Abstract
Aging is the main cause of decline in oocyte quality, which can further trigger the failure of assisted reproductive technology (ART). Exploring age-related genes in oocytes is an important way to investigate the molecular mechanisms involved in oocyte aging. To provide novel insight into this field, we performed a pooled analysis of publicly available datasets, using the overlapping results of two statistical methods on two Gene Expression Omnibus (GEO) datasets. The methods utilized in the current study mainly include Spearman rank correlation, the Wilcoxon signed-rank test, t-tests, Venn diagrams, Gene Ontology (GO), Protein–Protein Interaction (PPI), Gene Set Enrichment Analysis (GSEA), Gene Set Variation Analysis (GSVA), and receiver operating characteristic (ROC) curve analysis. We identified hundreds of age-related genes across different gene expression datasets of in vitro maturation-metaphase II (IVM-MII) oocytes. Age-related genes in IVM-MII oocytes were involved in the biological processes of cellular metabolism, DNA replication, and histone modifications. Among these age-related genes, FAM111A expression presented a robust correlation with age, seen in the results of different statistical methods and different datasets. FAM111A is associated with the processes of chromosome segregation and cell cycle regulation. Thus, this enzyme is potentially an interesting novel marker for the aging of oocytes, and warrants further mechanistic study.
1. Introduction
Female fertility decreases progressively with age. It has been revealed that the fertility of a woman in her mid-30s is only half of the fertility potential of a woman in her mid-20s; and this is reduced to less than 5% for a woman in her 40s [1]. The decrease in fertility is reflected in the lower quantity and quality of oocytes. Age-related decline in oocyte quality is an important non-male cause of assisted reproductive technology (ART) failures, and there is no successful treatment for the age-related decline in oocyte quality [2]. Increased aneuploidy is an important cause of the decline in oocyte quality [3]. In addition, oocyte quality decline has also been associated with nuclear and mitochondrial DNA damage [4,5], meiotic spindle abnormalities, chromosomal misalignment, mitochondrial dysfunction [6], telomere shortening [7], and abnormal structure of the zona pellucida [8,9] and nucleolus [10].
To date, several investigations have explored the influence of maternal age on oocyte gene expression, using microarray and RNA-seq technology. Of these, most have been single-center studies on one or two types of oocytes, with limited sample size. Different analysis methods were used in each study, and the results of these studies remain controversial. Some studies reported that gene expression data for the human germinal vesicle (GV) oocytes [11,12], from women across age groups, remained stable; while others revealed that gene expression among GV oocytes [13], in vivo ovulation-metaphase Ⅱ (IVO-MII) oocytes [14,15,16,17], and in vitro maturation-metaphase II (IVM-MII) oocytes [12,13], were significantly influenced by maternal age. To obtain more generalizable results, multi-center studies with large sample sizes are required.
Given the above research background, in this study, we performed an integrative analysis of the available datasets. The data were derived from two original Gene Expression Omnibus (GEO) datasets, which provide gene expression profiles of oocytes from women of various ages. These two datasets were deposited, respectively, by Reyes et al. [12] and Llonch et al. [13]. In the study of Reyes et al. [12], age was treated as a categorical variable, and differential gene expression analysis between young and old groups was performed using the DESeq2 package. In the study of Llonch et al. [13], age was treated as a continuous variable, and Pearson correlation was applied to investigate the association between gene expression and age. In the current study, age was treated as a categorical variable, and the overlapping results of limma moderated t-tests and Spearman correlation were obtained. The primary aim of this study was to explore oocyte age-related genes, which were validated in a second dataset, and to investigate the potential biological functions affected by these age-related genes using functional enrichment analysis. We believe our results provide more reliable insight into understanding the molecular mechanisms underlying the age-related decline in oocyte quality, which may facilitate the development of targeted treatments designed to improve oocyte quality.
2. Materials and Methods
2.1. Data Download and Processing
Gene expression profiles of oocytes (Accession No. GSE95477 [12], GSE158802 [13], and GSE87201 [17]) were obtained from the NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/, accessed on 9 August 2021) database. Data processing included quantile normalization and log2-transformation. All R packages were run in R software (version 4.1.0).
2.2. Age-Related Analysis
Next, as oocyte quality declines with age, we treated age as a categorical variable (using median age to stratify the oocytes), and examined the correlation between age and gene expression by performing Spearman rank correlation, using the R function ‘cor.test’. For each detected gene, the Spearman correlation value (r) and p-value were calculated. Genes with absolute value r > 0.3 and p-value < 0.05 were considered significantly correlated with age.
Differential gene expression analysis between young and old groups was performed using R package ‘limma’ (version 3.44.3) [18] and moderated t-tests. Genes with absolute fold change (FC) value > 2 and p-value < 0.05 were considered significant.
Then, Venn diagrams were generated using ‘jvenn’ [19] in order to visualize the overlapping results of Spearman rank correlation and t-tests, for IVM-MII oocytes from GSE95477 (referred to as Group I in the current study) and GSE158802 (referred to as Group II in the current study).
Based on the overlapping results presented in the Venn diagrams, the overlapping age-related genes in different IVM-MII oocytes were further validated for GV oocytes from GSE158802 (referred to as Group III in the current study) and IVO-MII oocytes from GSE87201 (referred to as Group IV in the current study), using the same methods and parameters as described above.
2.3. Gene Ontology (GO) Functional Enrichment Analysis
To gain an insight into the biological functions of the age-associated genes, Gene Ontology (GO) functional enrichment analysis was performed using package ‘ClusterProfiler’ (version 3.12.0) [20], with the overlapping age-related genes as the input gene list. The R packages ‘ggplot2’ (version 3.3.3) [21] and ‘GOplot’ (version 1.0.2) [22] were used to plot the results. GO categories with p-value < 0.05 were considered as significantly enriched.
