Abstract
In high-dimensional gene expression data analysis, the accuracy and reliability of cancer classification and selection of important genes play a very crucial role. To identify these important genes and predict future outcomes (tumor vs. non-tumor), various methods have been proposed in the literature. But only few of them take into account correlation patterns and grouping effects among the genes. In this article, we propose a rank-based modification of the popular penalized logistic regression procedure based on a combination of and penalties capable of handling possible correlation among genes in different groups. While the penalty maintains sparsity, the penalty induces smoothness based on the information from the Laplacian matrix, which represents the correlation pattern among genes. We combined logistic regression with the BH-FDR (Benjamini and Hochberg false discovery rate) screening procedure and a newly developed rank-based selection method to come up with an optimal model retaining the important genes. Through simulation studies and real-world application to high-dimensional colon cancer gene expression data, we demonstrated that the proposed rank-based method outperforms such currently popular methods as lasso, adaptive lasso and elastic net when applied both to gene selection and classification.
Keywords:
gene-expression data; ℓ2 ridge; ℓ1 lasso; adapative lasso; elastic net; BH-FDR; Laplacian matrix MSC:
62F03; 62F07; 62P10
1. Introduction
Microarrays are an advanced and widely used technology in genomic research. Tens of thousands of genes can be analyzed simultaneously with this approach [1]. Identifying the genes related to cancer and building high-performance prediction models of maximal accuracy (tumor vs. non-tumor) based on gene expression levels are among central problems in genomic research [2,3,4]. Typically, in high-dimensional gene expression data analysis, the number of genes is significantly larger than the sample size, i.e., . Hence, it is particularly challenging to identify those genes that are relevant to cancer disease and put forth prediction models. The main problem associated with high-dimensional data () is that of overfitting or overparametrization which leads to poor generalizability from training to test data.
Therefore, various researchers apply different types of regularization methods to overcome this “curse of dimensionality” in regression and other statistical and machine learning frameworks. These regularization approaches include, for example, the -penalty or lasso [5], which performs continuous shrinkage and feature selection simultaneously; smoothly clipped -penalty or SCAD [6], which is symmetric, non-concave and has singularities at the origin to produce sparse solutions; fussed lasso [7], which imposes the -penalty on the absolute difference of regression coefficients in order to enforce some smoothness of coefficients; or the adaptive lasso [8], etc. Unfortunately, -regularization sometimes perform inconsistently when used for variable selection [8]. In some situations, it introduces a major bias in estimated parameters in the logistic regression [9,10]. In contrast, the elastic net regularization procedure [11] as a combination of - and -penalties can successfully handle the highly correlated variables which are grouped together. Among the procedures mentioned above, elastic net and fussed lasso penalized methods are appropriate for gene expression data analysis. Unfortunately, when some prior knowledge needs to be utilized, e.g., when studying complex diseases such as cancer, those methods are not appropriate [4]. To account for a regulatory relationship between the genes and a priori knowledge about these genes, network-constrained regularization [4] is known to perform very well by incorporating a Laplacian matrix into the -penalty from the enet procedure. This Laplacian matrix represents a graph-structure of genes which are linked with each other. To select significant genes in high-dimensional gene expression data for classification, the graph-constrained regularization method is extended to logistic regression model [12].
Using penalized logistic regression methods [12,13] and graph-constrained procedures [4,12], we would build rank-based logistic regression method with variable screening procedure to improve the power of detecting most promising variables as well as classification capability.
The rest of this article is organized as follows. In Section 2, we describe variable screening procedure with adjusted p-values and regularization procedure for grouped and correlated predictors and present the computational algorithm. Further, we state the ranking criteria of four models and summarize the result of ranking procedure. In Section 3, we compare the proposed procedure with existing cutting edge regularization methods on simulation studies. Next, we apply four penalized logistic regression methods to the high dimensional gene expression data of colon cancer carcinoma to evaluation and comparison of the performance. Finally, we present a brief discussion of results and future research direction.
