Abstract
The utilization of nitrogen (N) is crucial for the optimal growth and development of plants. As the dominant form of nitrogen in temperate soil, nitrate (NO3−) is absorbed from the soil and redistributed to other organs through NO3− transporters (NRTs). Therefore, exploration of the role of NRTs in response to various NO3− conditions is crucial for improving N utilization efficiency (NUE). Here, we present a comprehensive genome-wide analysis and characterization of the NRT gene family in Korean pine, an invaluable tree species cultivated extensively in northeastern China. A total of 76 PkNRTs were identified in Korean pine and further divided into three subfamilies (NRT1/NPF, NRT2, and NRT3) based on phylogenetic analysis. All PkNRTs were distributed on 11 chromosomes, with multiple tandem duplications observed. The tissue-specific expression analysis indicated that most PkNRTs showed differential expression in six vegetative tissues. Furthermore, a significantly greater number of lateral roots was observed in seedlings under nitrogen-deficient conditions, accompanied by an increase in both total root biomass and root length. The temporal expression profiles of 16 PkNRTs in seedling roots revealed that four PkNRTs, PkNPF5.6, PkNPF5.13, PkNPF6.1, and PkNPF6.2, exhibited significantly upregulated expression under the NO3− deficiency condition, whereas robust induction was observed for PkNPF1.1, PkNRT2.6, and PkNRT3.3 upon the NO3− sufficiency condition. The expression patterns of the PkNRTs suggest their potential diverse roles as key participants in root NO3− uptake under varying NO3− conditions during root development. These findings would provide a theoretical foundation for further investigations into the functions of PkNRTs in Korean pine.
1. Introduction
Nitrogen (N) is an essential macronutrient for most of higher-plant growth and productivity, playing critical roles in various physiological and metabolic processes [1,2]. The dominant form of N in temperate soils is nitrate (NO3−), accompanied by ammonium (NH4+), amino acids, and other N-containing substances. The appropriate application of N can enhance crop yields, whereas the excessive use of N may have opposite effects on plants [3,4]. Therefore, the understanding of regulatory mechanisms underlying NO3− acquisition processes is crucial for optimizing plants’ N use efficiency (NUE) capacity.
Roots play a vital role in N absorption from the soil for plants. Plants adapt their N uptake and transport mechanisms through modification in root architecture and the optimization of metabolic processes [5]. The initial step in triggering nitrogen responses is activating N absorption mediated by transporters at the root surface. To adapt to different concentrations of NO3− in soil, plant roots have evolved a low-affinity transport system (LATS, >1 mM) and high-affinity transport system (HATS, <1 mM) for assisting NO3− uptake [6]. Most NO3− obtained by plants from soil is actively transported through a group of NO3− transporters (NRTs), which constitute a diverse family with many members and distinct functions [7,8]. Several genes from the NRT family have been identified in HATS and LATS with apparent function. Based on sequence homology and functional characteristics, NRTs are primarily grouped into three subfamilies: the Nitrate Transporter 1/Peptide Transporter (NRT1/PTR, also known as NPF), Nitrate Transporter 2 (NRT2), and NRT3 (as known as NAR) families [9,10]. NRT1 and NRT2 function as NO3− transporters responsible for LATS and HATS, respectively [11]. Unlike NRT1 and NRT2, NRT3 members lack the inherent ability for NO3− transportation; instead, they play important roles in high-affinity NO3− transport by regulating the activity of NRT2 proteins [12]. In addition to their involvement in NO3− transport, NRT proteins have been reported to participate in a diverse range of physiological processes in plants, including shoot/root development, hormone transportation, and stress signaling response [13,14].
Up to now, many members in the NRT family have been identified in higher plants, while only a few have been well characterized. Among the three subfamilies, the NRT1/NPF clade exhibits greater a abundance of members than the NRT2 and NRT3 subfamilies, indicating the significant roles of NRT1/NPF in NO3− absorption, translocation, and assimilation [15,16]. AtNRT1.1 (also known as CHL1/NPF6.3) was the first identified member of the NRT family in Arabidopsis, which has dual affinity (low and high affinity) for NO3− absorption and root-to-shoot transport [17,18]. Except for AtNRT1.1, most NRT1 members exhibited low affinity. For example, NPF4.6/NRT1.2 and NPF2.7/NAXT1 had low affinity for NO3− absorption, and were also shown to be involved in root NO3− uptake, with NPF4.6 acting on NO3− influx and NPF2.7 involved in NO3− efflux, respectively [19,20]. The other identified NRTs have been reported to play a crucial role in the internal transport of NO3−, including processes such as xylem and phloem loading, as well as translocation to leaves or seeds [21]. The OsNPF2.4 functions as a low-affinity transporter in the uptake of NO3− and N recycling in rice [22]. The mutations in AtNRT1.9/NPF2.9 resulted in a decrease in NO3− content within the root xylem and an increase in NO3− content within the shoot, thereby impacting the allocation of NO3− between the root and shoot [23]. In Arabidopsis, HATS is mediated by the interaction between NRT2 with NPF6.3. The expression of AtNRT2.4 is upregulated in both roots and shoots in response to N starvation. The overexpression of AtNRT2.4 enhances the facilitation of NO3− absorption by the roots and its subsequent translocation from the roots to the phloem [24]. Although the characterization of some NRTs has been accomplished, there remains a lack of comprehension regarding the functions of the majority of NRTs in N uptake, transport, and utilization. Therefore, further investigation and verification of NRT protein functions in plants are essential for advancements in N fertilization.
Korean pine (Pinus koraiensis) is an invaluable tree species in northeastern China, holding significant ecological, economic, and social importance [25,26]. Because of the high content of various nutrients in pine nuts and excellent wood properties, Korean pine is cultivated extensively in northeast China. The current limitation of forest growth in northern China is due to the low availability of nitrogen in the soil. Hence, N fertilization plays a vital role in expediting the growth rate of Korean pine, given its slow growth rate and lengthy reproductive cycle. Its NUE can be effectively improved by increasing the absorption capacity of NO3− in roots and the redistribution capacity of NO3− in other tissues [27]. Though great progress has been achieved for plant NRT genes, there is a lack of research on gymnosperms, particularly Korean pine.
