Abstract
The Apetala2/Ethylene Responsive Factor (AP2/ERF) family represents a critical group of transcription factors in plants, recognized for their roles in growth, development, fruit ripening, and postharvest processes. This study aimed to identify and characterize the AP2/ERF gene family in passion fruit (Passiflora edulis Sims) and investigate their potential roles in flavor enhancement. A total of 91 PeAP2/ERF genes were identified and classified into five subfamilies. Chromosome localization and collinearity analysis demonstrated their distribution across all nine chromosomes of passion fruit, with tandem duplication events identified as a key driver of family expansion. Exon–intron configurations and motif compositions were highly conserved among PeAP2/ERF genes. Promoter cis-acting element analysis indicated potential regulation by environmental signals, including abiotic and biotic stresses, as well as hormonal cues. Postharvest storage induced the expression of 59 PeAP2/ERF genes over time. Notably, PeAP2-10 was found to enhance the expression of PeSTP6, a gene associated with sugar transport, suggesting its potential influence on the flavor profile of passion fruit. These findings provide valuable insights into the functional roles of PeAP2/ERF genes in passion fruit, highlighting their significance in postharvest management and flavor quality enhancement strategies.
1. Introduction
Among the transcription factor families, the Apetala2/Ethylene Responsive Factor (AP2/ERF) family is recognized as one of the most conserved and plant-specific families, playing a crucial role in various plant species [1]. Members of the AP2/ERF family possess at least one highly conserved AP2 domain, which consists of 60 to 70 amino acids. Based on the number of AP2 domains and other DNA-binding domains, the AP2/ERF superfamily can be divided into five subfamilies: APETALA2 (AP2), dehydration responsive element binding (DREB), ethylene responsive element binding protein (ERF), related to ABI3/VP (RAV), and Soloist. The AP2 subfamily contains two AP2 domains, and the other subfamilies have a single AP2 domain [2,3].
AP2/ERF family genes are essential for plant development [4]. For instance, in Arabidopsis, AtERF-13 regulates lateral root growth and development by modulating long-chain fatty acid synthesis [5]; in poplar, ERF139 promotes xylem expansion following its expression in the stems [6]; in Liriodendron chinensis, AP2/ERF family genes are involved in the regulation of leaf growth and development [7]; in soybean, TOE4b regulates photoperiodic flowering [8]; in sweet cherry, PavRAV2 negatively regulates fruit size by directly inhibiting PavKLUH expression [9]; in Perilla seeds, three AP2/ERFs (WRI, ABI4, and RAVI) regulate oil production [10]. In addition to their roles in growth and development, AP2/ERF genes also respond to environmental stimuli and hormonal signals. In Brassica napus leaves, 118 AP2/ERF family genes are likely to enhance cold tolerance through various molecular pathways [11]. In Eschscholzia California (California poppy), AP2/ERF genes respond to MeJA, with 20 family members being up-regulated [12]. Recent studies have indicated that AP2/ERFs also play significant roles in fruit development and postharvest. For example, FcAP2/ERF regulates the rapid ripening of fig fruit [13]. In addition, PgAP2/ERF5, PgAP2/ERF36, PgAP2/ERF58, and PgAP2/ERF86 are associated with fruit hardness, and the PgAP2/ERF gene family is a potentially important regulator of pomegranate fruit development. It has been shown that AP2/ERFs are abundantly expressed in postharvest fruits and respond to environmental and hormonal responses. Notably, more than half of the PgAP2/ERFs are expressed at relatively lower levels in postharvest pomegranate fruits under low temperature storage. Furthermore, PgAP2/ERF4, PgAP2/ERF15, PgAP2/ERF26, PgAP2/ERF30, PgAP2/ERF35, and PgAP2/ERF45 genes are up-regulated in treatments that promote pomegranate postharvest preservation [14]. Studies on the function and regulatory mechanisms of AP2/ERF in fruit ripening and postharvest storage have found that CpERF9 specifically binds to the promoters of CpPME1/2 and CpPG5 in papaya fruits, represses promoter activity, and regulates fruit ripening [15]. In loquat, EjAP2-1 binds to EjMYB1/2 and depresses Ej4CL1 promoter activity, consequently regulating phenylpropanoid metabolism under cold stress [16]. However, the role of AP2/ERF genes in the postharvest storage of passion fruit remains unclear.
Passion fruit (Passiflora edulis Sims), a tropical and subtropical crop with year-round fruiting and high nutritional value, has recently gained attention for genomic and flavor metabolism studies. In recent years, biological research on passion fruit has increasingly focused on its genome and flavor metabolism. Among these, 174 MYB family members and 14 SBP family members have been identified, and both are primarily involved in abiotic stresses, such as temperature, drought, and salt stress [17,18]. The promoter regions of PebHLHs are involved in developmental regulation, hormonal responses, and stress responses. In the context of flavor metabolism in passion fruit, it has been discovered that the induced PeLOX4 expression results in an increase in LOX enzyme activity, which in turn, promotes the synthesis of volatile esters in the pulp and enhances the aroma of the fruit [19]. Transient overexpression of PeCWINV5 in passion fruit increases soluble sugar accumulation [20]. Multiomics analyses of passion fruit have revealed that the ACX, ADH, ALDH, and HPL gene families are key regulators for ester synthesis, especially ACX13/14/15/20, ADH13/26/33, ALDH1/4/21, and HPL4/6. The Terpene Synthase (TPS) gene family is crucial for terpenoid synthesis, especially PeTPS2/3/4/24 [21]. Recently, our work has indicated that low temperatures and MeJA effectively preserve the fruit and delay the aging process of passion fruit during postharvest storage [22]. Nevertheless, the role of AP2/ERFs in the postharvest preservation of passion fruit remains unknown.
Considering that AP2/ERF genes are responsive to hormones and low temperature, and given their biological functions in plant growth, especially in the postharvest senescence of fruit [23], we hypothesized that this gene family may play crucial roles in the postharvest preservation of passion fruit. In this study, we carried out a genome-wide characterization of AP2/ERF genes and investigated the regulation of downstream genes in postharvest passion fruit. Our results provide key information regarding the AP2/ERF genes of passion fruit and present important experimental data on the role of PeAP2/ERF genes in the postharvest preservation of passion fruit.
2. Results
2.1. Identification of Ninety-One PeAP2/ERF Genes in the Passion Fruit Genome
A BLASTp analysis utilizing AtAP2/ERF sequences as query sequences led to the identification of 91 AP2/ERF-encoding genes within the passion fruit genome. After removing redundant sequences and alternative transcripts corresponding to the same genes, the PeAP2/ERF genes were designated according to their order in the passion fruit genomic sequence database (Table 1). The lengths of PeAP2/ERF proteins ranged from 118 amino acids (PeERF-47) to 1209 amino acids (PeDREB-2), and their molecular weights (MW) ranged from 7.22 kDa (PeERF-47) to 13.31 kDa (PeDREB-2). The predicted isoelectric points (pI) of the PeAP2/ERF proteins ranged from 4.26 (PeDREB-19) to 10.87 (PeERF-47). Notably, 52 proteins exhibited pIs ≥ 7, indicating a positive charge in acidic solutions. The hydrophilic indices of all proteins ranged from −1.267 to 0.749, reflecting their varying degrees of hydrophobicity. In summary, the 91 AP2/ERF genes in passion fruit showed significant variations in their physical and chemical properties (Table 1).