2.4. The Capability of Hub Gene FAM111 Trypsin-like Peptidase A (FAM111A) for Distinguishing Young and Old Oocytes
The FAM111 Trypsin-Like Peptidase A (FAM111A) expression levels between young and old groups were compared using t-tests (for normally distributed expression data) or Wilcoxon signed-rank tests (for non-normally distributed expression data) in R package ‘ggpubr’ (version 0.4.0) [23].
Receiver operating characteristic (ROC) curves and the area under the curve (AUC) were obtained via R package ‘pROC’ (version 1.15.3) [24] and visualized using R package ‘ggplot2’ (version 3.3.3) [21]. The AUC had values generally ranging between 0.5 and 1. Typically, AUC results are categorized as ‘uninformative’ (AUC = 0.5), ‘less accurate’ (0.5 < AUC ≤ 0.7), ‘moderately accurate’ (0.7 < AUC ≤ 0.9), or ‘very accurate’ (0.9 < AUC < 1) [25].
2.5. Gene Set Enrichment Analysis (GSEA) and Gene Set Variation Analysis (GSVA)
To explore the association between FAM111A expression and biological processes/signaling pathways, Gene Set Enrichment Analysis (GSEA) and Gene Set Variation Analysis (GSVA) were carried out, with FAM111A as the input gene list. GSEA is a computational method for determining whether a given gene set exerts statistically significant and concordant differences between two cohorts/phenotypes [26]. GSEA analyses of IVM-MII oocytes from Group I (GSE95477) and Group II (GSE158802) were performed using ‘ClusterProfiler’ (version 3.12.0) [20] with default parameters. Terms with p-value < 0.05 were considered statistically significant. GSVA is a non-parametric and unsupervised algorithm that transforms genes in a sample matrix into pre-defined gene sets, without the need for knowing in advance the experiment design [27]. GSVA analyses of IVM-MII oocytes from Group II (GSE158802) were individually implemented via the R package ‘GSVA’ (version 1.30.0) [27] with default parameters. Terms with p-value < 0.05 and absolute value r > 0.3 were considered significant. All GO and KEGG related gene sets were obtained from the Molecular Signature Database (MsigDB, http://software.broadinstitute.org/gsea/msigdb/, accessed on 28 August 2021).
2.6. Protein–Protein Interaction (PPI) Network Based on FAM111A
Protein–Protein Interaction (PPI) networks were constructed for FAM111A and its interacting proteins using the GeneMANIA database [28]. GeneMANIA is a user-friendly and regularly updated web interface for predicting gene function, analyzing gene lists, and extending the given gene list with genes of similar function using available proteomics and genomics data [29]. It has been widely applied in the study of PPI relationships. The PPI networks were constructed in terms of co-expression, physical interaction, co-localization, genetic interaction, common pathways, shared protein domains, etc.
2.7. Correlation Analysis of FAM111A and Its Interacting Proteins
To explore the potential association between FAM111A and its interacting proteins, gene expression profiles of FAM111A and its interacting proteins were extracted from oocytes from Group I and Group II. R package ‘SVA’ (version 3.32.1) [30] was used to remove batch effects. The gene-gene correlation matrix between FAM111A and its interacting partners was generated using R package ‘corrplot’ (version 0.84) [31] with Spearman correlations. Genes with absolute value r > 0.3 and p-value < 0.05 were considered significantly correlated.
3. Results
A summary of the gene expression profiles included in the current analysis is shown in Figure 1A, and Figure 1B presents an overview of the analysis procedures of the study. Based on the median age of each group, the oocytes were stratified into young and old oocytes. The rationale for using the median age to stratify young and old oocytes is the same as that for female fertility, with a downward trend continuing with age. The major objective of our study is to explore the differences presented by oocytes that are already visible at the molecular level.
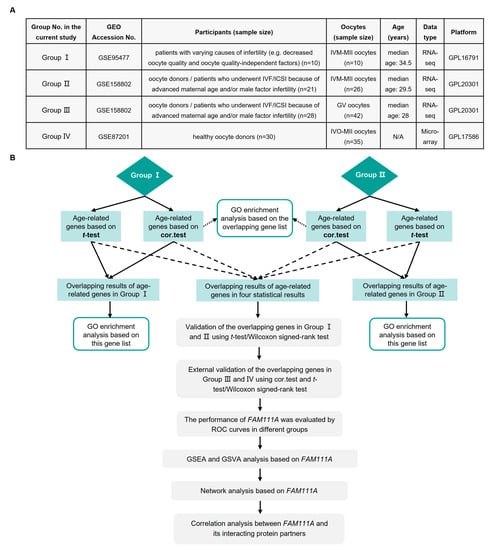
Figure 1.
Summary of the present study. (A) Groups included in the analysis of this study. (B) Flowchart of this study. GEO, Gene Expression Omnibus; IVF/ICSI, in vitro fertilization/intracytoplasmic sperm injection; IVM-MII, in vitro maturation-metaphase II; GV, germinal vesicle; IVO-MII, in vivo ovulation-metaphase II; N/A, not available; cor.test, Spearman rank correlation; GO, Gene Ontology; ROC, receiver operating characteristic; GSEA, Gene Set Enrichment Analysis; GSVA, Gene Set Variation Analysis; FAM111A, FAM111 Trypsin-Like Peptidase A.
3.1. Impact of Age on the Gene Expression of Human Oocytes
Based on Spearman correlation, IVM-MII oocytes from Group I (GSE95477) and Group II (GSE158802) presented 1322 and 933 genes, respectively, showing altered expression with age (Supplementary Tables S1 and S2). Based on t-tests, IVM-MII oocytes from Group I and Group II presented, respectively, 377 and 480 genes, with different expression levels between young and old oocytes (Supplementary Tables S3 and S4).