In the present study, genome-wide identification and comprehensive bioinformatics analyses of the NRTs in Korean pine were performed. A total of 76 NRTs were identified and characterized, including chromosomal location, genetic structure, and the phylogenetic relationship of the PkNRT gene family. The tissue-specific expression pattern of the PkNRTs was also explored. Furthermore, the phenotypic responses of Korean pine seedlings to nitrogen deficiency treatments were evaluated, along with the expression profiles of specific PkNRTs expressed in roots. These findings will facilitate the identification of PkNRT functions, thereby offering new insights into elucidating the molecular regulatory network of nitrogen uptake and transport in Korean pine.
2. Results
2.1. Characterization of the PkNRT Gene Family Members in Korean Pine
A total of 76 PkNRT proteins were identified that contained NRT or NRT-like repeats, including 66 NRT1/NPF, 6 NRT2, and 4 NRT3 members, in Korean pine (Table 1). The predicted number of amino acids varies widely among the three subfamilies of PkNRTs. The number of amino acids in the NPF and NRT2 subfamilies ranges from 533 (PkNRT2.4) to 661 (PkNPF2.1), with the corresponding molecular weight ranges from 57.87 kDa to 73.07 kDa. In contrast, the members in the NRT3 subfamily have notably shorter amino acid sequences (207aa–268aa) and lower molecular weights (22.58–23.98 kDa). The theoretical predicted isoelectric point (pI) range of NRT proteins is 5.25 to 9.73, and the pI values of most members (65/76) are greater than 7, indicating that most NRT proteins tend to be positively charged at a physiological pH. In terms of the hydrophobicity analysis, most PkNRTs are hydrophobic, and only PkNRT3.4 possesses the negative GRAVY value, indicating its potential as a hydrophilic protein. Additionally, the number of transmembrane domains (TMs) in the NPF and NRT2 proteins is between 8 and 12, except for the NRT3 protein, which has only 1 TM.

Table 1.
Characteristics of the PkNRT genes in Korean pine.
2.2. Phylogenetic Analysis of PkNRTs
The phylogenetic relationships among the NRT gene family members in Arabidopsis, Poplar, Ginkgo, and Korean pine were elucidated through the construction of a phylogenetic tree. As expected, the PkNRTs were divided into three subfamilies: NPF/NRT1, NRT2, and NRT3 (Figure 1). The NPF subfamily comprised the majority of the NPF/NRT1 members, and it was further classified into eight clusters (NPF1–NPF8). Among them, NPF5 has the most members (NPF5.1–NPF5.32), and NPF3 has the fewest member (NPF3.1). The members of NPF5 were further categorized into three distinct subgroups. The PkNPF1 and PkNPF2 subfamilies are closely positioned on the phylogenetic tree, sharing the same branch, which suggests that these subfamilies may be evolutionarily closely related. In addition, the PkNRTs were found to cluster more closely with those of Ginkgo than those of Arabidopsis, indicating that Korean pine and Ginkgo may be more closely related evolutionarily.
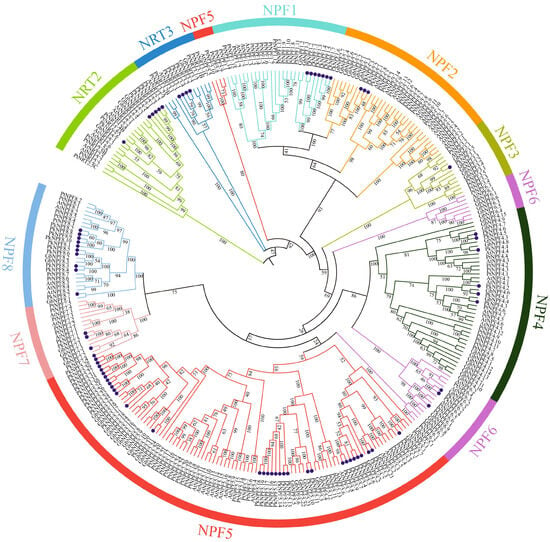
Figure 1.
Phylogenetic analysis of NRT proteins from Arabidopsis thaliana, Populus trichocarpa, Ginkgo biloba, and Pinus koraiensis. The NRT proteins were classified into three subfamilies: NPF/NRT1, NRT2, and NRT3. NPF/NRT1 can be further classified into eight subfamilies (NPF1–NPF8). The dark blue dots indicate the PkNRT proteins.
2.3. Motifs, Conserved Domain, and Gene Structure Analysis of PkNRTs
After analyzing the conserved motifs, it was observed that a majority of PkNRTs exhibited a consistent distribution pattern and contained all identified motifs (Figure 2A–C). Only a few genes displayed atypical motif distributions, such as PkNPF5.5 in the NPF subfamily, PkNRT2.6 in the NRT2 subfamily, and PkNRT3.1 in the NRT3 subfamily. Although the motifs in the NPF subfamily are highly conserved, gene structure analysis revealed that certain genes within this subfamily, such as PkNPF4.5, PkNPF4.6, PkNPF5.6, PkNPF8.9, PkNPF8.10, PkNPF8.1, and PkNPF8.8, possess much longer introns, which impact the length of their genomic sequences (Table S2). Through the analysis of conserved structural domains, it was found that the NPF subfamily has different superfamily structural domains, including MFS_NPF1_2(cd17416), MFS_NPF3(cd17415), MFS_NPF4(cd17414), MFS_NPF5(cd17417), MFS_NPF6(cd17413), MFS_NPF7(cd17419), and the MFS superfamily(cl28910) (Figure 2D). The NRT2 and NRT3 subfamilies have only one highly conserved structural domain, named the PLN00028(cl30556) and NAR2(cl25236) domains, respectively (Figure 2E,F).