2. Materials and Methods
2.1. Adjusted p-Values: Benjamini and Hochberg False Discovery Rate (BH-FDR)
Multiple hypohtesis testing methods have been playing an important role in selecting most promising features while controlling type I error in high-dimensional settings. One of the most popular methods is BH-FDR [14,15] which is concerned with the expected proportion of incorrect number of rejections among a total number of rejections. The formula is mathematically expressed as , where V is the number of false positives and R is the total number of rejections. In this paper, the FDR method is used both for the purpose of prelimary variable screening both in the simulation studies and real data analysis to be presented later. The procedure of the method is as follows:
- (1)
- Let be the p-values of m hypothesis tests and sort them with the increasing oder: .
- (2)
- Let for a given threshold q. If , then reject the null hypotheses associated with . Otherwise, no hypotheses are rejected.
2.2. Regularized Logistic Regression
In the following, we present the regularized logistic regression model used in this paper (cf. [12]). Since this model is an integral part of our computational algorithm to be outlined in the section to follow, presenting the formula with all appropriate notations is necessary for our purposes.
Let the matrix
denote the design matrix, where n is the sample size and m is the total number of predictor variables. Without loss of generality, we assume the data are standardized with respect to each variable. This step is also performed by the pclogit R-package used in the present paper. Define the parameter vector comprised of an intercept and m “slopes”, . The objective function then is written as
with the log-likelihood function
and resulting probabilities
Here, is the penalty function and the response variable takes the value 1 for cases and 0 for controls. The i-th individual is deemed case or control based on the probability . Following [4], statistical dependence among the m explanatory variables can be modeled by a graph, which, in turn, can be described by its m-dimensional Laplacian matrix with the entries
Here, is the degree of a vertex v, i.e., the number of edges through this vertex. If there is no link in v (i.e., v is isolated), then . The martix L is symmetric, positive semi-definite and has 0 as the smallest eigenvalue and 2 as the largest eigenvalue. In the following, we will write to refer to adjecent vertices. The penalty term in Equation (1) can is defined as
Here, and are tuning parameters meant to control the sparsity and smoothness, is the -norm and denotes the summation over all adjacent vertex pairs. When , the penalty reduces to that of lasso [5], and if is replaced by the -identity matrix , the penalty corresponds to that of an elastic net [11]. If and , we arrive at ridge regression. In Equation (2), the penalty consists of - and -components. The -penalty is a degree-scaled difference of coefficients between linked predictors. According to [4], the predictor variables with more connections have larger coefficients. That is why small change of expression in the variables can lead to large change in response. Thus, this imposes sparsity and smoothness as well as correlation and grouping effects among variables. In case-control DNA methylation data analysis, ring networks and fully connected networks (cf. Figure 1) are typically used to describe correlation pattern of CpG sites within genes [12]. The Laplacian matrix is sparse and tri-diagonal (except for two corner elements) for ring networks and has all non-zero elements for fully connected networks. Those variables with more links produce strong grouping effects and are more likely to be selected in both networks [12].
Figure 1.
The ring network (left) and F.con network (right) are shown for the case there are two genes consisting of 6 and 9 CpG sites, respectively.
2.3. Computational Algorithm
Li & Li (2010) [16] developed an algorithm for graph-constrained regularization motivated by a coordinate descent algorithm from [17] for solving the unconstrained minimization problem for the objective in Equation (1). The algorithm implementation from the pclogit R-package [12,13] replaced the identity matrix by Laplacian matrix in the elastic net algorithm from the glmnet R-package [18]. According to Equation (1), the objective function is
where
with and for some .
Following [18], we perform a second-order Taylor expansion of around the current estimate to approximate the objective in Equation (1) via
where
Now, if all other estimates for all are fixed, can be computed. To update the estimate from , we have to set the gradient of equal zero (strictly speaking, zero has to be included in the subgradient of ) and then solve for to obtain
where
and denotes the “soft threshholding” operator given by
If the u-th predictor has no links to other predictors, then in Equation (4) becomes zero, while Equation (3) takes the form
Thus, the regularization reduces to that of the elastic net (enet) procedure. In general, when the linkage is nontrivial, the term is added to the elastic net to get the desired grouping effect.