We used Venn diagrams (Figure 2) to identify age-related genes with overlapping Spearman correlation and t-test results. There were 249 age-related genes with overlapping results in oocytes from Group I (Figure 2B), 397 age-related genes with overlapping results in oocytes from Group II (Figure 2C), and three age-related genes with overlapping results (FAM111A, Isopentenyl-Diphosphate Delta Isomerase 1 (IDI1), and Lymphocyte Antigen 6 Family Member K (LY6K)) in all the analysis results (Figure 2D,E).
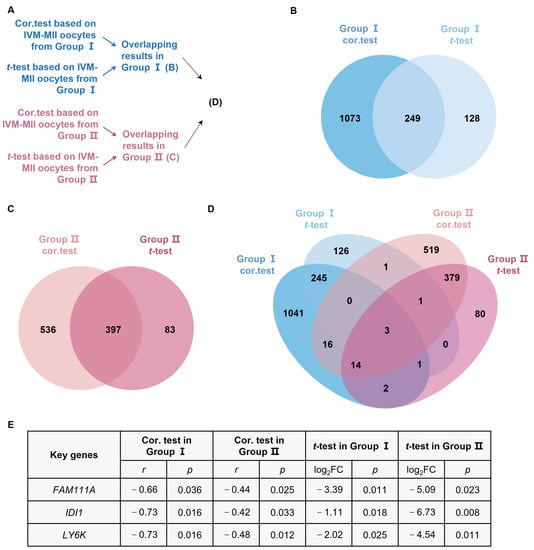
Figure 2.
Analysis of age-related genes. (A) Flowchart of the age-related analysis. (B) Overlapping results of cor.test and t-test for IVM-MII oocytes from Group I. (C) Overlapping results of cor.test and t-test for IVM-MII oocytes from Group II. (D) Overlapping results of age-related genes in Group I and Group II. (E) Based on the cor.test and t-test, FAM111A, IDI1, and LY6K are the statistically significant age-related genes in both Group I and Group II. IDI1, Isopentenyl-Diphosphate Delta Isomerase 1; LY6K, Lymphocyte Antigen 6 Family Member K. FC, fold change.
To test the validity of our findings (i.e., three overlapping age-related genes), we analyzed GV oocytes from Group III and IVO-MII oocytes from Group IV, and confirmed the correlation of FAM111A with age. The correlation coefficients of FAM111A, IDI1, and LY6K were, respectively, −0.32 (p = 0.039), −0.11 (p = 0.470), and 0.08 (p = 0.602), in Group III. The correlation coefficients of FAM111A, IDI1, and LY6K in Group Ⅳ were, respectively, 0.34 (p = 0.046), 0.05 (p = 0.796), and −0.09 (p = 0.605).
3.2. Cellular Metabolism, DNA Replication, and Histone Modifications Are the Biological Processes Enriched by Age-Related Genes in Two Datasets
GO enrichment analysis, based on the age-related genes with overlapping Spearman correlation and t-test results, revealed that cellular metabolism, DNA replication, and histone modifications were the biological processes significantly affected by age in both Group I (Figure 3A) and Group II (Figure 3B). GO enrichment analysis, based on the age-related genes with overlapping Spearman correlation results from Group I and Group II, also revealed that biological processes, such as the oxidation of fatty acids/lipids, fatty acid catabolic process, and histone modifications, were significantly influenced by age (Figure 4).
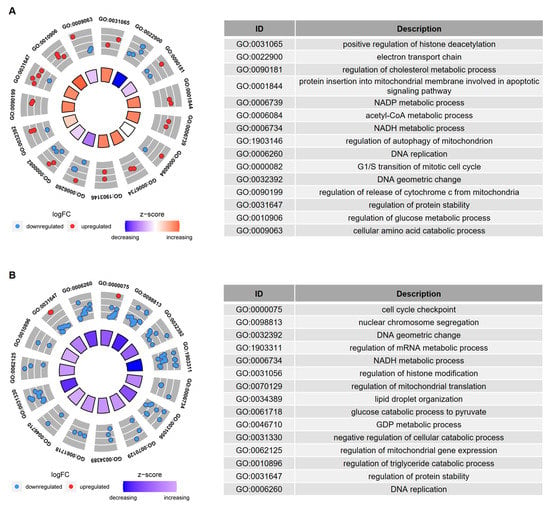
Figure 3.
GO enrichment analysis of the age-related genes in IVM-MII oocytes from (A) Group I and (B) Group II. The age-related genes in this analysis were obtained from the overlapping cor.test and t-test results. The inner circle in the left figure indicates the Z-score value. The outer ring represents the significantly enriched terms, and the most upregulated and downregulated genes. The blue dot indicates the downregulated gene and the red dot indicates the upregulated gene. The right figure is the description of the enriched GO terms.
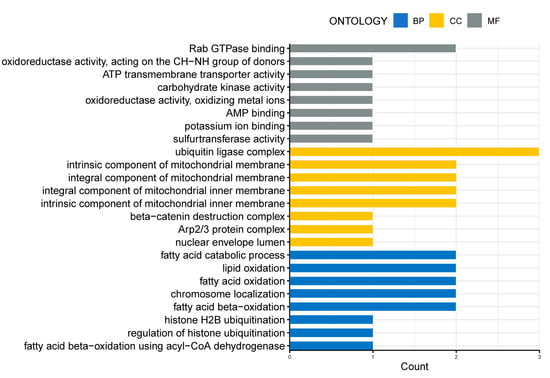
Figure 4.
GO enrichment analysis of the overlapping age-related genes in IVM-MII oocytes from Group I and Group II. The age-related genes in this analysis were obtained from the overlapping results of cor.test in Group I and Group II. The x-axis represents the counts of the genes enriched in GO terms. The y-axis represents the significantly enriched terms in BP, CC, and MF. BP, biological process; CC, cellular component; MF, molecular function.