Table 1.
Identification and characterization of PeAP2/ERF genes and their predicted proteins in passion fruit.
2.2. PeAP2/ERF Proteins Are Classified into Five Subfamilies
To investigate the evolutionary relationships among the passion fruit PeAP2/ERF family members, a phylogenetic tree was constructed by aligning the full-length protein sequences of 141 A. thaliana and 91 passion fruit sequences (Figure 1). The AP2/ERF family members were grouped into five subfamilies. The ERF subfamily comprised 53 members, the DREB subfamily contained 25 members, and the AP2 subfamily consisted of 10 members, together accounting for approximately 96.7% of the PeAP2/ERF family members; the Soloist and RAV subfamilies contained 2 and 1 member, respectively (Figure 1). Notably, the similarity among members of the same subfamily was higher in both passion fruit and A. thaliana. This accurate classification of AP2/ERF protein family members, based on their shared structural characteristics, highlights their high similarity across different species.
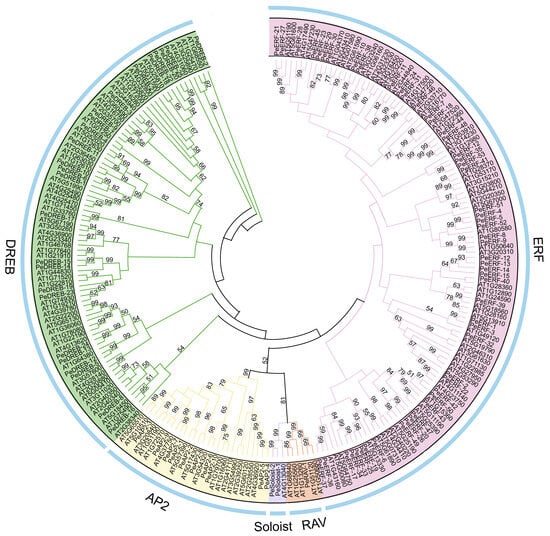
Figure 1.
Phylogenetic relationships of AP2/ERF proteins from Arabidopsis and passion fruit. A total of 141 AtAP2/ERFs proteins from Arabidopsis and 91 PeAP2/ERFs from passion fruit were utilized to construct a phylogenetic tree using the Neighbor-Joining (NJ) method implemented in MEGA12 software. The numbers on the branches represent bootstrap values calculated from 1000 replicates. All AP2/ERF proteins were classified into five subfamilies: AP2, DREB, ERF, RAV, and Soloist, which are represented by yellow, green, pink, tawny, and purple, respectively.
2.3. Chromosome Localization and Collinearity Analysis of the PeAP2/ERF Genes
The 91 PeAP2/ERF genes were distributed across all nine chromosomes of the passion fruit. Specifically, PeERF-1 to PeERF-11, PeDREB-1 to PeDREB-3, PeSoloist-1 to PeSoloist-2, and PeAP2-1 were located on chromosome 1; PeERF-12, PeRAV-1, and PeAP2-2 were located on chromosome 2; PeERF-13 to PeERF-17, and PeAP2-3 to PeAP2-4 were located on chromosome 3; PeERF-18 to PeERF-24, PeDREB-4 to PeDREB-7, and PeAP2-5 were located on chromosome 4; PeERF-25 to PeERF-31, PeDREB-8 to PeDREB-12, and PeAP2-6 were located on chromosome 5; PeERF-32 to PeERF-39, PeDREB-13 to PeDREB-16, and PeAP2-7 to PeAP2-8 were located on chromosome 6; PeERF-40 to PeERF-44, PeDREB-17 to PeDREB-21, and PeAP2-9 were located on chromosome 7; PeERF-45 to PeERF-46 and PeAP2-10 were located on chromosome 8; PeERF-47 to PeERF-51 and PeDREB-22 to PeDREB-23 were located on chromosome 9. Additionally, PeERF-52 to PeERF-53 and PeDREB-24 to PeDREB-25 were located on unanchored scaffolds (Figure 2).
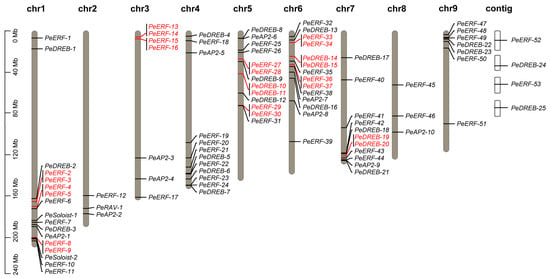
Figure 2.
Chromosomal distribution and tandem duplication analysis of PeAP2/ERF genes. Each bar represents a chromosome, and tandemly duplicated gene pairs are highlighted in red. The approximate locations of each PeAP2/ERF gene on each chromosome are indicated.
Further collinear analysis using MCScanX (v1.0.0) revealed that many gene members had collinearity within or between passion fruit chromosomes. The results indicated the presence of 12 tandem duplication gene pairs: PeERF-2 and PeERF-3, PeERF-4 and PeERF-5, PeERF-8 and PeERF-9, PeERF-13 and PeERF-14, PeERF-15 and PeERF-16, PeERF-27 and PeERF-28, PeERF-29 and PeERF-30, PeERF-33 and PeERF-34, PeERF-36 and PeERF-37, PeDREB-10 and PeDREB-11, PeDREB-14 and PeDREB-15, and PeDREB-19 and PeDREB-20. These tandem duplication events significantly contribute to the diversity and evolutionary history of gene families and play a crucial role in understanding the adaptive evolution of species (Figure 2).
Furthermore, a multicollinearity analysis was carried out to identify robust orthologs of passion fruit AP2/ERF genes in the genomes of other species, specifically A. thaliana (Figure 3). The analysis revealed that the collinear gene pairs among these species had undergone lineage-specific expansions during evolution. The results showed that the highest collinearity was observed between passion fruit and tomato, followed by passion fruit and Arabidopsis. Chromosome 1 exhibited the highest number of orthologs with all other species. Overall, the maximum number of collinear orthologs was found between passion fruit and Arabidopsis, suggesting that the AP2/ERF genes were conserved and likely shared common ancestors, with the exception of instances of duplication or loss (Figure 3).
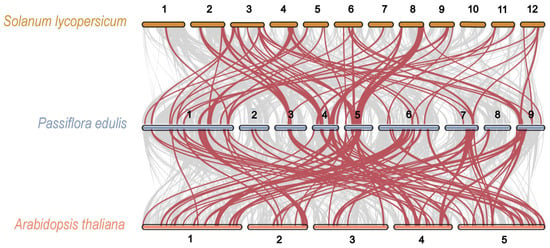
Figure 3.
Collinearity analysis of PeAP2/ERF genes between passion fruit and two other species (A. thaliana and Solanum lycopersicum). The grey lines in the background represent collinear blocks between passion fruit and other plant genomes, while the wine-red lines highlight the collinear PeAP2/ERF gene pairs. The chromosomes of Arabidopsis, passion fruit, and tomato are represented by tangerine, blue-grey, and tan bars, respectively, with numbers representing the respective chromosomes.