2.4. Adaptive Link-Constrained Regularization
When there is a link between two predictors but their regression coefficients have different signs, the coefficients cannot be expected to be smooth [16]—even locally. To resolve this problem, we first need to estimate the sign of the coefficients and then refit the model with estimated signs. When the number of predictor variables is smaller than that of sample points, ordinary least squares are performed, while ridge estimates are computed, otherwise. We have to modify the Laplacian matrix in the penalty function:
and then update the -function in Equation (4) via
2.5. Accuracy, Sensitivity, Specificity and Area under the Receiver Operating Curve (AUROC)
We evaluated four metrics of binary classification for each of lasso, adaptive lasso, elastic net and the proposed rank based logistic regression methods to compare the performance. These metrics are accuracy, sensitivity, specificity and AUROC.

Table 1.
Confusion table: a is the number of true positives, b the number of false positives, c the number of false negatives and d the number of true negatives.
The last metric AUROC is related to the probability that the classifier under consideration will rank a randomly selected positive case higher than a randomly selected negative case [19]. The values of all these fours metrics—accuracy, sensitivity, specificity and AUROC—range from 0 to 1. The value of 1 represents a perfect model whereas the value of 0.5 corresponds to “coin tossing”. The class prediction for each individual in binary classification is made based on a continuous random variable z. Given a threshold k as a tuning parameter, an individual is classified as “positive” if and “negative”, otherwise. The random variable z follows a probability density if the individual belongs to “positives” and , otherwise. So, the true positive and true negative rates are given by
Now, the AUROC statistic can be expressed as
where and are the values of positive or negative instances, respectively.
2.6. Ranking and Best Model Selection
The penalty function in Equation (3) has two tuning parameters, namely, and . The "limiting” cases and correspond to ridge and lasso regression, respectively. For a fixed value of , the model selects more variables for smaller ’s and fewer variables for larger ’s. Theoretically, the result continuosly depends on and should not significantly change under small perturbations of the latter [12,13]. Empirically, however, we discovered that the results produced by pclogit significantly vary with . In pclogit, the Laplacian matrix determines the group effects of predictors and is calculated from adjacency matrix via
where is the degree matrix and is the adjacency matrix. The degree-scaled difference of predictors in Equation (3) is computed from the normalized Laplacian matrix
We computed the adjacency matrix by using the information from the correlation matrix obtaining
Here, is a specific cut-off value for correlation. So, is another tuning parameter in our model which needs to be optimally selected. In summary, to find an optimal combination of parameters and , we make the combination of tuning parameter and , where the total number of combinations is given by
with K and L being the number of and values, respectively. We compared the performance for each of different combinations with T resamplings. The (negative) measure of performance for each combination is the misclassification or error rate. The pair producing the smallest misclassification rate is declared optimal and used in the next step. The sparse coefficient matrix with dimensions ( number of ’s) is used in pclogit (cf. [12,13]). By default, . We extracted all predictors with non-zero coefficients for each of values. Then we built 100 logistic regression models. Given estimated parameter values , we have the estimated class probability for a predictor vector at each of values.
Using the “naïve” Bayesian approach, we infer if and , otherwise. The values of accuracy, sensitivity, specificity and AUROC statistics are computed for each of 100 models and ranked in an increasing order by their values. Note that AUROC method does not use a fixed cut-off value, e.g., , but rather describes the overall performance with all possible cut-off values in the decision rule. Let , , , comprise the ranking matrix . The first row, i.e., , displays the ranking of models with respect to their accuracy. Similarly, ranks the models with respect to their sensitivity, , in terms of specificity and by AUROC. Suppose, . Then in the 1st row (i.e., in terms of accuracy), model 5 outperforms model 8. We calculate the column means () of the matrix. The column with the highest overall mean value of accuracy, sensitivity, specificity and AUROC will be chosen as the resulting optimal model. Note that there is a one-to-one correspondence between columns and the 100 competing models. In (the unlikely) case of two or more columns producing the same mean, the column with a smaller index j is selected since the model represented by such column is more parsimonious. Formally, suppose p and q, , are two column indices in the matrix. If , the q-th column will be selected and the associated model becomes our proposed rank-based penalized logistic regression model.
