. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature 2004, 430, 992–998. [Google Scholar] [CrossRef]
- Martin, A.; Scharff, M.D. Somatic hypermutation of the AID transgene in B and non-B cells. Proc. Natl. Acad. Sci. USA 2002, 99, 12304–12308. [Google Scholar] [CrossRef]
- Maul, R.W.; Gearhart, P.J. AID and somatic hypermutation. Adv. Immunol. 2010, 105, 159–191. [Google Scholar]
- Ohm-Laursen, L.; Barington, T. Analysis of 6912 unselected somatic hypermutations in human VDJ rearrangements reveals lack of strand specificity and correlation between phase II substitution rates and distance to the nearest 3′ activation-induced cytidine deaminase target. J. Immunol. 2007, 178, 4322–4334. [Google Scholar] [CrossRef]
- Delker, R.K.; Fugmann, S.D.; Papavasiliou, F.N. A coming-of-age story: Activation-induced cytidine deaminase turns 10. Nat. Immunol. 2009, 10, 1147–1153. [Google Scholar] [CrossRef]
- Saunders, K.O. Conceptual approaches to modulating antibody effector functions and circulation half-life. Front. Immunol. 2019, 10, 1296. [Google Scholar] [CrossRef]
- Robbie, G.J.; Criste, R.; Dall’Acqua, W.F.; Jensen, K.; Patel, N.K.; Losonsky, G.A.; Griffin, M.P. A novel investigational Fc-modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. Antimicrob. Agents Chemother. 2013, 57, 6147–6153. [Google Scholar] [CrossRef]
- Schlothauer, T.; Herter, S.; Koller, C.F.; Grau-Richards, S.; Steinhart, V.; Spick, C.; Kubbies, M.; Klein, C.; Umaña, P.; Mössner, E. Novel human IgG1 and IgG4 Fc-engineered antibodies with completely abolished immune effector functions. Protein Eng. Des. Sel. 2016, 29, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Bournazos, S.; Corti, D.; Virgin, H.W.; Ravetch, J. V Fc-optimized antibodies elicit CD8 immunity to viral respiratory infection. Nature 2020, 588, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Riechmann, L. Affinity improvement of single antibody VH domains: Residues in all three hypervariable regions affect antigen binding. Immunotechnology 1996, 2, 169–179. [Google Scholar] [CrossRef]
- Hermann, P.C. Computational design of antibody-affinity improvement beyond in vivo maturation. Nat. Rev. Drug Discov. 2007, 6, 1171–1176. [Google Scholar]
- Sircar, A.; Gray, J.J. SnugDock: Paratope structural optimization during antibody-antigen docking compensates for errors in antibody homology models. PLoS Comput. Biol. 2010, 6, e1000644. [Google Scholar] [CrossRef]
- Sefid, F.; Payandeh, Z.; Azamirad, G.; Abdolhamidi, R.; Rasooli, I. In silico engineering towards enhancement of bap—VHH monoclonal antibody binding affinity. Int. J. Pept. Res. Ther. 2019, 25, 273–287. [Google Scholar] [CrossRef]
- Das, N.C.; Chakraborty, P.; Bayry, J.; Mukherjee, S. In silico analyses on the comparative potential of therapeutic human monoclonal antibodies against newly emerged SARS-CoV-2 variants bearing mutant spike protein. Front. Immunol. 2021, 12, 782506. [Google Scholar] [CrossRef]
- Zhao, J.; Nussinov, R.; Wu, W.-J.; Ma, B. In silico methods in antibody design. Antibodies 2018, 7, 22. [Google Scholar] [CrossRef]
- Zhu, K.; Day, T.; Warshaviak, D.; Murrett, C.; Friesner, R.; Pearlman, D. Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction. Proteins Struct. Funct. Bioinform. 2014, 82, 1646–1655. [Google Scholar] [CrossRef]
- Marcatili, P.; Rosi, A.; Tramontano, A. PIGS: Automatic prediction of antibody structures. Bioinformatics 2008, 24, 1953–1954. [Google Scholar] [CrossRef]
- Martin, A.C.; Cheetham, J.C.; Rees, A.R. Modeling antibody hypervariable loops: A combined algorithm. Proc. Natl. Acad. Sci. USA 1989, 86, 9268–9272. [Google Scholar] [CrossRef]
- Sircar, A.; Kim, E.T.; Gray, J.J. RosettaAntibody: Antibody variable region homology modeling server. Nucleic Acids Res. 2009, 37, W474–W479. [Google Scholar] [CrossRef]
- Weitzner, B.D.; Kuroda, D.; Marze, N.; Xu, J.; Gray, J.J. Blind prediction performance of Rosetta Antibody 3.0: Grafting, relaxation, kinematic loop modeling, and full CDR optimization. Proteins Struct. Funct. Bioinform. 2014, 82, 1611–1623. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