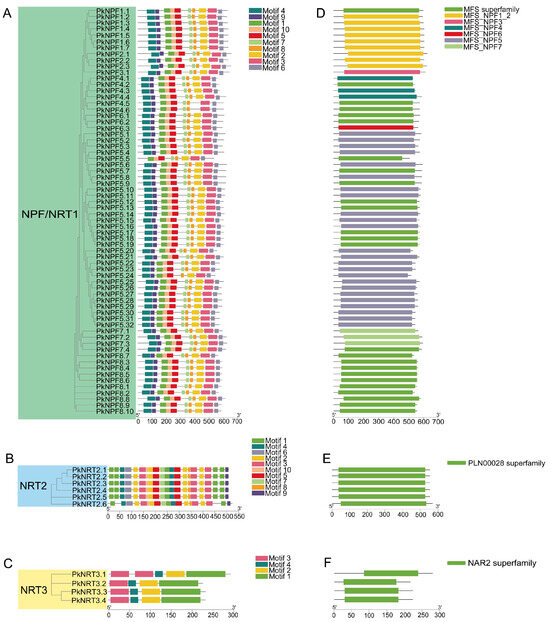
Figure 2.
The motif, gene structure, and conserved domains of PkNRTs. (A–C) The motif analysis of NRT1/NPF, NRT2, and NRT3 proteins. (D–F) The conserved domains of PkNRTs. All detailed information of identified conserved motif sequences is provided in Table S1. Gene structure detailed information is provided in Table S2. A scale bar is provided at the bottom.
2.4. Analysis of Chromosomal Distribution and Collinearity of PkNRTs
According to the chromosomal location analysis, it was found that the PkNRTs were distributed on 11 chromosomes of Korean pine, except for Chr03 (Figure 3). The density of PkNRTs also exhibited variation among the chromosomes. Chr09 possessed the highest number of PkNRT genes, whereas Chr01 had only three PkNRTs. For the NPF subfamily members, most members of PkNPF5 were found on Chr02, Chr06, and Chr07, which aligns with the three subgroups identified in the phylogenetic analysis. The distribution of PkNPF8 proteins was observed on Chr05 and Chr09, whereas all members of PkNPF1 and PkNPF2 were found to be located on Chr04 and Chr10, respectively. The members of the NRT3 subfamily, similarly to those of PkNPF1 and PkNPF2, exhibited exclusive distribution on Chr09. Additionally, for the NRT2 subfamily, four out of the six NRT2 members were distributed on Chr12. It is worth noting that a large number of PkNRTs were clustered in adjacent positions on the chromosome (such as the PkNPF2, PkNPF5, PkNPF8, PkNRT2, and PkNRT3 subfamilies), indicating that the PkNRTs may have undergone frequent duplication events during evolution.
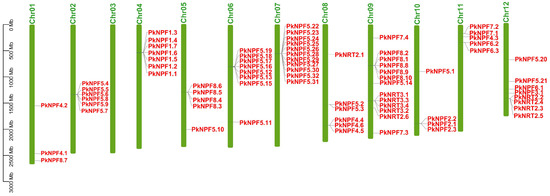
Figure 3.
Chromosomal location of PkNRTs. The chromosome numbers are shown above. The unit of scale is in megabases (Mb).
The analysis of collinearity between species is of great significance for further studying the evolutionary process of the NRT family. Through collinearity analysis between Korean pine, Poplar and Ginkgo, it was found that Korean pine had 14 direct collinear gene pairs with Ginkgo, and these genes were included in the PkNPF and PkNRT2 subfamilies (Figure 4, Table S3). In contrast, the evolutionary relationship between Korean pine and Poplar was relatively distant, with only one collinear gene pair belonging to the NPF8 subfamily in common. Furthermore, we conducted a collinearity analysis comparing Korean pine with rice and Arabidopsis, two model plants in angiosperms. The results revealed no presence of a collinear gene pair in NRTs among Korean pine, rice, and Arabidopsis (Figure S1). In addition, the ratio of non-synonymous (Ka) to synonymous substitution (Ks) rates was calculated for the 15 direct collinear gene pairs to determine whether there was selective pressure acting on these protein-coding genes (Table S3). The Ka/Ks ratios of the NRT gene pairs in these species are less than 1, indicating that these genes primarily underwent purifying selection during evolution. This purifying selection suggested that deleterious non-synonymous mutations are selectively eliminated to preserve their function, thereby maintaining their crucial roles in NO3− absorption and transport. In addition, we calculated the Ka/Ks ratios of tandemly duplicated gene pairs in PkNRTs (Table S4). The results revealed that most gene pairs have undergone purifying selection (Ka/Ks < 1), indicating that these NRT genes were relatively conserved during the evolutionary process.
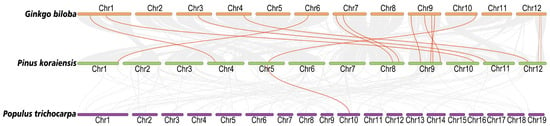
Figure 4.
Collinearity analysis of NRTs between Ginkgo biloba and P. koraiensis and P. trichocarpa. The gray lines represent the collinearity between P. koraiensis and other two species. Collinear gene pairs in the NRT gene family are marked with red lines.
2.5. Characterization of CREs in the Promoter Regions of PkNRTs
To further investigate the potential functions of the NRT gene family, the cis-regulatory elements (CREs) were analyzed (Figure S2). The results showed that CREs identified in the promoter of PkNRT genes were mainly divided into three categories: plant development, hormones, and stress (Table S5). Among them, CREs related to plant development were CCAAT-box, which plays an important role in early plant growth and tissue differentiation. The CREs associated with abiotic stress responses, such as the MBS element implicated in drought induction and the LTR element involved in cold response, were also found in PkNRT promoters. Additionally, the hormone-responsive elements were enriched in PkNRT promoters (Figure S2), including auxin response elements (AuxRR-core, TGA-box, and TGA-element), gibberellin response elements (GARE-motif and P-box), abscisic acid response elements (ABRE), salicylic acid response elements (SARE and TCA-element), and jasmonic acid response elements (CGTCA-motif and TGACG-motif), which suggests that hormones may play a potential role in regulating the expression of PkNRTs.