3. Results
3.1. Analysis of Simulated Data
We conducted extensive simulation studies to compare the performance in terms of accuracy, sensitivity, specificity and AUROC as well as the power of detecting true important variables by the proposed method with the performance of such three prominent regularized logistic regression methods as lasso, adaptive lasso and elastic net. We decided to focus on these (meanwhile) classical methods due to their popularity both in the literature and applications. Some of their very recently developed comptetitors such as [20] (R-package SelectiveInference) and [21] (R-package islasso) are currently gaining attention from the community and will be used as benchmarks in our future research.
Continuing with the description of our simulation study, all predictors were generated from a multivariate normal distribution with the following probability density function
with an m-dimensional mean vector and an -dimensional covariance matrix . Writing out the covariance matrix
the correlation matrix can be expressed as
The binary response variable is generated using Bernoulli distribution with individual probability () defined as
is the matrix of true important variables and is the associated preassigned regression coefficients. Next, we present the details of the three different simulation scenarios considered.
- Under scenario 1, each of the simulated datasets has 200 observations and 1000 predictors. Here, for all vectors, we let and . Pairwise correlation of was applied to the first eight variables, while the remaining 992 variables were left uncorrelated. The -vector was chosen asEach of the datasets was split into training and test sets with equal proportions.
- The datasets under scenario 2 also have 200 observations and 1000 predictors. Again, and . Now, the first five variables were assumed to have a correlation of . The remaining 995 variables were independent. The -vector was selected asEach of the datasets was split into training and test sets with equal proportions.
- Under the last scenario 3, each of the datasets has 150 observations and 1000 predictors. We let and . The first five variables were assigned into a correlation value of , while the variables with indices from 11 to 30 were chosen to have the correlation value of . Outside of these two blocks, the variables were assumed uncorrelated. The -vector was chosenThe dataset was split into training and test sets with ratio of 70 to 30.
We compared the proposed rank-based penalized logistic regression method with lasso, adaptive lasso and elastic net methods from the glmnet R-package [11]. Algorithm 1 summarizes the procedure to calculate the average value of accuracy, sensitivity, specificity and AUROC based on a given number of iterations for each of the three simulation scenarios.
| Algorithm 1 Calculation of overall mean and standard deviation on simulation studies |
|
In Table 2, we compare the estimated mean and standard deviation of accuracy, sensitivity, specificity and AUROC values based on 200 iterations under correlation structure of in the simulation of scenario 1. The proposed rank-based penalized method shows the highest accuracy of 0.963 with the standard deviation of 0.02, sensitivity of 0.961 with standard deviation of 0.03, specificity of 0.965 with standard deviation of 0.03. In addition, it yields the same AUROC of 0.995 with standard deviation of 0.01 as elastic net and adaptive lasso.

Table 2.
Comparison of the performance among the four methods over 200 replications under simulation scenario 1. The values in parentheseses are the standard deviations.
In Table 3, we compare estimated mean and standard deviation of accuracy, sensitivity, specificity and AUROC values using 200 iterations under correlation structure of in the simulation of scenario 2. The proposed rank-based method also shows highest accuracy of 0.831 with standard deviation 0.04, sensitivity of 0.833 with standard deviation of 0.06, specificity of 0.829 with standard deviation of 0.05. In addition, the proposed method produces AUROC of 0.913 with standard deviation of 0.03. This is the second highest value which is slightly lower than the AUROC value of the elastic net.

Table 3.
Comparison of the performance among the four methods over 200 replications under simulation scenario 2. The values in parentheseses are the standard deviations.