2.4. The Structure of PeAP2/ERFs Is Highly Conserved
We analyzed the structure of the PeAP2/ERF genes to gain further insights into their functional and evolutionary diversification. The intron–exon structures were plotted according to subfamily order in the phylogenetic tree (Figure 4). Based on sequence similarity, all PeAP2/ERF genes comprised 5′- or 3′-noncoding regions (UTRs), 1 to 10 exons, and zero to nine introns (Figure 4). Specifically, PeAP2-4 and PeAP2-5 in the subfamily AP2 contained 10 exons. Other PeAP2 members had six to nine exons. PeERF members had one to seven exons. In the PeERF subfamily, 28 genes contained one exon, and none of these members had UTRs. PeDREB genes typically lacked introns and UTRs, except for PeDREB-2, PeDREB-8, and PeDREB-16, which had UTRs and at least two exons. PeRAV-1 contained three exons. PeSoloist-1 and PeSoloist-2 contained three and six exons, respectively (Figure 4). Additionally, PeAP2/ERFs in the duplication pairs exhibited similar exon and intron lengths and numbers (Figure 4).
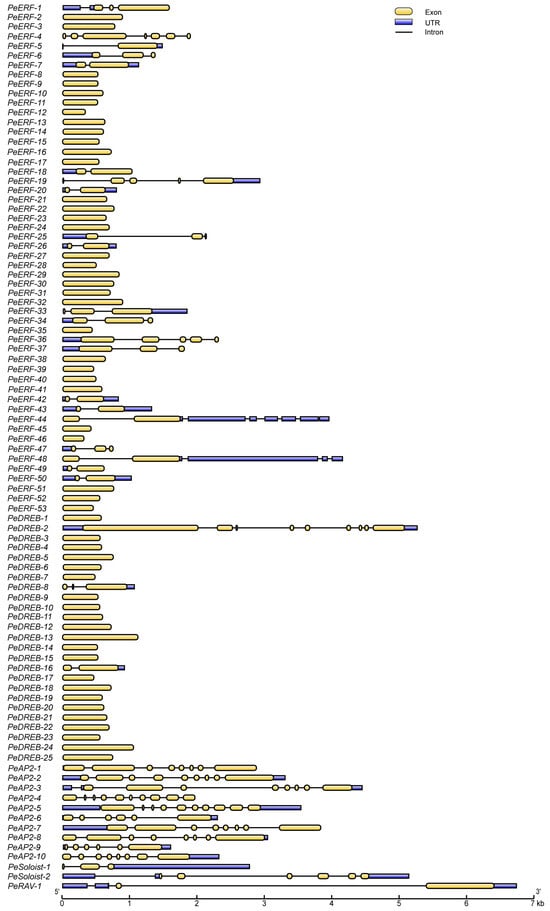
Figure 4.
Structure of PeAP2/ERF genes. Exons and introns are represented with yellow boxes and black lines, respectively. Blue boxes refer to the untranslated regions (UTRs). The exon and intron lengths are calculated according to the scale at the bottom.
Using TBtools software (v2.121), we identified 10 motifs in PeAP2/ERF proteins, with lengths ranging from 8 to 41 amino acids (Figure 5 and Figure S1). Specifically, the PeAP2 subfamily contained seven motifs, including motifs 1, 2, 3, 5, 7, 9, and 10. Among them, the subfamily members shared the conserved motifs of 2, 5, and 9. The PeERF subfamily contained motifs 1, 2, 3, 4, and 8. The PeDREB subfamily contains motifs 1, 2, 3, 4, 6, 7, and 10. Among these, the PeERF and PeDREB subfamilies shared the conserved motifs of motifs 1 and 2; however, the conserved motif unique to the PeERF subfamily was motif 4, while the conserved motif unique to the PeDREB subfamily was motif 3. The PeSoloist subfamily contained motifs 1, 2, 3, and 4. The PeRAV subfamily contained motifs 1 and 2. Both the PeSoloist and PeRAV subfamilies had only one conserved motif (motif 1), which may serve as AP2 domain (Figure 5).
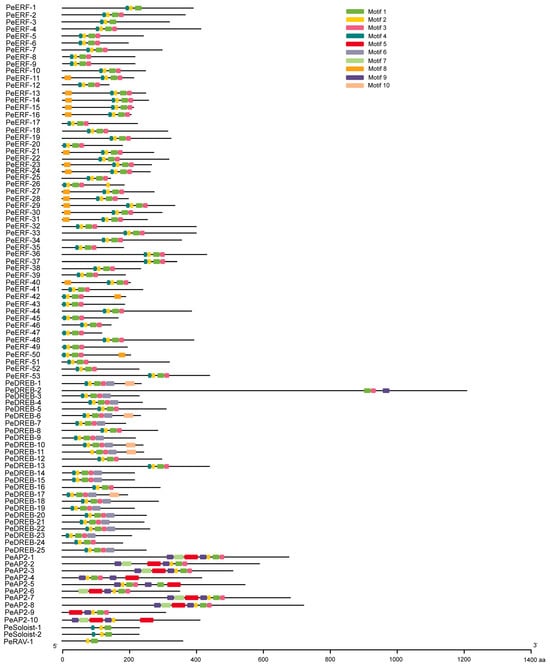
Figure 5.
The conserved motifs of PeAP2/ERF proteins. Motifs are shown in the order in which they are arranged in the original phylogenetic tree. Each motif is represented by a colored box and the nonconserved sequences are presented with black lines. The length of the motif is indicated by the width of the image (unit: amino acids).
2.5. Identification of Cis-Acting Elements in PeAP2/ERF Promoters
Transcription factor-binding sites and regulatory cis-elements in the promoter region are pivotal for regulating gene expression. We analyzed the putative regulatory cis-elements in the promoters of PeAP2/ERF genes using the PlantCARE database 5.0. In addition to the common and core cis-elements, such as CAAT and TATA boxes, several cis-acting elements related to environmental responses, hormones, and growth and development were identified in the promoters of PeAP2/ERF genes (Figure 6). Most of the PeAP2/ERF genes showed cis-elements associated with environment responses, indicating that PeAP2/ERF genes may play a vital role in responding to various abiotic stresses (Figure 6). Moreover, at least five phytohormone responsive cis-elements were observed in the promoter of each PeAP2/ERF gene, suggesting that PeAP2/ERF genes are involved in various hormone signaling pathways in passion fruit (Figure 6). In addition, the number of cis-elements responsive to environmental stress and hormones far exceeded those related to growth and development.
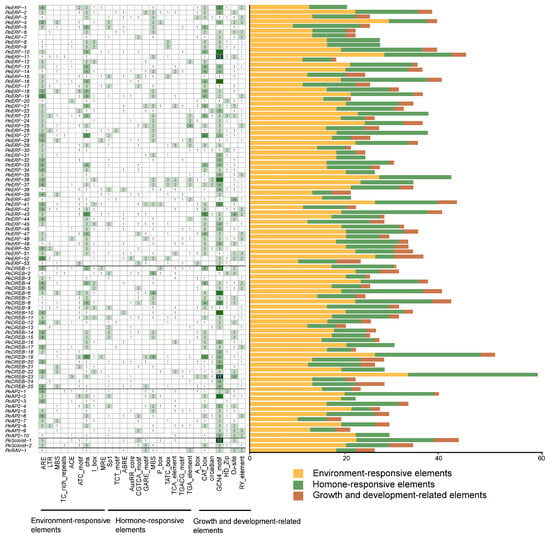
Figure 6.