2.6. Transcript Abundance Analysis for Different Tissues in PkNRTs
To explore the tissue-specific expression of PkNRTs, we conducted further investigation into their expression pattern by utilizing transcriptome data obtained from six different tissues (needle, phloem, xylem, bud, shoot, and root) (Table S6). The expression patterns of certain genes in specific tissues were observed (Figure 5A). For example, PkNPF1.1, PkNPF1.2, PkNPF1.3, PkNPF4.3, PkNPF5.6, PkNPF5.7, PkNPF5.18, and PkNRT3.2 exhibited high expression levels specifically in the root. A total of 13 NRTs were highly expressed in the needle, including PkNPF1.5, PkNPF2.1, PkNPF2.2, PkNPF3.1, PkNPF5.2, PkNPF5.9, PkNPF5.15, PkNPF5.21, PkNPF6.2, PkNPF7.3, PkNPF8.5, PkNRT2.5, and PkNRT3.4. As the only member of the PkNPF3 family, the expression of PkNPF3.1 was found to be highest in the bud, followed by the root and needle. In addition, the majority of PkNPF4 members exhibited specific expression in phloem and xylem. PkNPF4.2 and PkNPF4.4 showed high expression only in the xylem, while the expression of PkNPF4.1 and PkNPF4.5 was detected in both the phloem and xylem, indicating their crucial roles in the internal NO3− transport in xylem and phloem loading or unloading. The expression analysis of PkNRTs, which ranked among the top 12 in each tissue, revealed that PkNPF5.2, PkNPF5.21, PkNPF6.1, and PkNPF8.5 showed high expression in at least five tissues (Figure 5B), suggesting potential roles in governing the fundamental function of N element transport in Korean pine. In contrast, the expression levels of nine genes (PkNPF1.4, PkNPF1.6, PkNPF4.6, PkNPF5.24, PkNPF6.3, PkNPF8.2, PkNPF8.3, PkNRT2.1, and PkNRT2.3) were consistently low in each tissue (Figure 5C). These findings suggest potential roles of PkNRTs in the absorption and transport of NO3− within specific tissues, thereby facilitating N utilization during the growth of Korean pine.
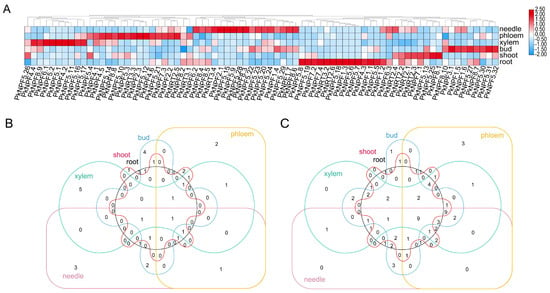
Figure 5.
Transcriptome-based expression profiling of PkNRTs. (A) Expression of PkNRTs from six Korean pine tissues including root, shoot, bud, xylem, phloem, and needle. (B) The Venn diagram of the top 12 most highly expressed PkNRTs involved in each tissue. (C) The Venn diagram of PkNRTs with the lowest expression levels (FPKM < 0.5) between different tissues.
2.7. Effects of N Treatment on Growth of Korean Pine Seedlings
To investigate the impact of N on the growth of Korean pine seedlings, 1-month-old seedlings were treated with either a N-containing Hoagland nutrient solution (referred to as ‘NS’ below) or a N-deficient solution (referred to as ‘ND’ below) for 12 weeks. No morphological changes were visible in the needle and hypocotyl of seedlings in response to different N treatments, whereas the lateral root growth of the seedlings under ND treatment exhibited a significant increase (Figure 6A). The biomass of the seedlings was further evaluated under ND and NS treatments. The biomass of the needle and hypocotyl significantly increased under the NS condition, whereas the biomass of the root exhibited a significant increase under the ND condition (Figure 6B–D). The phenotypes of the seedling roots were measured to confirm the impact of N on the formation of lateral roots. The number of lateral roots, total root length, and root surface area of the seedlings were significantly increased under the ND condition (Figure 6E–G). However, the root diameter of the seedlings decreased upon nitrogen deprivation (Figure 6H).
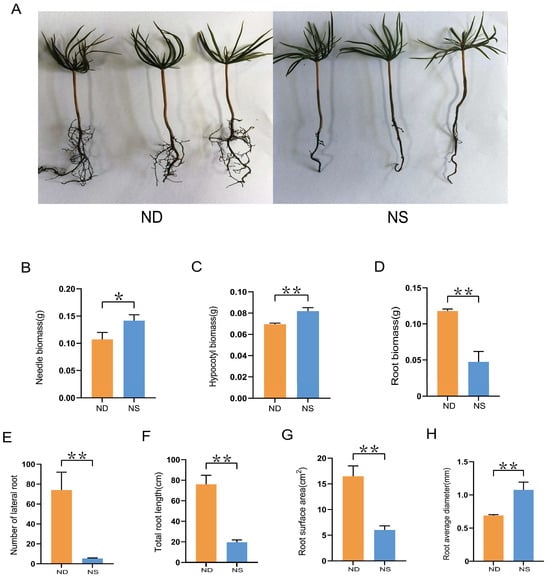
Figure 6.
Effect of nitrogen treatment on the growth of Korean pine seedlings. (A) Phenotype of seedlings treated with or without nitrogen for 12 weeks. (B–D) Biomass of needle, hypocotyl, and root under ND and NS conditions. (E) Number of lateral roots. (F) Total root length of seedlings. (G) Root surface area. (H) Root mean diameter. Bars show means of three biological replicates ± standard error. Asterisk indicates significant difference between nitrogen treatment using Student’s t-test (* p < 0.05; ** p < 0.01).