In Table 4, we compare estimated mean and standard deviation of accuracy, sensitivity, specificity and AUROC values with 150 iterations under correlation structure of and in simulation of scenario 3. The proposed method shows highest accuracy of 0.916 with standard deviation of 0.04, sensitivity of 0.919 with standard deviation of 0.06, specificity of 0.912 with standard deviation of 0.06 and AUROC of 0.977 with standard deviation of 0.02.

Table 4.
Comparison of the performance among the four methods over 150 replications under simulation scenario 3. The values in parentheseses are the standard deviations.
Furthermore, we compared the performance in terms of selecting the number of true important variables by each of the four methods under three different simulation scenarios. First, we performed multiple hypothesis testing with BH-FDR [15] to reduce the dimensionality of the data. After performing a screening step to retain the relevant variables, we used them as input for the proposed rank-based penalized method with the regularization step outlined in Section 2.3. We illustrate the performance of variable selection with boxplots in Figure 2, Figure 3 and Figure 4 for simulation scenarios 1, 2, and 3. Each figure displays two boxplots, which, in turn, depict the distribution of the number of variables selected (NVS) and the number of true important variables (NTIV) within the number of variables selected (NVS) with each of the four methods computed based on the given number of iterations in each of the three simulation scenarios.
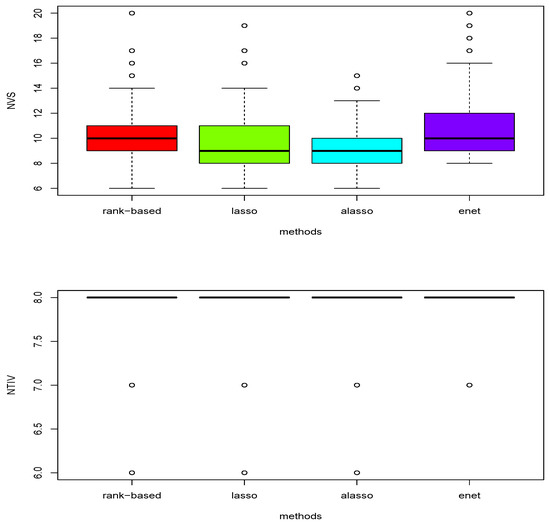
Figure 2.
Boxplots of total number of variables (NVS) selected and the number of true important variables (NTIV) within the number of variables selected with four different models under scenario 1 based on 200 replications.
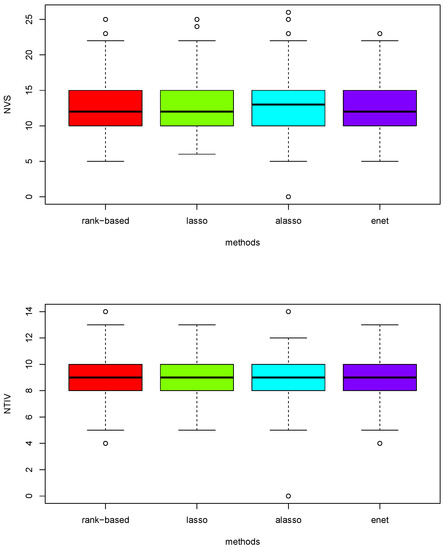
Figure 3.
Boxplots of total number of variables (NVS) selected and the number of true important variables (NTIV) within the number of variables selected with four different models on scenario 2 based on 200 replications.
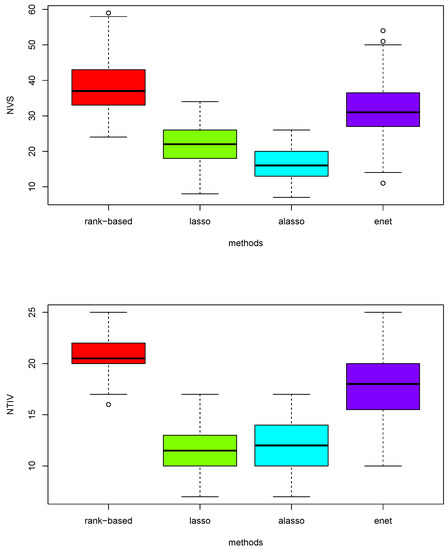
Figure 4.