3.3. FAM111A Is the Hub Age-Related Gene in In Vitro Maturation-Metaphase II (IVM-MII) Oocytes and FAM111A Expression Is also Correlated with Age in Germinal Vesicle (GV) and In Vivo Ovulation-Metaphase II (IVO-MII) Oocytes
Based on t-test and Wilcoxon signed-rank test results, we found that FAM111A expression not only showed significant differences between young and old oocytes from Group I (Figure 5A) and Group II (Figure 5B), but also showed significant differences in GV oocytes from Group III (Figure 5C) and IVO-MII oocytes from Group Ⅳ (Figure 5D). Based on the ROC analysis, FAM111A showed good discrimination between young and old oocytes (Figure 5E–H).
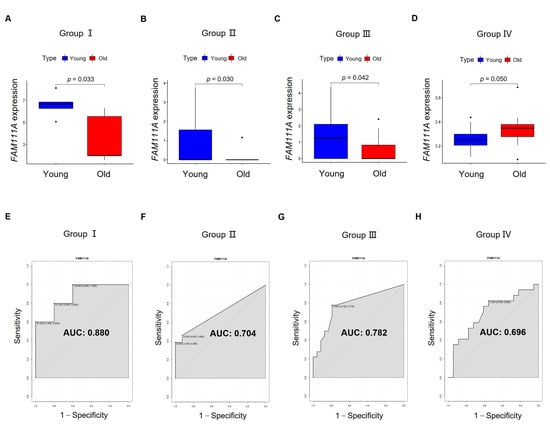
Figure 5.
The capability of FAM111A for distinguishing young and old oocytes. The expression of FAM111A in the oocytes from young and old groups is visualized in (A) Group I and (B) Group II, and further validated in (C) Group III and (D) Group IV. The x-axis represents young and old oocytes. The y-axis represents FAM111A expression. p-values were calculated using a t-test or Wilcoxon signed-rank test. ROC curves are visualized in (E) Group I, (F) Group II, (G) Group III, and (H) Group Ⅳ. The x-axis of the ROC graph represents 1—specificity, and the y-axis represents sensitivity. AUC, area under the curve.
3.4. Chromosome Segregation and Regulation of Cell Cycle Are the Main Pathways Regulated by FAM111A in IVM-MII Oocytes
Based on the overlapping results of the GSEA analysis of IVM-MII oocytes from Group I (Supplementary Table S5) and Group II (Supplementary Table S6), and GSVA analysis of IVM-MII oocytes from Group II (Supplementary Table S7), meiotic chromosome segregation, the regulation of the cell cycle, the positive regulation of the WNT signaling pathway, actin filament organization, and microtubule-associated complex, were terms significantly regulated by FAM111A (Table 1).

Table 1.
The terms overlapping in the results of GSEA and GSVA based on IVM-MII oocytes.
3.5. Predicted FAM111A-Associated Protein Includes Zinc Finger Protein 226 (ZNF226), Which Functions in Transcriptional Regulation
We used the GeneMANIA database to analyze the FAM111A-associated proteins (Figure 6A). These proteins were predicated based on a highly adaptive algorithm and hundreds of datasets, which were collected from ten publicly available databases, such as GEO, BioGRID, Interologous Interaction Database (I2D), and Pathway Commons [29]. The results reveal that FAM111A interacted with Proliferating Cell Nuclear Antigen (PCNA), Mediator Complex Subunit 18 (MED18), Zinc Finger Protein 226 (ZNF226), Nicastrin (NCSTN), and Kringle Containing Transmembrane Protein 2 (KREMEN2), etc. Further, Spearman correlation analysis of the FAM111A-associated proteins, based on the gene expression profiles of the oocytes, revealed a borderline correlation between FAM111A and ZNF226 (correlation coefficient r = 0.32; p = 0.054) (Figure 6B).
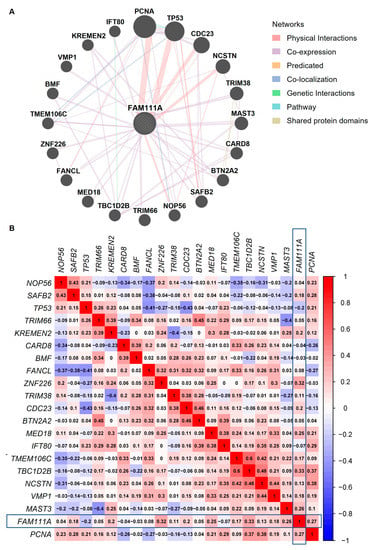
Figure 6.
The PPI network and correlation analysis between FAM111A and its interacting protein partners. (A) The PPI network of FAM111A and its interacting protein partners. (B) Spearman gene-gene correlation matrix between FAM111A and its interacting partners. In the matrix, red and blue represent positive and negative correlations, respectively. The number in each square indicates the Spearman correlation coefficient (r-value). PPI, Protein–Protein Interaction.
4. Discussion
In this study, we explored the influence of age on oocytes at the gene expression level. To increase the robustness of our findings, we explored the age-related genes using two statistical methods; moreover, we attempted to identify overlapping results by combining two independent datasets. We found hundreds of genes with significantly altered expression levels between oocytes from young and old age groups; the biological processes influenced by these genes mainly included cellular metabolism, DNA replication, and histone modifications. Among these age-related genes, FAM111A expression displayed robust correlations with age in the different statistical methods and different datasets. To our knowledge, this is the first report of a potential gene marker for human oocyte aging. We found that FAM111A may be involved in the biological processes associated with chromosome segregation and the regulation of the cell cycle. FAM111A protein could potentially interact with regulators of transcription (i.e., ZNF226) and transcription factors (i.e., PCNA).