Putative cis-elements enriched in the promoter regions of PeAP2/ERF family genes. Number of cis-acting elements in the promoter regions of individual PeAP2/ERF genes. The colors in the left figure indicate the numbers of cis-elements. Darker green shades correspond to higher numbers.
2.6. Analysis of PeAP2/ERFs Expression Pattern in Postharvest Passion Fruit
Previously, we performed a transcriptome analysis in postharvest passion fruit under various storage conditions [22]. To further explore the potential roles of PeAP2/ERFs during postharvest storage, we analyzed the expression patterns. PeAP2-1, PeAP2-8, PeAP2-9, PeDREB-7, PeDREB-20, PeERF-27, PeERF-39, PeERF-50, and PeERF-53 were not expressed in postharvest fruit. Specifically, 6 ERF genes, 12 DREB genes, 4 AP2 genes, and 1 Soloist gene were down-regulated during postharvest. The remaining 59 family genes would be up-regulated at least at one time point (Figure 7). However, each individual gene exhibited a distinct expression pattern. For instance, PeERF-17 peaked in expression at 1 day postharvest (dph); PeDREB-3, PeDREB-23, PeRAV-1, PeERF-8, PeERF-9, and PeERF-44 reached their dominant expression levels at 2 dph; PeAP2-7, PeDREB-12, PeERF-4, PeERF-10, PeERF-28 were mainly expressed at 4 dph; at 5 dph, PeDREB-1, PeDREB-6, PeERF-7, PeERF-12, PeERF-22, PeERF-41, PeERF-48 showed the highest expression; on the 6th day, the expression of PeAP2-6, PeERF-13, PeERF-15, PeERF-16, PeERF-5, PeERF-6, PeERF-24, PeERF-33, PeERF-36, PeERF-37, PeERF-38, PeERF-46, PeERF-52 had the highest expression; PeDREB-24 and PeERF-40 attained the highest expression at 8 dph (Figure 7 and Figure S2).
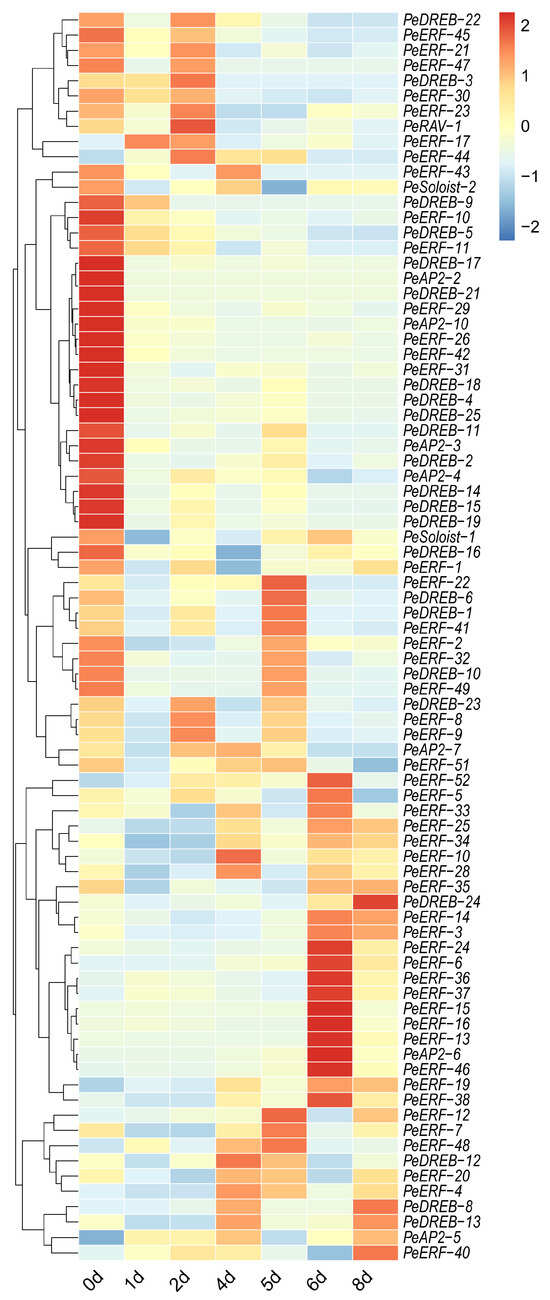
Figure 7.
Hierarchical clustering analyses of the PeAP2/ERFs’ expression in postharvest passion fruits. The relative gene expression levels were determined using the passion fruit housekeeping gene PeACTIN as an internal control. The varying expression values of PeAP2/ERF genes were analyzed with RNA-seq data. Different colors correspond to expression values.
2.7. PeAP2-10 Directly Regulates PeSTP6
To explore the regulation network of PeAP2/ERF family genes, we performed co-expression analysis using RNA-seq data described in our previous work. Based on the expression patterns, the genes were divided into 11 modules. Specifically, we focused on PeAP2-10, the only member containing two motif 5 sequences. The expression of PeAP2-10 was tightly correlated with several genes, including PeSTP6 (P_edulia040010232.g), a sugar transporter-encoding gene homologous to SlSTP1 (Figure 8 and Figures S2–S4).
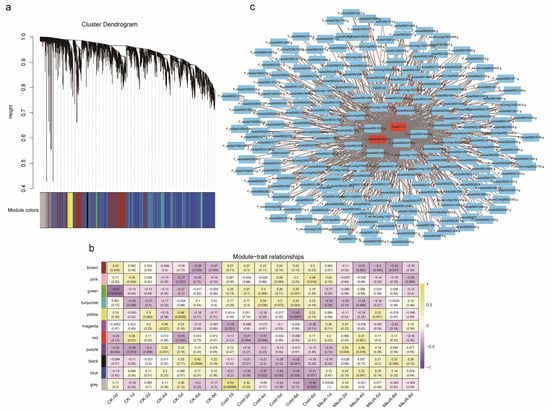
Figure 8.
Co-expression analysis. (a) Co-expression clustering dendrogram based on expression data generated by a synthetic Gene Regulatory Network (GRN) after 1000 times (iterations). (b) Association heatmap between modules and phenotypic traits (cold and MeJA). Each cell contains the correlation coefficient and p-value. (c) The co-expression network of 218 genes in brown module. The red labeled genes were PeAP2-10 and P_edulia040010232.g (PeSTP6), respectively.
The promoter of PeSTP6 contains two GCC_box cis-acting elements (Figure 9a and Figure S5), suggesting that PeSTP6 serves as a candidate target of AP2 proteins. Subsequently, we performed yeast one-hybrid assays and found that PeAP2-10 bound to the promoter of PeSTP6 (Figure 9b). A dual luciferase assay confirmed that PeAP2-10 induced the promoter activity of PeSTP6 (Figure 9c,d). These data indicate that PeAP2-10 regulates the expression of PeSTP6.