Boxplots of total number of variables (NVS) selected and the number of true important variables (NTIV) within the number of variables selected with four different models under scenario 3 based on 150 replications.
Figure 2 reports that the proposed rank-based method has a slightly higher median number of variables selected (displayed as a thick line in the upper boxplots) than lasso, adaptive lasso and elastic net under scenario 1. The lower boxplots show that all four methods performed head-to-head for selection of true important variables under scenario 1 with 200 replications. Table 5 compares the mean and the standard deviation (in parentheseses) of the number of variables (NVS) selected and the number of true important variables (NTIV) in NVS for each of the four methods over 200 replications. The proposed rank-based method and elastic net performed head-to-head while slightly outperforming lasso and adaptive lasso.

Table 5.
Estimated mean and standard deviation of number of variables (NVS) selected and the number of true important variables (NTIV) among NVS with four different models under simulated scenario 1 with 200 replications. The values in parentheseses are standard deviations.
Figure 3 suggests the proposed method has a marginally higher median number of variable selected compared to the other three methods in the upper boxplot. It is also clear that the proposed method has a slightly higher median number of true important variables in the lower boxplot on scenarios 2 computed with 200 replications. Table 6 confirms that the rank-based penalized method has the highest mean both for selecting the number of variables and important variables.

Table 6.
Estimated mean and standard deviation of number of variables (NVS) selected and the number of true important variables (NTIV) among NVS in four different models under simulation scenario 2 with 200 replications. The values in parentheseses are standard deviations.
In Figure 4, the upper boxplot demonstrates that the proposed rank-based method has the highest median number of variables selected, elastic net has second highest median, lasso has third largest median and adaptive lasso has the smallest median under scenario 3 based on 150 replications. The lower boxplots also show that the proposed rank-based method has the highest median number of true important variables selected. However, adaptive lasso has a higher median number of true important variables than lasso unlike the upper boxplots. Thus, the proposed rank based-method clearly outperforms other three methods under high-correlation settings among variables.
Table 7 summarizes the number of variables selected and true important variables selected across the four methods under the high-correlation setting among variables computed from 150 replications. The proposed rank-based method has the highest mean number of overall variables selected and true important variables selected.

Table 7.
Estimated mean and standard deviation of number of variables (NVS) selected and the number of true important variables (NTIV) among NVS in four different models under simulation scenario 3 with 150 replications. The values in parentheseses are standard deviations.
3.2. Real Data Example
We applied four logistic regression methods to select differentially expressed genes and assess their discrimination capability between colon cancer cases and healthy controls using high-dimensional gene expression data [22]. The colon cancer gene expression dataset is available at [23]. It contains 2000 genes with the highest minimal intensity across 62 tissues. The data were measured on 40 colon tumor samples and 22 normal colon tissue samples. We split the data set into training and testing sets with proportions 70% and 30%, respectively. To detect significantly differentially expressed genes for high-dimensional colon cancer carcinoma and measure classification prediction, we adapted two step procedures of filtering and variable selection. First, we applied BH-FDR [15] to select most promising candidates of genes as a preprocessing step and then used the screened genes as input to the proposed rank-based method and three other popular methods—lasso, adaptive lasso and elastic net. The performance in terms of accuracy, sensitivity, specificity and AUROC as well as the selection probabilities for the four methods are reported in Table 8 and Table 9, respectively.

Table 8.
Estimated mean values and standard deviations for the four metrics across the four competing penalized logistic regression models computed from 100 resamplings. The values in parentheseses are standard deviations.

Table 9.
List of top 5 ranked genes across rank-based, lasso, adaptive and elastic net. An extra asterix (*) sign is put next to a gene each time the gene is selected by one of four methods.