We identified 249 overlapping age-related genes in oocytes from Group I, and 397 overlapping age-related genes in oocytes from Group II. Based on GO enrichment analysis, we found these two gene lists were both enriched in biological processes associated with cellular metabolism, DNA replication, and histone modifications. The overlapping genes, from Spearman rank correlation, in oocytes from Group I and Group II were also enriched in the terms of the oxidation of fatty acids/lipids, fatty acid catabolic process, and histone modifications. Similarly, Duncan et al. found that age-associated gene signatures in mouse oocytes were involved in the alteration of protein metabolism [10]. Another study, based on liquid and gas chromatography coupled to mass spectrometry, also found that total free fatty acids were decreased in old equine oocytes compared with young oocytes. Furthermore, through the quantification of aerobic and anaerobic metabolism, these researchers found that the metabolic activity of equine oocytes was impaired by maternal aging [32]. Fatty acids are vital substrates for early reproductive events (e.g., oocyte maturation [33,34] and embryo implantation [35]). Upon demand, the metabolism of lipids provides a good source of energy during oocyte maturation. For example, during pig oocyte maturation, fatty acids in lipid droplets (LDs) provide adenosine triphosphate (ATP) via β-oxidation, serving as an energy source for oocytes [36]. The lipid modulators have been investigated to improve the developmental competence of oocytes and embryos in animal models [37]. Modifications to LD morphology and lipid metabolism during the period of oocyte maturation have been revealed to possibly interfere with monospermic fertilization and embryo development in the pig model [38,39]. It has been revealed that the degradation of LDs tends to be via the lipolysis of triacylglycerols to fatty acyl-CoA by lipases at the surface of the LDs, which is often in preparation for mitochondrial metabolism [40].
Corroborating our results, the alteration of histone methylation [41,42] and histone acetylation [43,44,45] has been reported in old mammalian oocytes. Altered histone modifications usually correlated with changes in chromatin configuration and gene transcription activity [46], which may affect oocyte fertilization and embryo development [44]. For example, decreased histone 4 lysine 20 tri-methylation (H4K20me3) results in chromosomal segregation defects in cell culture [47]. The inhibitor of histone deacetylase during IVM contributes to the reduced developmental potential of bovine oocytes [48]. Notably, epigenetic changes have been regarded as a hallmark of aging, besides telomere shortening [49]. However, studies of age-related epigenetic changes in oocytes are still finite, and, therefore, further studies on this topic are highly awaited.
We identified FAM111A as the most robust gene that presented altered gene expression during aging. The overlapping results of GSEA and GSVA analysis based on FAM111A revealed that chromosome segregation and regulation of the cell cycle were the main pathways regulated by FAM111A in IVM-MII oocytes from both datasets. Other studies on human oocytes [13,50] have come to similar conclusions, that transcripts associated with chromosome segregation and cell cycle regulation are altered during oocyte aging. Fine et al. revealed that FAM111A expression is cell-cycle-dependent in cell-cycle-synchronized T98G cells [51]. FAM111A is a chymotrypsin-like serine protease, and displays autocleavage activity in vivo [52]. It has been revealed that FAM111A is important for DNA replication [52,53] and plays a role in viral restriction [51,54]. Furthermore, it is hypothesized that FAM111A might engage in DNA binding [52], as it localizes to replication forks [55]. Nevertheless, until now, the functions of FAM111A remain largely unclarified and require further investigation.
We also found that the WNT signaling pathway was positively regulated by FAM111A in IVM-MII oocytes, and that FAM111A expression was decreased in aging IVM-MII oocytes. Similarly, a recent study also found that the WNT signaling was downregulated in low-quality rescue IVM (rIVM) oocytes, and revealed that down-regulation of the GATA-1/CREB1/WNT signaling axis might result in the poor maturation performance of rIVM oocytes [56]. This might indicate the existence of a potential link between FAM111A expression and WNT signaling, which needs to be further investigated. The WNT signaling pathway is a highly conserved pathway. It plays a major role in cell and tissue maintenance, polarity, and differentiation [57], as well as in maintaining female germ cell survival [58]; furthermore, it is frequently dysregulated in human cancers [59]. The canonical WNT pathway is initiated by WNT proteins [60], which have been revealed as being responsible for regulating the cell cycle and deciding cell fate [61].
Furthermore, based on our PPI network results, we found that FAM111A protein may have interactions with ZNF226 and PCNA. The Spearman correlation analysis based on the gene expression profiles from Group I and Group II also revealed a borderline positive correlation between FAM111A expression and ZNF226 expression. PCNA is a transcription factor that promotes DNA polymerase delta binding to DNA [62], and plays an essential role in DNA replication, DNA repair [63], and cell cycle regulation [62]. It has also been reported that ZNF226 plays a vital role in transcriptional regulation [64]. Our finding reveals a potential link between transcriptional regulation and oocyte aging.
Though the same statistical methods were used in different datasets, there was still variation in age-related genes between the different datasets, with relatively few overlapping results. This could be due to differences in sequencing methods and coverage of the sequencing platforms, as well as differing age ranges and sources of the study participants. In addition, for the in vitro maturation of oocytes, no unified IVM protocols are established across different in vitro fertilization (IVF) centers [65]. However, differences in methods of follicular priming, time of oocyte retrieval, choices of IVM medium and supplementation, and duration of in vitro culture, may also contribute towards differential gene expression in IVM oocytes. Another limitation is that, in our differential gene expression analysis, p-values (and not adjusted p-values) less than 0.05 were considered statistically significant. This lack of compensation may increase the frequency of false-positive results. However, we extracted the overlapping results with different analytical methods and datasets, which to some extent could reduce variability and provide relatively reliable results.