2.8. Expression Profiles of PkNRTs Under Different N Conditions
To investigate the roles of PkNRTs under N treatment, we subjected seedlings to a 28-day N treatment to elucidate the time-dependent expression patterns of root-specific expressed PkNRTs (Figure 7). The following genes showed increased expression in response to NS treatment: PkNPF1.1, PkNPF3.1, PkNPF5.18, PkNPF7.2, PkNPF7.3, PkNRT2.4, PkNRT2.6, PkNRT3.2, and PkNRT3.3. Meanwhile, the expression levels of PkNPF5.27 and PKNPF6.1 were observed to decrease. Conversely, some genes were induced under the ND condition (e.g., PkNPF3.1, PkNPF5.6, PkNPF5.13, PkNPF5.18, PKNPF6.1, PkNPF6.2, PkNPF8.5, PkNRT2.4, PkNRT2.6, and PkNRT3.2), whereas reduced expression was observed for PkNPF1.1, PkNPF5.27, and PKNRT3.3. Based on a qRT-PCR analysis, we observed the induction of PkNPF3.1, PkNPF5.18, PkNPF7.2, PkNPF7.3, PkNRT2.6, and PkNRT3.2 through both ND and NS treatment during root development. In addition, the expression levels of PkNPF5.6, PkNPF5.13, PkNPF6.1, and PkNPF6.2 were sharply upregulated in response to ND treatment, whereas the expression levels of PkNPF1.1, PkNRT2.6, and PkNRT3.3 were robustly induced in response to N supply and higher than those observed under the ND condition. Notably, the expression of PkNPF6.1 was induced by N starvation and repressed by N supply, whereas PkNPF1.1 and PkNRT3.3 exhibited the opposite expression pattern during the prolonged N treatment. The expression patterns of PkNRTs varied under ND and NS treatment, implying the potential diverse roles of PkNRTs in the process of lateral root development.
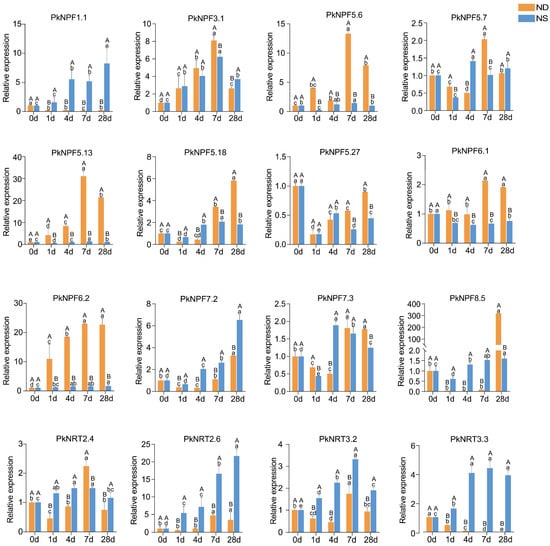
Figure 7.
Relative expression of 16 selected PkNRTs under different nitrogen conditions. The expression levels of selected PkNRTs in roots of Korean pine seedlings after 1 d, 4 d, 7 d, and 28 d under NS and ND treatment was calculated by qRT-PCR, with PkActin as the reference gene. Capital letters indicate significant differences in expression levels between the two different treatments (ND and NS) at the same time (p < 0.05), while lowercase letters denote significant differences in expression levels at different times (0 d, 1 d, 4 d, 7 d, and 28 d) under the same treatments (p < 0.05). Bars show the means of three biological replicates ± standard error.
3. Discussion
N is widely used not only in the cultivation of agricultural crops, but also in plantation silviculture. Forest growth in northern China is currently limited by drought and low N availability [28]. Korean pine plays a dominant role in the formation of broad-leaved Korean pine mixed forest (BKF), which is a typical type of forest vegetation found in the northeast of Asia [29]. The response of Korean pine growth to climate change, specifically in relation to continuous N deposition and drought, has been extensively investigated [30,31]. However, the molecular mechanism underlying N absorption and transportation in Korean pine remains poorly understood. The majority of plants acquire N through their roots, with NO3− serving as the primary N source in soil. The NRT proteins have been identified as crucial NO3− transporters involved in the uptake and translocation of NO3− in plants. Here, we performed the first comprehensive characterization of the NRT gene family and provided novel insights into its functional roles under different N conditions.
In higher plants, the members of the NRT family are abundantly present throughout the genome, with the majority belonging to the NRT1/NPF subfamily and exhibiting a high sequence similarity [32,33,34]. Our genome-wide analysis revealed a total of 76 PkNRT genes in Korean pine, with the majority (66/76) belonging to the NPF/NRT1 subfamily (Figure 1). The members of the NPF subfamily were further classified into eight clusters based on distinct structural domains identified in Ginkgo [35]. According to the phylogenetic analysis, the NRT proteins in Korean pine exhibited a closer clustering with those in Ginkgo, while PkNRTs exhibited a more distant evolutionary relationship with three model plants in angiosperms in the collinearity analysis (Figure 4 and Figure S1). The homology of NRT genes between Ginkgo and Poplar is closer than that between Ginkgo and Arabidopsis [35]. The findings indicate that there may be functional diversity within the NRT family in gymnosperms, as evidenced by significant differences in evolutionary trajectories between gymnosperms and angiosperms.
The expansion of gene families invariably leads to the tandem duplication of gene clusters within the genome. Variations in the number of PkNPF subfamily members were observed. For instance, the PkNPF5 clade comprises 32 members, indicating a twofold increase compared to Arabidopsis [36]. Conversely, the PkNPF2 and PkNPF3 clades consist of only three members and one member, respectively, which is significantly fewer than in Arabidopsis and other plants [16,37,38]. Additionally, high expression levels of NPF5 subfamily members across a wide range of tissues suggest that the NPF5 family plays multiple roles in nitrogen uptake and transport. Additionally, the functional diversification of the NPF5 family in Korean pine was complemented by the expansion and diversification of the NPF2 and NPF3 families in Arabidopsis during evolution. This evolutionary process resulted in a relative reduction in the number of NPF5 members and an expansion of the NPF2 and NPF3 families in Arabidopsis. This suggests that gene expansion and deletion events have also occurred within the PkNPF subfamily. Similar findings were also observed in other plants, such as Ginkgo [35], Pineapple [9], wild soybean [39], and radish [16]. Moreover, we found that most PkNRTs exhibited a tandem distribution pattern across the chromosomes, including members in NPF1, NPF2, NPF5, NPF8, NRT2, and NRT3 (Figure 3). The members involved in tandem regions exhibited a more closely related evolutionary relationship in the phylogenetic analysis, indicating potential functional redundancy among these genes.