Algorithm 2 outlines above protocols the procedure of calculating the average values of accuracy, sensitivity, specificity and AUROC through 100 bootstrap iterations applied to the colon cancer gene expression data. In Table 8, the performance of all four metrics are computed based on 100 iterations of resampled subsets of individuals.
| Algorithm 2 Calculation of mean with standard deviation on colon cancer data |
|
The average AUROC of 0.853 with standard deviation of 0.06 in the proposed rank-based method has the highest value compared to other three methods. Also the accuracy of 0.853 with standard deviation of 0.08 are optimal among the four methods. The values of sensitivity (0.860) and specificity (0.840) are also better than those of the other three methods. In summary, it is fair to conclude that the proposed rank-based method outperforms the other three popular penalized logistic regression methods. Table 9 shows top 5 ranked genes with highest selection probabilities for the proposed rank-based method, lasso, adaptive lasso and elastic net. An expressed sequence tag (EST) of Hsa.1660 associated with colon cancer carcinoma is found by all four methods. Hsa.36689 [24,25] is shown and top ranked by the proposed method, lasso and elastic net. Hsa692 also appeared and is second ranked by the proposed method, lasso and elastic net. In addition, Hsa.37937 is shown and is third and second ranked by the proposed method and elastic net, respectively.
4. Discussion
In this paper, we proposed a new rank-based penalized logistic regression method to improve classification performance and the power of variable selection in high-dimensional data with strong correlation structure.
Our simulation studies demonstrated that the proposed method improves not only the performance of classification or class prediction but also the detection of true important variables under various correlation settings among features when compared to existing popular regularization methods such as lasso, adaptive lasso, and elastic net. As demonstrated by simulation studies, if the true important variables are not passed through the filtering method such as BH-FDR, their chance of being selected in the final model decreases signficantly, thus, leading to reduction in variable selection and classification performance. Therefore, effective filtering methods which are likely to retain as many most promising variables as possible are indispensable.
Applied to high-dimensional colon gene expression data, the proposed rank-based logistic regresson method with BH-FDR screening produced the highest average AUROC value of 0.917 with standard deviation of 0.06 and accuracy of 0.853 with standard deviation of 0.08 using 100 resampling steps. The proposed method produced a good balance between sensitivity and specificity in contrast to other methods. Elastic net demonstrated the second best peformance with an average AUROC value of 0.903 with standard deviation of 0.07. A probable reason is that elastic net accounts for group correlation effects. In addition, we compared top 5 ranked ESTs across the proposed method, lasso, adaptive lasso and elastic net [12]. They had a common EST of Hsa.1660 associated to colon cancer data. We also found that Hsa.36689 was both deemed important and top ranked by the proposed method, lasso and elastic net. This also applied to Hsa.692, which was deemed important and second top ranked by the proposed method and lasso, whereas it was only third-ranked by the elastic net. Hsa.37937 was detected by both the proposed method and the elastic net. Hence, the four ESTs mentioned appear to be promising candidate biomarkers associated with colon cancer carcinoma. The function of the genes corresponding to ESTs is summarized in Table 9.
5. Conclusions
In this study the proposed rank-based classifier demonstrated the superiority in not only classification prediction but also the power of detecting true important variables when compared to lasso, adaptive lasso, and elastic net through the extensive simulation studies. Besides, in the application of high-dimensional colon cancer gene expression data, the proposed classifier showed the best performance in terms of accuracy and AUROC among the four classifiers considered in the paper. As a future research, we would develop the methodology of variable selection and compare the performance with those of most recent competitors such as [20,21,26,27], etc.
Author Contributions
All authors have equally contributed to this work. All authors wrote, read, and approved the final manuscript.
Funding
This research received no external funding.