5. Conclusions
In summary, based on two independent datasets of IVM-MII oocytes, we identified hundreds of genes that were influenced by age, and that might be involved in the biological processes associated with cellular metabolism, DNA replication, and histone modifications. Among these age-related genes, FAM111A may act as a biomarker for oocyte aging, and deserves further investigation. Our findings could advance current knowledge of the molecular mechanisms involved in oocyte aging, and potentially facilitate the development of targeted treatments designed to improve oocyte quality.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10020257/s1. Table S1. Results of Spearman correlation on IVM-MII oocytes from Group I. IVM-MII, in vitro maturation-metaphase II; Table S2. Results of Spearman correlation on IVM-MII oocytes from Group II; Table S3. Results of t-test on IVM-MII oocytes from Group I; Table S4. Results of t-test on IVM-MII oocytes from Group II; Table S5. Results of GSEA analysis on IVM-MII oocytes from Group I. GSEA, Gene Set Enrichment Analysis; Table S6. Results of GSEA analysis on IVM-MII oocytes from Group II; Table S7. Results of GSVA analysis on IVM-MII oocytes from Group II. GSVA, Gene Set Variation Analysis.
Author Contributions
Conceptualization, U.J., V.v.S., T.K., M.K., S.M. (Sarah Meister); data analysis and visualization, H.Y., C.P., J.v.D., S.E.; Writing—original draft preparation, H.Y., C.P., J.v.D., S.E.; Writing—review and editing, M.K., C.K., M.R., C.H., S.G.F., G.W.; supervision, U.J., V.v.S., S.M. (Sven Mahner). All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Ethical review and approval were waived for this study, because all of the data used in the current study were anonymized data from GEO database.
Informed Consent Statement
Patient consent was waived because all of the data used in the current study were anonymized data from GEO database.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: GEO: https://www.ncbi.nlm.nih.gov/geo/ (accessed on 15 November 2021).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dohle, G.R.; Colpi, G.M.; Hargreave, T.B.; Papp, G.K.; Jungwirth, A.; Weidner, W. EAU guidelines on male infertility. Eur. Urol. 2005, 48, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Turan, V.; Oktay, K. BRCA-related ATM-mediated DNA double-strand break repair and ovarian aging. Hum. Reprod. Update 2020, 26, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Baird, D.T.; Collins, J.; Egozcue, J.; Evers, L.H.; Gianaroli, L.; Leridon, H.; Sunde, A.; Templeton, A.; Van Steirteghem, A.; Cohen, J.; et al. Fertility and ageing ESHRE Capri Workshop Group. Hum. Reprod. Update 2005, 11, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Marangos, P.; Stevense, M.; Niaka, K.; Lagoudaki, M.; Nabti, I.; Jessberger, R.; Carroll, J. DNA damage-induced metaphase i arrest is mediated by the spindle assembly checkpoint and maternal age. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Keefe, D.L.; Niven-Fairchild, T.; Powell, S.; Buradagunta, S. Mitochondrial deoxyribonucleic acid deletions in oocytes and reproductive aging in women. Fertil. Steril. 1995, 64, 577–583. [Google Scholar] [CrossRef]
- Selesniemi, K.; Lee, H.J.; Muhlhauser, A.; Tilly, J.L. Prevention of maternal aging-associated oocyte aneuploidy and meiotic spindle defects in mice by dietary and genetic strategies. Proc. Natl. Acad. Sci. USA 2011, 108, 12319–12324. [Google Scholar] [CrossRef]
- Keefe, D.L.; Liu, L. Telomeres and reproductive aging. Proc. Reprod. Fertil. Dev. 2009, 21, 10–14. [Google Scholar] [CrossRef]
- Valeri, C.; Pappalardo, S.; De Felici, M.; Manna, C. Correlation of oocyte morphometry parameters with woman’s age. Proc. J. Assist. Reprod. Genet. 2011, 28, 545–552. [Google Scholar] [CrossRef][Green Version]
- Omidi, M.; Khalili, M.A.; Nahangi, H.; Ashourzadeh, S.; Rahimipour, M. Does women’s age influence zona pellucida bi-refringence of metaphase ΙΙ oocytes in in-vitro maturation program? Int. J. Reprod. Biomed. 2013, 11, 823–828. [Google Scholar]
- Duncan, F.E.; Jasti, S.; Paulson, A.; Kelsh, J.M.; Fegley, B.; Gerton, J.L. Age-associated dysregulation of protein metabolism in the mammalian oocyte. Aging Cell 2017, 16, 1381–1393. [Google Scholar] [CrossRef]
- Smits, M.A.J.; Wong, K.M.; Mantikou, E.; Korver, C.M.; Jongejan, A.; Breit, T.M.; Goddijn, M.; Mastenbroek, S.; Repping, S. Age-related gene expression profiles of immature human oocytes. Mol. Hum. Reprod. 2018, 24, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.M.; Silva, E.; Chitwood, J.L.; Schoolcraft, W.B.; Krisher, R.L.; Ross, P.J. Differing molecular response of young and advanced maternal age human oocytes to IVM. Hum. Reprod. 2017, 32, 2199–2208. [Google Scholar] [CrossRef] [PubMed]