Given that all members of the NRT gene family are transmembrane proteins, the tissue-specific expression of NRTs provides a comprehensive understanding of their functional roles across diverse tissues and developmental stages [21]. Here, we elucidated the gene expression patterns of PkNRTs across different tissues. The majority of PkNRTs exhibited high expression levels in needles or roots, indicating their crucial role in NO3− uptake by the roots and subsequent redistribution to the needles for various physiological processes in plants (Figure 5). Notably, the expression of members within the NPF4 clade predominantly occurred in the xylem and phloem, whereas the NPF2 clade exhibited high expression in both the needle and phloem, indicating that these two clades of PkNPFs function in NO3− transport in aboveground tissues [40]. Furthermore, the expression of several of these genes is significantly higher in almost all tissues, indicating their potential fundamental roles in NO3− uptake and transport. In contrast, nine PkNRTs showed low expression levels in all detected tissues. Some of these genes, such as PkNPF4.6, PkNPF6.3, and PkNPF8.2 possess fewer cis-regulatory elements in their promoters. The promoters of other genes such as PkNPF1.4, PkNPF1.6, and PkNPF8.3 contain numerous MeJA-responsive elements, while promoters of PkNPF2.1 and PkNPF2.3 harbor drought-responsive elements, indicating that these genes may play roles in plant stress responses or hormonal regulation. However, it should be noted that the expression of NRTs may be regulated by many other factors, such as soil pH, drought, cold, and salinity, as well as hormones [41,42,43,44]. In fact, we have identified numerous hormone-, drought-, and cold-responsive CREs in the promoter region of PkNRTs (Figure S2). Therefore, the lower expression of PkNRTs may also indicate their function in other undetected tissues, such as the callus or bark, or in response to hormones or cold. Further investigations will be conducted to elucidate the expression pattern of PkNRTs in response to abiotic stress and hormone stimuli. Although further functional analysis is required to identify the roles of PkNRTs, their expression patterns may provide insight into their potential functions in NO3− acquisition and transport within Korean pine.
The presence of NO3− is crucial for root growth, development, and architecture, especially in relation to lateral root development [45]. A high and consistent concentration of NO3− in the growth medium inhibits the development of lateral roots, which is associated with the accumulation of NO3− and N metabolites within the plants [46]. In many plants, the stimulation of lateral root growth was observed under low NO3− conditions, with a significant increase in lateral root number, total root length, and root biomass [47,48,49]. Similarly, we observed that N deficiency promoted lateral root growth in Korean pine seedlings, as evidenced by an increase in their number and biomass (Figure 6). Expression changes in response to different N concentrations reveal the potential roles of NRT proteins for NO3− absorption and transport. The expression of high-affinity NO3− transporters, such as NRT2.1, NRT2.4, NRT2.5, and NPF6.3, was induced in response to N starvation to enhance root NO3− acquisition [11,18]. The majority of members in the NPF/NRT1 subfamily, such as NPF7.3, NPF4.6, and NPF2.7, which function as low-affinity NO3− transporters, exhibited an increased expression in response to N supply [50,51]. In the present work, the expression of 16 PkNRTs, which have been identified to exhibit high or specific expression in the root, was further detected under the ND and NS conditions. The expression levels of PkNRTs in seedling roots displayed differential patterns (Figure 7). Among the PkNRTs, the significant upregulation of PkNPF1.1, PkNRT2.6, and PkNRT3.3 expression under NS conditions suggested their critical roles as low-affinity NO3− transporters in root NO3− uptake. Meanwhile, PkNPF5.6, PkNPF5.13, PkNPF6.1, and PkNPF6.2 were identified as potential members of HATS for efficient NO3− uptake due to their robustly upregulated expression levels in response to ND treatment, which exhibited a strong correlation with lateral root development.
4. Materials and Methods
4.1. Identification of NRT Gene Family in Korean Pine
In order to identify all NRT gene family members in Korean pine, a BLASTP search was conducted in the full protein sequence isolated from P. koraiensis genome by using the NRT protein sequences of Arabidopsis thaliana, Ginkgo biloba, and Populus trichocarpa as a query retrieved from the TAIR (https://www.arabidopsis.org/, accessed on 17 July 2024), Ginkgo DB (https://ginkgo.zju.edu.cn/genome/ftp/, accessed on 17 July 2024), and phytozome databases (https://phytozome-next.jgi.doe.gov/, accessed on 17 July 2024), respectively. The complete genome of P. koraiensis and annotation information were obtained from our group. The protein sequences of P. koraiensis utilized in this study can be found in Table S8. The sequences were submitted to the Pfam database (http://pfam.xfam.org/, accessed on 18 July 2024) for screening and identification of the PTR2 (PF00854) core domain in the NPF subfamily, the MSF_1 (PF07690) core domain in the NRT2 subfamily, and the NAR2 (PF16974) core domain in the NRT3 subfamily. After eliminating the redundant sequences, the biophysical properties, such as amino acid length, theoretical isoelectric point (pI), molecular weight (Mw), and grand average hydropathy (GRAVY) for each PkNRT was calculated using the ProtParam tool available on the ExPASy Server (https://web.expasy.org/protparam/, accessed on 18 July 2024). The TMHMM-2.0 tool (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0, accessed on 19 July 2024) was employed to predict the transmembrane regions of the putative PkNRTs.
4.2. Phylogenetic Analysis and Structural Characterization of PkNRTs
To investigate the phylogenetic relationships of the PkNRTs, multiple sequence alignment of the NRT protein sequences from A. thaliana, P. trichocarpa, G. biloba, and P. koraiensis was carried out using ClustalW. The phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replicates in MEGA 11.0 software. The integrity of the conservative domains of the PkNRTs was verified and analyzed using the batch CD-search tool at the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 19 July 2024). MEME Version 5.5.0 (https://meme-suite.org/meme/tools/meme, accessed on 19 July 2024) was utilized for conservative motif analysis, with a maximum cardinal number of 10 set for NPF and NRT2, and 4 set for NRT3. The detailed sequence information of these motifs is presented in Table S1. The General Feature Format (GFF) annotation files were used for gene structure analysis. TBtools visualizes PkNRT gene structures, conservative motifs, and domains [52].