Acknowledgments
We would like to thank the reviewers for their valuable comments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Houwelingen, H.C.V.; Bruinsma, T.; Hart, A.A.M.; Veer, L.J.V.; Wessels, L.F.A. Cross-validated Cox regression on microarray gene expression data. Stat. Med. 2006, 25, 3201–3216. [Google Scholar] [CrossRef] [PubMed]
- Lofti, E.; Keshavarz, A. Gene expression microarray classification using PCA–BEL. Comput. Biol. Med. 2014, 54, 180–187. [Google Scholar]
- Algamal, Z.Y.; Lee, M.H. Penalized logistic regression with the adaptive LASSO for gene selection in high-dimensional cancer classification. Expert Syst. Appl. 2015, 42, 9326–9332. [Google Scholar] [CrossRef]
- Li, C.; Li, H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics 2008, 24, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Fan, J.; Li, R. Variable selection via nonconcave penalized likelihood and its oracle properties. J. Am. Stat. Assoc. 2001, 96, 1175–1182. [Google Scholar] [CrossRef]
- Tibshirani, R.; Saunders, M. Sparsity and smoothness via the fused lasso. J. R. Stat. Soc. B 2005, 67, 91–108. [Google Scholar] [CrossRef]
- Zou, H. The adaptive Lasso and its oracle properties. J. Am. Stat. Assoc. 2006, 101, 1418–1429. [Google Scholar] [CrossRef]
- Meinshausen, N.; Yu, B. Lasso-type recovery of sparse representations for high-dimensional data. Ann. Stat. 2009, 37, 246–270. [Google Scholar] [CrossRef]
- Huang, H.; Liu, X.Y.; Liang, Y. Feature selection and cancer classification via sparse logistic regression with the hybrid L1/2+2 regularization. PLoS ONE 2009, 11, e0149675. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Sun, H.; Wang, S. Penalized logistic regression for high-dimensional DNA methylation data with case-control studies. Bioinformatics 2012, 28, 1368–1375. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wang, S. Network-based regularization for matched case-control analysis of high-dimensional DNA methylation data. Stat. Med. 2012, 32, 2127–2139. [Google Scholar] [CrossRef] [PubMed]
- Reiner, H.; Yekutieli, D.; Benjamin, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 2003, 19, 368–375. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Li, C.; Li, H. Variable selection and regression analysis for graph-structured covariates with an application to genomics. Ann. Appl. Stat. 2010, 4, 1498–1516. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Hofling, H.; Tibshirani, R. Pathwise coordinate optimization. Ann. Appl. Stat. 2007, 1, 302–332. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Lee, J.D.; Sun, D.L.; Sun, Y.; Taylor, J.E. Exact post-selection inference, with application to the lasso. Ann. Stat. 2016, 44, 907–927. [Google Scholar] [CrossRef]
- Cilluffo, G.; Sottile, G.; La Grutta, S.; Muggeo, V.M.R. The Induced Smoothed lasso: A practical framework for hypothesis testing in high dimensional regression. Stat. Methods Med. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Alon, U.; Barakai, N.; Notterman, D.A.; Gish, K.; Ybarra, S.; Mack, D.; Levine, A.J. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 1999, 96, 6745–6750. [Google Scholar] [CrossRef]
- Available online: http://genomics-pubs.princeton.edu/oncology/affydata/index.html (accessed on 25 April 2019).
- Ding, Y.; Wilkins, D. A simple and efficient algorithm for gene selection using sparse logistic regression. Bioinformatics 2003, 19, 2246–2253. [Google Scholar]
- Li, Y.; Campbell, C.; Tipping, M. Bayesian automatic relevance determination algorithms for classfifying gene expression data. Bioinformatics 2002, 18, 1332–1339. [Google Scholar] [CrossRef]
- Frost, H.R.; Amos, C.I. Gene set selection via LASSO penalized regression (SLPR). Nucleic Acids Res. 2017. [Google Scholar] [CrossRef]
- Boulesteix, A.L.; De, B.R.; Jiang, X.; Fuchs, M. IPF-LASSO: Integrative L1-Penalized Regression with Penalty Factors for Prediction Based on Multi-Omics Data. Comput. Math. Methods Med. 2017. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).