- Llonch, S.; Barragán, M.; Nieto, P.; Mallol, A.; Elosua-Bayes, M.; Lorden, P.; Ruiz, S.; Zambelli, F.; Heyn, H.; Vassena, R.; et al. Single human oocyte transcriptome analysis reveals distinct maturation stage-dependent pathways impacted by age. Aging Cell 2021, 20, e13360. [Google Scholar] [CrossRef] [PubMed]
- Grøndahl, M.L.; Yding Andersen, C.; Bogstad, J.; Nielsen, F.C.; Meinertz, H.; Borup, R. Gene expression profiles of single human mature oocytes in relation to age. Hum. Reprod. 2010, 25, 957–968. [Google Scholar] [CrossRef]
- Zhang, J.J.; Liu, X.; Chen, L.; Zhang, S.; Zhang, X.; Hao, C.; Miao, Y.L. Advanced maternal age alters expression of ma-ternal effect genes that are essential for human oocyte quality. Aging 2020, 12, 3950–3961. [Google Scholar] [CrossRef]
- Yuan, L.; Yin, P.; Yan, H.; Zhong, X.; Ren, C.; Li, K.; Chin Heng, B.; Zhang, W.; Tong, G. Single-cell transcriptome analysis of human oocyte ageing. J. Cell. Mol. Med. 2021, 25, 6289–6303. [Google Scholar] [CrossRef]
- Barragán, M.; Pons, J.; Ferrer-Vaquer, A.; Cornet-Bartolomé, D.; Schweitzer, A.; Hubbard, J.; Auer, H.; Rodolosse, A.; Vassena, R. The transcriptome of human oocytes is related to age and ovarian reserve. Mol. Hum. Reprod. 2017, 23, 535–548. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis (2nd ed.). Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A. ggpubr: “ggplot2” Based Publication Ready Plots. R Packag. Version 0.4.0 2020. Available online: https://CRAN.R-project.org/package=ggpubr (accessed on 9 August 2021).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Greiner, M.; Pfeiffer, D.; Smith, R.D. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev. Vet. Med. 2000, 45, 23–41. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide ex-pression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Wei, T.; Simko, V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J. R package “corrplot”: Visualization of a Correlation Matrix. Statistician 2017, 56, 316–324. [Google Scholar]
- Catandi, G.D.; Obeidat, Y.M.; Broeckling, C.D.; Chen, T.W.; Chicco, A.J.; Carnevale, E.M. Equine maternal aging affects oocyte lipid content, metabolic function and developmental potential. Reproduction 2021, 161, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Sturmey, R.G.; Reis, A.; Leese, H.J.; McEvoy, T.G. Role of fatty acids in energy provision during oocyte maturation and early embryo development. Reprod. Domest. Anim. 2009, 44, 50–58. [Google Scholar] [CrossRef] [PubMed]
- McKeegan, P.J.; Sturmey, R.G. The role of fatty acids in oocyte and early embryo development. Reprod. Fertil. Dev. 2012, 24, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Norwitz, E.R.; Schust, D.J.; Fisher, S.J. Implantation and the Survival of Early Pregnancy. N. Engl. J. Med. 2001, 345, 1400–1408. [Google Scholar] [CrossRef]
- Sturmey, R.G.; Leese, H.J. Energy metabolism in pig oocytes and early embryos. Reproduction 2003, 126, 197–204. [Google Scholar] [CrossRef]
- Prates, E.G.; Nunes, J.T.; Pereira, R.M. A role of lipid metabolism during cumulus-oocyte complex maturation: Impact of lipid modulators to improve embryo production. Mediat. Inflamm. 2014, 2014, 692067. [Google Scholar] [CrossRef] [PubMed]
- Prates, E.G.; Marques, C.C.; Baptista, M.C.; Vasques, M.I.; Carolino, N.; Horta, A.E.M.; Charneca, R.; Nunes, J.T.; Pereira, R.M. Fat area and lipid droplet morphology of porcine oocytes during in vitro maturation with trans-10, cis-12 conjugated linoleic acid and forskolin. Animal 2013, 7, 602–609. [Google Scholar] [CrossRef]
- Prates, E.G.; Alves, S.P.; Marques, C.C.; Baptista, M.C.; Horta, A.E.M.; Bessa, R.J.B.; Pereira, R.M. Fatty acid composition of porcine cumulus oocyte complexes (COC) during maturation: Effect of the lipid modulators trans-10, cis-12 conjugated linoleic acid (t10, c12 CLA) and forskolin. Vitr. Cell. Dev. Biol.-Anim. 2013, 49, 335–345. [Google Scholar] [CrossRef]
- Bradley, J.; Swann, K. Mitochondria and lipid metabolism in mammalian oocytes and early embryos. Int. J. Dev. Biol. 2019, 63, 93–103. [Google Scholar] [CrossRef]
- Manosalva, I.; González, A. Aging changes the chromatin configuration and histone methylation of mouse oocytes at germinal vesicle stage. Theriogenology 2010, 74, 1539–1547. [Google Scholar] [CrossRef]
- Shao, G.B.; Wang, J.; Zhang, L.P.; Wu, C.Y.; Jin, J.; Sang, J.R.; Lu, H.Y.; Gong, A.H.; Du, F.Y.; Peng, W.X. Aging alters histone H3 lysine 4 methylation in mouse germinal vesicle stage oocytes. Reprod. Fertil. Dev. 2015, 27, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.C.; Yan, L.Y.; Lei, Z.L.; Miao, Y.L.; Shi, L.H.; Yang, J.W.; Wang, Q.; Ouyang, Y.C.; Sun, Q.Y.; Chen, D.Y. Changes in histone acetylation during postovulatory aging of mouse oocyte. Biol. Reprod. 2007, 77, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Suo, L.; Meng, Q.G.; Pei, Y.; Yan, C.L.; Fu, X.W.; Bunch, T.D.; Zhu, S.E. Changes in acetylation on lysine 12 of histone H4 (acH4K12) of murine oocytes during maternal aging may affect fertilization and subsequent embryo development. Fertil. Steril. 2010, 93, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Manosalva, I.; González, A. Aging alters histone H4 acetylation and CDC2A in mouse germinal vesicle stage oocytes. Biol. Reprod. 2009, 81, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ma, Z.; Peng, L.; Kuhn, C.; Rahmeh, M.; Mahner, S.; Jeschke, U.; von Schönfeldt, V. Comparison of histone h3k4me3 between ivf and icsi technologies and between boy and girl offspring. Int. J. Mol. Sci. 2021, 22, 8574. [Google Scholar] [CrossRef]