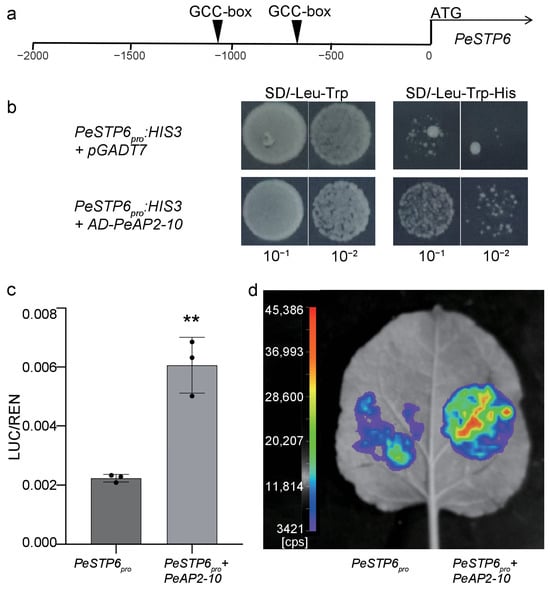
Figure 9.
PeAP2-10 regulates PeSTP6. (a) The position of the GCC_box identified through PeSTP6 promoter analysis. (b) Yeast one-hybrid (Y1H) assays demonstrated that PeAP2-10 binds to the PeSTP6 promoters. Dual-luciferase (Dual-LUC) assays (c) and imaging (d) confirmed that PeAP2-10 activated PeSTP6 transcription. In these experiments, PeSTP6pro and PeAP2-10 represent the reporter and effector constructs, respectively. Values represent means and standard deviations of three biological replicates, with three technical replicates for each biological replicate. Double asterisks (**) indicate p values < 0.01.
3. Discussion
The biological and molecular functions of AP2/ERF genes in passion fruit remain unexplored. To elucidate the potential roles of PeAP2/ERF genes in passion fruit, we identified 91 PeAP2/ERF genes distributed across all 9 chromosomes of its genome (Table 1 and Figure 2). Consistent with findings in Arabidopsis [24], cucumber [25], barley (Hordeum vulgare) [26], and tomato [27], PeAP2/ERFs in passion fruit were classified into five subfamilies based on their phylogenetic relationships (Figure 1). Most AP2/ERF members were found in the AP2, ERF, and DREB subfamilies. This distribution aligns with observations in desert legumes (153 members: with 24 AP2, 59 DREB, and 68 ERF) [28]. Rhododendron has a total of 120 AP2/ERF family members, including 17 AP2s, 44 DREBs, and 53 ERFs, which constitute approximately 95% of the total number of AP2/ERF members [29]. This indicates that the AP2, ERF, and DREB subfamilies account for a large proportion of the total AP2/ERF family members and may play a broad role.
Tandem and segmental duplication events, which arise from whole genome duplication (WGD) through polyploidization or local chromosomal rearrangements, contribute to gene family expansion and evolution [30]. The maximum number of collinear orthologs was identified between passion fruit and Arabidopsis, indicating that PeAP2/ERF genes are conserved and probably share common ancestors (Figure 3). Duplication events likely occurred in a common ancestor, and duplicated genes tend to exhibit a closer relationship. Our phylogenetic tree revealed that 10 out of the 12 gene pairs in tandem duplication events were derived from the ERF subfamily (Figure 2).
The number of PeAP2/ERF genes in passion fruit exceeds that in Brassica napus (87 members) [31] and Penniseagraria (78 members) [32], yet it is less than that in tomato (134 members) [27], rice (170 members) [33], and wheat (322 members) [34]. This variation highlights the diversity in the distribution of PeAP2/ERF genes among different species. It is estimated that passion fruit diverged from the Euphorbiaceae lineage approximately 65 million years ago (MYA) and from Salicaceae approximately 60 MYA [35]. Moreover, genomic analysis has demonstrated that passion fruit experienced a recent whole-genome duplication (WGD) event after diverging from both Euphorbiaceae and Salicyliaceae [36,37]. The relatively small number of AP2/ERF genes implies the occurrence of gene loss events, which often follow WGD, during the evolutionary process of passion fruit. Similar assumptions have been made for Pennisetum glaucum, which naturally has a strong tolerance to various environmental stresses but contains a small number of AP2/ERF superfamily members [32]. However, the specific mechanisms by which different AP2/ERF family members contribute to the domestication and adaptation of passion fruit remain to be clarified.
Passion fruit is cultivated in tropical and subtropical areas, and its fruits are harvested at the early age [21,35]. Therefore, understanding the fruit characteristics during the postharvest maturation period is essential. Previous studies have reported that AP2/ERF genes play crucial roles in the preservation of postharvest fruits. For instance, the expression of ERFs is important in the ripening and senescence of peach during postharvest storage [38]. In red-fruit loquat, EjERF39 responds to environmental conditions, which is involved in the lignification of immature loquat fruits [39]. The specific expression of AP2/ERF genes had been noticed during the rapid softening of postharvest papaya pulp [40]. Recently, we discovered that the quality and flavor of postharvest passion fruit were significantly enhanced following low-temperature and MeJA treatment [22]. In this study, we characterized the induced expression of PeAP2-10 and identified the PeAP2-10-PeSTP6 expression cascade. However, the specific functions of AP2/ERF genes in this process remain unclear.
Postharvest preservation is essential for fruit quality and flavor. In this study, most members of the PeAP2/ERF family responded to postharvest storage, exhibiting varying degrees of response, except for PeAP2-1, PeAP2-8, PeAP2-9, PeDREB-7, PeDREB-20, PeERF-27, PeERF-39, PeERF-50, and PeERF-53 (Figure 7). More than 60% (59 out of 91) of family members were up-regulated with the extension of postharvest time (Figure 7). Notably, PeAP2-10 expression correlated with PeSTP6 (Figure S2), a putative ortholog of the tomato sucrose transporter gene governing soluble solid content [41]. Transactivation assays confirmed PeAP2-10’s role to induce PeSTP6 (Figure 9), implicating it in sugar transport and flavor modulation. These findings highlight PeAP2/ERFs as potential regulators of postharvest metabolic dynamics in passion fruit.
4. Materials and Methods
4.1. Plant Materials
Mature fruits of passion fruit (Passiflora edulis Sims) cultivar Tainong 1 were collected from the Passion Fruit Science and Technology Backyard in Baisha, Hainan, China. The fruits were stored for 0, 1, 2, 4, 5, 6, or 8 days. Three biological replicates were utilized, with each consisting of six fruits [22].
4.2. Identification of the PeAP2/ERF Family Members in Passion Fruit
The genome information of passion fruit was obtained from a previous study [21]. The amino acid sequences of 141 AtAP2/ERF proteins in the A. thaliana were downloaded from the TAIR database (https://www.arabidopsis.org/, accessed on 28 July 2023), and the sequences of 134 SlAP2/ERF proteins in the Solanum lycopersicum were retrieved from the Sol Genomics Network database (https://solgenomics.net/, accessed on 28 July 2023). These sequences were subsequently employed as queries for a BLAST search against the passion fruit genome, utilizing an E-value threshold of <e−10 and a sequence identity of >50% [42]. Meanwhile, Hidden Markov Model (HMM) (version 3.0, http://hmmer.janelia.org/) was used with the default parameters for the identification of AP2/ERF proteins. The AP2 domain (PF00847) was retrieved from the Pfam database (http://pfam.xfam.org, accessed on 30 July 2023). The presence of the AP2 domain in each protein was confirmed using the SMART (http://smart.embl-heidelberg.de/, accessed on 30 July 2023).