4.3. Colinearity Analysis and Calculation of Ka/Ks Values of PkNRTs
To study the collinearity relationship of the PkNRTs, the chromosomal locations of these candidate PkNRTs were obtained from the GFF annotation files, and they were generated using the TBtools 2.1 software. Gene repeat events and collinearity relationships were analyzed using the Multiple Collinear Scan Toolkit (MCScanX 1.0) [53]. The O. sativa genome was downloaded from the Rice Genome DB (https://rice.uga.edu/, accessed on 21 July 2024). The Ka/Ks calculator program in TBtools was utilized to compute the non-synonymous substitution rate (Ka), synonymous substitution rate (Ks), and Ka/Ks ratios among collinear gene pairs and tandemly duplicated gene pairs.
4.4. Analysis of CREs in the Promoters of PkNRTs
The 2000 bp promoter region, located upstream of the transcription start site (TSS) of PkNRTs, was loaded into the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 22 July 2024) database to identify potential cis-regulatory elements (CREs).
4.5. Transcriptome Sequencing for Tissue-Specific Abundance Analysis
Current-year needles, buds, shoots, phloem, xylem, and roots were collected from a 200-year-old Korean pine. The phloem and xylem were obtained from the primary branches of the tree, while root samples were partially extracted from the soil at depths ranging from 10 to 20 cm. Each part’s samples comprised three biological replicates. All collected materials were frozen in liquid nitrogen and subsequently sent to BGI (Shenzhen, China) for transcriptome sequencing. FPKM (fragments per kilobase of exon per million reads mapped) values were calculated based on the RNA-seq data, and the average FPKM of the three replicates in each tissue was calculated. Heat maps were generated using TBtools.
4.6. Plant Materials and Treatments
The Korean pine seeds were obtained from the Hongwei Seed Orchard of the Lushuihe Forestry Bureau, Jilin Province, China, and their dormancy was released using the variable temperature stratification method [54]. The seeds underwent variable temperature stratification treatment (20 °C, 60% humidity for 60 days; 5 °C, 60% humidity for 90 days) to break seed dormancy. Then, they were sown in 3 × 7 pots filled with sterilized sand and placed in the flower greenhouse at 25 °C ± 2 under a 16 h photoperiod. During one month of seed germination, distilled water was used for irrigation. Then, the seedlings were subjected to a 12-week treatment with Hoagland nutrient solution (with 10 mM NO3− + 1 mM NH4+) and a N-deficient solution (with 0 mM NO3− and NH4+). The seedlings were watered with two solutions every 5 days. The needles, stems, and roots were collected at 12 weeks for phenotypic and physiological assays. The roots were harvested for RNA isolation at 1 day, 4 days, 7 days, and 28 days. All samples were collected in liquid N and stored at −80 °C until used.
4.7. Seedling Biomass and Root Morphology
The biomass of needle, hypocotyl, and root was determined by measuring their weight after being thoroughly dried at 65 °C until a constant weight was achieved. The roots were scanned with an Epson Expression 10000XL color scanner (SeikoEpson Corporation, Nagano-ken, Japan). The morphological traits of the roots were assessed through an analysis of 10 seedlings per treatment. Total length, lateral root number, root surface area, and mean diameter were determined using the rootsystem analyzer software WinRHIZO Pro 2016 (Regent Instruments Inc., Montreal, Québec, QC, Canada) [55].
4.8. Quantitative Real-Time PCR Analysis
According to the manufacturer’s protocol, the total RNA was extracted from the roots of Korean pine seedlings in response to N treatments using a TransZol Up Reagent (TransGen Biotech, Beijing, China). The purity and integrity of the isolated total RNA were assessed using agarose gel electrophoresis and a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, NC, USA). The samples were then stored at −80 °C until further analysis. For reverse transcription, removal of genomic DNA and first-strand cDNA synthesis using oligo (dT) were carried out using a TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (TransGen Biotech, Beijing, China). Quantitative real-time PCR (qRT-PCR) was performed with the Perfectstart Green qPCR SuperMix (TransGen Biotech, Beijing, China) by using a LightCycler® 480 System (Roche, Basel, Switzerland). The relative expression level was determined through the 2−∆∆CT method [56] with PkActin used as the internal control. All primers used for qRT-PCR analysis are listed in Table S7. Three biological and three technical replicates were performed for each analysis.
4.9. Statistical Analysis
Statistical analysis was carried out through a two-way analysis of variance (ANOVA) at the p < 0.05 level of significance using SPSS 27.0 software (SPSS Inc., Chicago, IL, USA). The values were determined using the mean ± standard deviation (SD) from three biological replicates. Figures were generated using Graphpad Prism 9.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants14020238/s1: Figure S1: Collinearity analysis of NRTs between Korean pine, rice, and Arabidopsis; Figure S2: The distribution and number of cis-regulatory elements (CREs) in the PkNRT gene promoter; Table S1: Detailed information about motif sequences; Table S2: Gene structure information of PkNRTs; Table S3: Selective pressure on gene pairs between Korean pine, Poplar, and Ginkgo; Table S4: Selective pressure on the tandem duplicated gene pairs in PkNRTs; Table S5: The potential cis-regulatory elements in the promoter region of 76 PkNRTs; Table S6: Expression levels of PkNRTs in different tissues; Table S7: List of qRT-PCR primers in selected PkNRTs; Table S8: The NRT genes of Pinus koraiensis.
Author Contributions
J.Y. and H.S. are mainly responsible for designing this work. J.Y. and X.Z. primarily carried out the bioinformatics analyses. X.Z. and H.W. performed experiments, collected the data, and wrote the original draft. P.W. and B.L. participated in experiments and handling figures and tables. P.Z. participated in the draft and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by grants from the National Key R&D Program of China (2022YFD2201100) and the Fundamental Research Funds for the Central Universities (2572023CT02).