- Gonzalo, S.; Blasco, M.A. Role of Rb family in the epigenetic definition of chromatin. Cell Cycle 2005, 4, 752–755. [Google Scholar] [CrossRef]
- Pontelo, T.P.; Franco, M.M.; Kawamoto, T.S.; Caixeta, F.M.C.; de Oliveira Leme, L.; Kussano, N.R.; Zangeronimo, M.G.; Dode, M.A.N. Histone deacetylase inhibitor during in vitro maturation decreases developmental capacity of bovine oo-cytes. PLoS ONE 2021, 16, e0247518. [Google Scholar] [CrossRef]
- Cypris, O.; Eipel, M.; Franzen, J.; Rösseler, C.; Tharmapalan, V.; Kuo, C.C.; Vieri, M.; Nikolić, M.; Kirschner, M.; Brümmendorf, T.H.; et al. PRDM8 reveals aberrant DNA methylation in aging syndromes and is relevant for hematopoietic and neuronal differentiation. Clin. Epigenetics 2020, 12, 125. [Google Scholar] [CrossRef]
- Steuerwald, N.M.; Bermúdez, M.G.; Wells, D.; Munné, S.; Cohen, J. Maternal age-related differential global expression profiles observed in human oocytes. Reprod. Biomed. Online 2007, 14, 700–708. [Google Scholar] [CrossRef]
- Fine, D.A.; Rozenblatt-Rosen, O.; Padi, M.; Korkhin, A.; James, R.L.; Adelmant, G.; Yoon, R.; Guo, L.; Berrios, C.; Zhang, Y.; et al. Identification of FAM111A as an SV40 Host Range Restriction and Adenovirus Helper Factor. PLoS Pathog. 2012, 8, e1002949. [Google Scholar] [CrossRef]
- Kojima, Y.; Machida, Y.; Palani, S.; Caulfield, T.R.; Radisky, E.S.; Kaufmann, S.H.; Machida, Y.J. FAM111A protects replication forks from protein obstacles via its trypsin-like domain. Nat. Commun. 2020, 11, 1318. [Google Scholar] [CrossRef] [PubMed]
- Rios-Szwed, D.O.; Garcia-Wilson, E.; Sanchez-Pulido, L.; Alvarez, V.; Jiang, H.; Bandau, S.; Lamond, A.; Ponting, C.P.; Alabert, C. FAM111A regulates replication origin activation and cell fitness. BioRxiv 2020, 1–26. [Google Scholar] [CrossRef]
- Nie, M.; Oravcová, M.; Jami-Alahmadi, Y.; Wohlschlegel, J.A.; Lazzerini-Denchi, E.; Boddy, M.N. FAM111A induces nu-clear dysfunction in disease and viral restriction. EMBO Rep. 2021, 22, e50803. [Google Scholar] [CrossRef] [PubMed]
- Alabert, C.; Bukowski-Wills, J.C.; Lee, S.B.; Kustatscher, G.; Nakamura, K.; De Lima Alves, F.; Menard, P.; Mejlvang, J.; Rappsilber, J.; Groth, A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014, 16, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.W.T.; Ng, J.K.W.; Liao, J.; Luk, A.C.; Suen, A.H.C.; Chan, T.T.H.; Cheung, M.Y.; Chu, H.T.; Tang, N.L.S.; Zhao, M.P.; et al. Single-cell RNA sequencing identifies molecular targets associated with poor in vitro maturation performance of oocytes collected from ovarian stimulation. Hum. Reprod. 2021, 36, 1907–1921. [Google Scholar] [CrossRef] [PubMed]
- Pamarthy, S.; Kulshrestha, A.; Katara, G.K.; Beaman, K.D. The curious case of vacuolar ATPase: Regulation of signaling pathways. Mol. Cancer 2018, 17, 41. [Google Scholar] [CrossRef]
- Liu, C.F.; Parker, K.; Humphrey, Y. WNT4/β-catenin pathway maintains female germ cell survival by inhibiting activin βB in the mouse fetal ovary. PLoS ONE 2010, 5, e10382. [Google Scholar] [CrossRef]
- Polakis, P. Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol. 2012, 4, 9. [Google Scholar] [CrossRef]
- Miller, J.R. The Wnts. Genome Biol. 2002, 3, reviews3001.1. [Google Scholar] [CrossRef]
- Cadigan, K.M.; Nusse, R. Wnt signaling: A common theme in animal development. Genes Dev. 1997, 11, 3286–3305. [Google Scholar] [CrossRef]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA, the Maestro of the Replication Fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Cazzalini, O.; Sommatis, S.; Tillhon, M.; Dutto, I.; Bachi, A.; Rapp, A.; Nardo, T.; Scovassi, A.I.; Necchi, D.; Cardoso, M.C.; et al. CBP and p300 acetylate PCNA to link its degradation with nucleotide excision repair synthesis. Nucleic Acids Res. 2014, 42, 8433–8448. [Google Scholar] [CrossRef] [PubMed]
- Persikov, A.V.; Rowland, E.F.; Oakes, B.L.; Singh, M.; Noyes, M.B. Deep sequencing of large library selections allows computational discovery of diverse sets of zinc fingers that bind common targets. Nucleic Acids Res. 2014, 42, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Kolben, T.; Meister, S.; Paul, C.; van Dorp, J.; Eren, S.; Kuhn, C.; Rahmeh, M.; Mahner, S.; Jeschke, U.; et al. Factors Influencing the In Vitro Maturation (IVM) of Human Oocyte. Biomedicines. 2021, 9, 1904. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).