4.3. Phylogenetic Analysis
Phylogenetic analysis was conducted using amino acid sequences of the identified PeAP2/ERFs and AtAP2/ERFs from A. thaliana. A phylogenetic tree was constructed using the Neighbor-Joining (NJ) method as implemented in MEGA12 software, with a bootstrap of 1000 replicates. Subsequently, the tree was subsequently visualized using iTol software v7 (https://itol.embl.de/, accessed on 30 July 2023) [43].
4.4. Chromosomal Distribution and Collinearity Analysis
The chromosomal locations of PeAP2/ERF genes were visualized using MapChart (version 2.32) [44]. MCScanX (v1.0.0) analysis [45] was employed to identify gene families containing PeAP2/ERF copies, and CIRCOS [46] was utilized to construct the collinearity map of PeAP2/ERF genes.
4.5. Gene Structure and Conserved Motifs of PeAP2/ERF
The gene structures of PeAP2/ERF were visually mapped based on the passion fruit genome annotation using the GSDS online tool (https://gsds.gao-lab.org/, accessed on 30 July 2023) [47]. Additionally, the protein sequences of PeAP2/ERF were submitted to TBtools [48] for motif prediction, with the motif discovery number set to 10 and all other parameters set to default.
4.6. Prediction of Cis-Elements in PeAP2/ERF Genes Promoter
The 2000-bp upstream of the start codon of the PeAP2/ERF family genes were submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 4 August 2023) for the prediction of cis-acting elements. Subsequently, the results from PlantCARE were subsequently sorted and simplified [49].
4.7. Yeast One-Hybrid
The promoter of the PeSTP6 gene was recombined into the pHIS2 plasmid to generate the PeSTP6pro:HIS3 construct. The CDS sequence of PeAP2-10 was inserted into the pGADT7 to generate the AD:PeAP2-10 construct. The recombinant plasmids were co-transformed into yeast strain Y187. Positive single colonies were selected. After identification, the SD/-Leu-Trp and SD/-Leu-Trp-His concentration determined by screening was used for dot-plate and interaction verification. The cells were incubated at 30 °C for 3–4 d and then photographed and recorded.
4.8. Transient Dual Luciferase Reporter Assays
The promoter of PeSTP6 was cloned into the PJG077 vector [22], and the CDS of PeAP2-10 was inserted into the pEAQ-HT-DEST2 vector to generate the reporter and the effector constructs, respectively. Then, these constructs were co-expressed in the tobacco leaves. After incubation for 44 h, the transfected cells were collected and homogenized in 300 μL of passive lysis buffer. A 10 μL aliquot of the crude extract was mixed with 50 μL of luciferase assay buffer, and the Firefly Luciferase (LUC) activity was measured using a Varioskan LUX (Thermo Scientific, Waltham, MA, USA). Stop and Glow Buffer (50 μL) was subsequently added to the reaction solution and the Renilla Luciferase (REN) activity was measured. The LUC/REN ratio was calculated to represent the relative activity of the transcription factors [22].
4.9. RNA Extraction and qRT-PCR
Total RNA in each sample was extracted, using the TRIzol reagent kit (Vazyme, Nanjing, China). All RNA was purified, using the Hifair® II 1st Strand cDNA Synthesis Kit (gDNA digester plus) (Yeasen, Shanghai, China) for first-strand cDNA synthesis. Quantitative RT-PCR was performed using the 2×SYBR Green qPCR Mix (SparkJade, Jinan, China). The Quantstudio™ 7 Flex Real-time PCR system (Applied Biosystems, Carlsbad, CA, USA) was used for detection. Samples were normalized using the PeActin gene (LOC8268098/XP_002531173.1) as an endogenous control. qRT-PCR primers were designed using Primer 3 based on the CDS sequences of the PeAP2/ERF genes (Table S1). Three biological replicates were analyzed, each containing six technical replicates. The 2−ΔCt comparative method was used to calculate the relative transcript levels in the samples [50,51].
4.10. Co-Expression Analysis
Gene co-expression network analysis was performed using pairwise correlations between quantile-normalized expression profiles derived from published RNA-seq data [22]. Scale-free weighted gene co-expression networks were constructed using the “WGCNA” package. The WGCNA analysis was conducted with the WGC-NAshiny plugin in TBtools, with the following parameters: RNA-seq data from passion fruit peel treated at 26 °C, 8 °C, and 100 mM MeJA for 0, 1, 2, 4, 5, 6, and 8 days; sample percentage = 0.9; expression cutoff = 1; filter method = MAD (Median Absolute Deviation); reserved genes = 20,000; final power selection = 14; minimum module size = 30; and module cuttree height = 0.4. The analysis was performed using the RNA-seq data to generate the final results.
Pearson’s correlations were employed to avoid assuming linear relationships between co-expressed genes, and genes displaying standard deviation equal to zero were eliminated in both empirical and simulated datasets. Unsupervised hierarchical clustering was applied to detect modules of highly co-expressed genes following the method described [52]. Association heatmaps and co-expression network analyses were performed using TBtools software (v2.121) and RNA-seq data.
4.11. Statistical Analysis
All results were presented as means ± standard deviation (SD) of at least three biological replicates. Student’s t-test was applied to analyze the significant differences (*, p < 0.05; **, p < 0.01).
5. Conclusions
In conclusion, this study provides a comprehensive characterization of the PeAP2/ERF gene family in passion fruit, revealing its significant role in regulating postharvest processes and flavor enhancement. The identification of 91 PeAP2/ERF genes and their classification into distinct subfamilies underscores the evolutionary conservation and functional diversity of this transcription factor family. Our findings indicate that PeAP2/ERF genes, particularly PeAP2-10, are responsive to postharvest conditions and may play a pivotal role in sugar transport, thereby influencing the flavor profile of passion fruit. The insights gained from this research not only enhance our understanding of the molecular mechanisms underlying fruit quality but also provide a foundation for future studies aimed at improving postharvest management practices. Ultimately, elucidating the specific functions of PeAP2/ERF genes will be essential for developing strategies to optimize the quality and marketability of passion fruit, contributing to the sustainability of this important crop in tropical and subtropical regions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants14050645/s1. Figure S1: The conserved motifs of amino acid sequences identified by MEME 5.5.7 software; Figure S2: qRT-PCR validation of the differentially expressed genes and the relative expression level of PeAP2-10 and PeSTP6; Figure S3: Phylogenetic relationships of STP proteins from Arabidopsis and passion fruit; Figure S4: Amino acid alignment analysis of SlSTP1 and PeSTP6; Figure S5. Promoter sequence of PeSTP6. Table S1: Primers sequences used in this study.
Author Contributions
Formal analysis, L.L., R.G., S.N., J.Y. and Y.G.; investigation, L.L., L.Z., R.G., S.N. and Y.G.; data curation, L.L. and Y.G.; visualization, L.L., L.Z. and Y.G.; writing—original draft preparation, J.Y., L.L., Y.G. and C.F.; writing—review and editing, Y.G. and C.F.; supervision, C.F.; project administration, C.F.; funding acquisition, J.Y. and C.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Project of Sanya Yazhou Bay Science and Technology City (SCKJ-JYRC-2024-29 and SYND-2022-02) and the Key R&D Project of Baoting Li and Miao Autonomous County (BTZDYF2025001).
Data Availability Statement
Data are contained within the article and Supplementary Material.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA/Ethylene Responsive Factor (AP2/ERF) Transcription Factors: Mediators of Stress Responses and Developmental Programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Wei, X.; Li, D.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; Boshou, L.; Diouf, D.; Cisse, N.; et al. Insight into the AP2/ERF Transcription Factor Superfamily in Sesame and Expression Profiling of Dreb Subfamily under Drought Stress. BMC Plant Biol. 2016, 16, 171. [Google Scholar] [CrossRef] [PubMed]
- Dipp-Alvarez, M.; Cruz-Ramirez, A. A Phylogenetic Study of the Ant Family Points to a Preant Gene as the Ancestor of Basal and Euant Transcription Factors in Land Plants. Front. Plant Sci. 2019, 10, 17. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.-L.; Xing, G.-M.; Liu, J.-X.; Duan, A.-Q.; Xu, Z.-S.; Li, M.-Y.; Zhuang, J.; Xiong, A.-S. Advances in AP2/ERF Super-Family Transcription Factors in Plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Wei, K.; Hu, K.; Tian, T.; Zhang, F.; Yu, Z.; Zhang, D.; Su, Y.; Sang, Y.; Zhang, X.; et al. MPK14-Mediated Auxin Signaling Controls Lateral Root Development Via ERF13-Regulated Very-Long-Chain Fatty Acid Biosynthesis. Mol. Plant 2021, 14, 285–297. [Google Scholar] [CrossRef]
- Wessels, B.; Seyfferth, C.; Escamez, S.; Vain, T.; Antos, K.; Vahala, J.; Delhomme, N.; Kangasjarvi, J.; Eder, M.; Felten, J.; et al. An AP2/ERF Transcription Factor ERF139 Coordinates Xylem Cell Expansion and Secondary Cell Wall Deposition. New Phytol. 2019, 224, 1585–1599. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Hao, Z.; Tu, Z.; Shen, Y.; Zhang, C.; Wen, S.; Yang, L.; Ma, J.; Li, H. Genome-Wide Survey and Identification of AP2/ERF Genes Involved in Shoot and Leaf Development in Liriodendron chinense. BMC Genom. 2021, 22, 807. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Du, H.; Huang, Z.; He, M.; Kong, L.; Fang, C.; Chen, L.; Yang, H.; Zhang, Y.; Liu, B.; et al. The AP2/ERF Transcription Factor Toe4b Regulates Photoperiodic Flowering and Grain Yield Per Plant in Soybean. Plant Biotechnol. J. 2023, 21, 1682–1694. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Liu, L.; Liu, C.; Song, L.; Dong, Y.; Chen, L.; Li, M. Sweet Cherry AP2/ERF Transcription Factor, PAVRAV2, Negatively Modulates Fruit Size by Directly Repressing PavKLUH Expression. Physiol. Plant. 2023, 175, e14065. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhang, K.; Li, C.-Y.; Xie, G.-W.; Lu, M.-T.; Qian, Y.; Shu, Y.-P.; Shen, Q. Genome-Wide Comprehensive Characterization and Transcriptomic Analysis of AP2/ERF Gene Family Revealed Its Role in Seed Oil and Ala Formation in Perilla (Perilla frutescens). Gene 2023, 889, 147808. [Google Scholar] [CrossRef]
- Du, C.; Hu, K.; Xian, S.; Liu, C.; Fan, J.; Tu, J.; Fu, T. Dynamic Transcriptome Analysis Reveals AP2/ERF Transcription Factors Responsible for Cold Stress in Rapeseed (Brassica napus L.). Mol. Genet. Genom. 2016, 291, 1053–1067. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Nishida, S.; Shitan, N.; Sato, F. Genome-Wide Identification of AP2/ERF Transcription Factor-Encoding Genes in California Poppy (Eschscholzia californica) and Their Expression Profiles in Response to Methyl Jasmonate. Sci. Rep. 2020, 10, 18066. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhai, Y.; He, J.; Song, M.; Flaishman, M.A.; Ma, H. AP2/ERF Genes Associated with Superfast Fig (Ficus carica L.) Fruit Ripening. Front. Plant Sci. 2022, 13, 1040796. [Google Scholar] [CrossRef]
- Wan, R.; Song, J.; Lv, Z.; Qi, X.; Han, X.; Guo, Q.; Wang, S.; Shi, J.; Jian, Z.; Hu, Q.; et al. Genome-Wide Identification and Comprehensive Analysis of the AP2/ERF Gene Family in Pomegranate Fruit Development and Postharvest Preservation. Genes 2022, 13, 895. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.-C.; Han, Y.-C.; Qi, X.-Y.; Shan, W.; Chen, J.-Y.; Lu, W.-J.; Kuang, J.-F. Papaya CPERF9 Acts as a Transcriptional Repressor of Cell-Wall-Modifying Genes CPPME1/2 and CPPG5 Involved in Fruit Ripening. Plant Cell Rep. 2016, 35, 2341–2352. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.-K.; Li, X.; Xu, Q.; Chen, J.-Y.; Yin, X.-R.; Ferguson, I.B.; Chen, K.-S. EjAP2-1, an AP2/ERF Gene, Is a Novel Regulator of Fruit Lignification Induced by Chilling Injury, Via Interaction with EjMYB Transcription Factors. Plant Biotechnol. J. 2015, 13, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yuan, J.; Zhang, D.; Deng, K.; Chai, G.; Huang, Y.; Ma, S.; Qin, Y.; Wang, L. Genome-Wide Identification and Characterization of the SBP Gene Family in Passion Fruit (Passiflora edulis Sims). Int. J. Mol. Sci. 2022, 23, 14153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-S.; Xu, Y.; Xing, W.-T.; Wu, B.; Huang, D.-M.; Ma, F.-N.; Zhan, R.-L.; Sun, P.-G.; Xu, Y.-Y.; Song, S. Identification of the Passion Fruit (Passiflora edulis Sims) MYB Family in Fruit Development and Abiotic Stress, and Functional Analysis of PeMYB87 in Abiotic Stresses. Front. Plant Sci. 2023, 14, 1124351. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Ma, F.; Wu, B.; Lv, W.; Xu, Y.; Xing, W.; Chen, D.; Xu, B.; Song, S. Genome-Wide Association and Expression Analysis of the Lipoxygenase Gene Family in Passiflora edulis Revealing PeLOX4 Might Be Involved in Fruit Ripeness and Ester Formation. Int. J. Mol. Sci. 2022, 23, 12496. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Wu, B.; Chen, G.; Xing, W.; Xu, Y.; Ma, F.; Li, H.; Hu, W.; Huang, H.; Yang, L.; et al. Genome-Wide Analysis of the Passion Fruit Invertase Gene Family Reveals Involvement of PeCWINV5 in Hexose Accumulation. BMC Plant Biol. 2024, 24, 836. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Huang, D.; Zhang, S.; Wang, W.; Ma, F.; Wu, B.; Xu, Y.; Xu, B.; Chen, D.; Zou, M.; et al. Chromosome-Scale Genome Assembly Provides Insights into the Evolution and Flavor Synthesis of Passion Fruit (Passiflora edulis Sims). Hortic. Res. 2021, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Li, K.; Cheng, Y.; Li, M.; Liu, L.; Mur, L.A.J.; Luo, J.; Fang, C. PeWRKY20 Represses PeMDH1 to Modulate Malic Acid Metabolism and Flavor Formation in Postharvest Passion Fruit. Postharvest Biol. Technol. 2024, 218, 113164. [Google Scholar] [CrossRef]
- Thirugnanasambantham, K.; Durairaj, S.; Saravanan, S.; Karikalan, K.; Muralidaran, S.; Islam, V.I.H. Role of Ethylene Response Transcription Factor (ERF) and Its Regulation in Response to Stress Encountered by Plants. Plant Mol. Biol. Report. 2015, 33, 347–357. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Dan, Y.; Niu, Y.; Wang, C.; Yan, M.; Liao, W. Genome-Wide Identification and Expression Analysis of the Trehalose-6-Phosphate Synthase (TPS) Gene Family in Cucumber (Cucumis sativus L.). PeerJ 2021, 9, e11398. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Wei, Y.; Xu, R.; Lin, S.; Luan, H.; Lv, C.; Zhang, X.; Song, X.; Xu, R. Genome-Wide Analysis of Apetala2/Ethylene-Responsive Factor (AP2/ERF) Gene Family in Barley (Hordeum vulgare L.). PLoS ONE 2016, 11, e0161322. [Google Scholar] [CrossRef]
- Yang, H.; Sun, Y.; Wang, H.; Zhao, T.; Xu, X.; Jiang, J.; Li, J. Genome-Wide Identification and Functional Analysis of the ERF2 Gene Family in Response to Disease Resistance against Stemphylium lycopersici in Tomato. BMC Plant Biol. 2021, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Haxim, Y.; Liang, Y.; Qiao, S.; Gao, B.; Zhang, D.; Li, X. Genome-Wide Investigation of AP2/ERF Gene Family in the Desert Legume Eremosparton songoricum: Identification, Classification, Evolution, and Expression Profiling under Drought Stress. Front. Plant Sci. 2022, 13, 885694. [Google Scholar] [CrossRef]
- Guo, Z.; He, L.; Sun, X.; Li, C.; Su, J.; Zhou, H.; Liu, X. Genome-Wide Analysis of the Rhododendron AP2/ERF Gene Family: Identification and Expression Profiles in Response to Cold, Salt and Drought Stress. Plants 2023, 12, 994. [Google Scholar] [CrossRef]
- Jarambasa, T.; Regon, P.; Jyoti, S.Y.; Gupta, D.; Panda, S.K.; Tanti, B. Genome-Wide Identification and Expression Analysis of the Pisum sativum (L.) Apetala2/Ethylene-Responsive Factor (AP2/ERF) Gene Family Reveals Functions in Drought and Cold Stresses. Genetica 2023, 151, 225–239. [Google Scholar] [CrossRef]
- Zhuang, J.; Zhu, B. Analysis of Brassica napus Ests: Gene Discovery and Expression Patterns of AP2/ERF-Family Transcription Factors. Mol. Biol. Rep. 2014, 41, 45–56. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Zhao, B.-Y.; Ye, X.; Du, J.; Song, J.-L.; Wang, W.-J.; Huang, X.-L.; Ouyang, K.-X.; Zhang, X.-Q.; Liao, F.-X.; et al. Genome-Wide Analysis of the AP2/ERF Gene Family in Pennisetum glaucum and the Negative Role of PgRAV_01 in Drought Tolerance. Plant Physiol. Biochem. 2024, 216, 109112. [Google Scholar] [CrossRef]
- Rashid, M.; He, G.; Yang, G.; Hussain, J.; Yan, X. AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots. Evol. Bioinform. 2012, 8, 321–355. [Google Scholar] [CrossRef]
- Riaz, M.W.; Lu, J.; Shah, L.; Yang, L.; Chen, C.; Mei, X.D.; Xue, L.; Manzoor, M.A.; Abdullah, M.; Rehman, S.; et al. Expansion and Molecular Characterization of AP2/ERF Gene Family in Wheat (Triticum aestivum L.). Front. Genet. 2021, 12, 632155. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.-Y.; Chen, L.-H.; Fan, B.-L.; Xu, Z.; Wang, Q.; Zhao, B.-Y.; Gao, M.; Yuan, M.-H.; ul Qamar, M.T.; Jiang, Y.; et al. Integrative Multiomics Profiling of Passion Fruit Reveals the Genetic Basis for Fruit Color and Aroma. Plant Physiol. 2024, 194, 2491–2510. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.P.; Crabtree, J.; Zhao, Q.; Lorenzi, H.; Orvis, J.; Puiu, D.; Melake-Berhan, A.; Jones, K.M.; Redman, J.; Chen, G.; et al. Draft Genome Sequence of the Oilseed Species Ricinus communis. Nat. Biotechnol. 2010, 28, 951–953. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Cai, B.; Peng, R.-H.; Zhu, B.; Jin, X.-F.; Xue, Y.; Gao, F.; Fu, X.-Y.; Tian, Y.-S.; Zhao, W.; et al. Genome-Wide Analysis of the AP2/ERF Gene Family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Han, S.; Wang, H.; Yu, M.; Ma, R.; Yu, Z. The Regulation of 1-Methylcyclopropene Treatment on the Subfamily Genes Expression of Ethylene Response Factors in Peaches during Storage. Acta Sci. Pol. Technol. Aliment. 2021, 20, 313–323. [Google Scholar] [CrossRef]
- Zhang, J.; Yin, X.-R.; Li, H.; Xu, M.; Zhang, M.-X.; Li, S.-J.; Liu, X.-F.; Shi, Y.-N.; Grierson, D.; Chen, K.-S. Ethylene Response Factor39-Myb8 Complex Regulates Low-Temperature-Induced Lignification of Loquat Fruit. J. Exp. Bot. 2020, 71, 3172–3184. [Google Scholar] [CrossRef]
- Soares, C.G.; do Prado, S.B.R.; Andrade, S.C.S.; Fabi, J.P. Systems Biology Applied to the Study of Papaya Fruit Ripening: The Influence of Ethylene on Pulp Softening. Cells 2021, 10, 2339. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shi, C.; Ge, P.; Li, F.; Zhu, L.; Wang, Y.; Tao, J.; Zhang, X.; Dong, H.; Gai, W.; et al. A 21-bp Indel in the Promoter of STP1 Selected during Tomato Improvement Accounts for Soluble Solid Content in Fruits. Hortic. Res. 2023, 10, uhad009. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped Blast and Psi-Blast: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) V3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCSCANX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Li, K.; Cheng, Y.; Fang, C. OsDWARF10, Transcriptionally Repressed by OsSPL3, Regulates the Nutritional Metabolism of Polished rice. Front. Plant Sci. 2023, 14, 1322463. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Guo, Z.; Wang, K.; Wang, R.; Fang, C. Comparative Metabolomic Analysis Reveals the Role of OsHPL1 in the Cold-Induced Metabolic Changes in Rice. Plants 2023, 12, 2032. [Google Scholar] [CrossRef] [PubMed]
- Jessica, B.; Leif, V.; Christine, W.; Russel, P.B.; Craig, P.H.; Qi, L.; Lynn, E.; Sandra, E.S. HIP: A Method for High-Dimensional Multi-View Data Integration and Prediction Accounting for Subgroup Heterogeneity. Brief. Bioinform. 2024, 25, bbae470. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).