Data Availability Statement
The data that support the findings of this study are available in the Supplementary Materials of this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gaudinier, A.; Rodriguez-Medina, J.; Zhang, L.; Olson, A.; Liseron-Monfils, C.; Bågman, A.-M.; Foret, J.; Abbitt, S.; Tang, M.; Li, B.; et al. Transcriptional regulation of nitrogen-associated metabolism and growth. Nature 2018, 563, 259–264. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, Y.; Xu, G. How does nitrogen shape plant architecture? J. Exp. Bot. 2020, 71, 4415–4427. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.-Q.; Lin, H.-X. Higher yield with less nitrogen fertilizer. Nat. Plants 2020, 6, 1078–1079. [Google Scholar] [CrossRef]
- You, L.; Ros, G.H.; Chen, Y.; Shao, Q.; Young, M.D.; Zhang, F.; de Vries, W. Global mean nitrogen recovery efficiency in croplands can be enhanced by optimal nutrient, crop and soil management practices. Nat. Commun. 2023, 14, 5747. [Google Scholar] [CrossRef]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-Regulated Auxin Transport by NRT1.1 Defines a Mechanism for Nutrient Sensing in Plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Y.; Hsu, P.-K.; Tsay, Y.-F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, P.; Liu, P.; Song, Y.; Zhang, D. Genetic Effects and Expression Patterns of the Nitrate Transporter (NRT) Gene Family in Populus tomentosa. Front. Plant Sci. 2021, 12, 661635. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Li, M.; Xu, W.; Liu, J.-H.; Li, C. Genome-Wide Identification of NRT Gene Family and Expression Analysis of Nitrate Transporters in Response to Salt Stress in Poncirus trifoliata. Genes 2022, 13, 1115. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yan, M.; Hu, B.; Priyadarshani, S.V.G.N.; Hou, Z.; Ojolo, S.P.; Xiong, J.; Zhao, H.; Qin, Y. Characterization and the Expression Analysis of Nitrate Transporter (NRT) Gene Family in Pineapple. Trop. Plant Biol. 2018, 11, 177–191. [Google Scholar] [CrossRef]
- Tahir, M.M.; Wang, H.; Ahmad, B.; Liu, Y.; Fan, S.; Li, K.; Lei, C.; Shah, K.; Li, S.; Zhang, D. Identification and characterization of NRT gene family reveals their critical response to nitrate regulation during adventitious root formation and development in apple rootstock. Sci. Hortic. 2021, 275, 109642. [Google Scholar] [CrossRef]
- Li, W.; Wang, Y.; Okamoto, M.; Crawford, N.M.; Siddiqi, M.Y.; Glass, A.D.M. Dissection of the AtNRT2.1:AtNRT2.2 Inducible High-Affinity Nitrate Transporter Gene Cluster. Plant Physiol. 2007, 143, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Kotur, Z.; Mackenzie, N.; Ramesh, S.; Tyerman, S.D.; Kaiser, B.N.; Glass, A.D.M. Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 2012, 194, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Walch-Liu, P.; Forde, B.G. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises l-glutamate-induced changes in root architecture. Plant J. 2008, 54, 820–828. [Google Scholar] [CrossRef]
- Tal, I.; Zhang, Y.; Jørgensen, M.E.; Pisanty, O.; Barbosa, I.C.R.; Zourelidou, M.; Regnault, T.; Crocoll, C.; Erik Olsen, C.; Weinstain, R.; et al. The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 2016, 7, 11486. [Google Scholar] [CrossRef]
- Akbudak, M.A.; Filiz, E.; Çetin, D. Genome-wide identification and characterization of high-affinity nitrate transporter 2 (NRT2) gene family in tomato (Solanum lycopersicum) and their transcriptional responses to drought and salinity stresses. J. Plant Physiol. 2022, 272, 153684. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; He, M.; Zhang, W.; Han, Y.; Zhang, X.; Zhang, X.; Zhu, Y.; Wang, Y.; Liu, L.; Xu, L. Genome-wide identification and expression analysis of RsNRT gene family reveals their potential roles in response to low-nitrogen condition in radish (Raphanus sativus L.). Sci. Hortic. 2023, 321, 112273. [Google Scholar] [CrossRef]
- Guo, F.-Q.; Wang, R.; Chen, M.; Crawford, N.M. The Arabidopsis Dual-Affinity Nitrate Transporter Gene AtNRT1.1 (CHL1) Is Activated and Functions in Nascent Organ Development during Vegetative and Reproductive Growth. Plant Cell 2001, 13, 1761–1777. [Google Scholar] [CrossRef]
- Guo, F.Q.; Wang, R.; Crawford, N.M. The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is regulated by auxin in both shoots and roots. J. Exp. Bot. 2002, 53, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Segonzac, C.C.; Boyer, J.-C.; Ipotesi, E.; Szponarski, W.; Tillard, P.; Touraine, B.; Sommerer, N.; Rossignol, M.; Gibrat, R.M. Nitrate Efflux at the Root Plasma Membrane: Identification of an Arabidopsis Excretion Transporter. Plant Cell 2007, 19, 3760–3777. [Google Scholar] [CrossRef]
- von Wittgenstein, N.J.J.B.; Le, C.H.; Hawkins, B.J.; Ehlting, J. Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol. Biol. 2014, 14, 11. [Google Scholar] [CrossRef] [PubMed]
- Kant, S. Understanding nitrate uptake, signaling and remobilisation for improving plant nitrogen use efficiency. Semin. Cell Dev. Biol. 2018, 74, 89–96. [Google Scholar] [CrossRef]
- Xia, X.; Fan, X.; Wei, J.; Feng, H.; Qu, H.; Xie, D.; Miller, A.J.; Xu, G. Rice nitrate transporter OsNPF2.4 functions in low-affinity acquisition and long-distance transport. J. Exp. Bot. 2015, 66, 317–331. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Tsay, Y.-F. Arabidopsis Nitrate Transporter NRT1.9 Is Important in Phloem Nitrate Transport. Plant Cell 2011, 23, 1945–1957. [Google Scholar] [CrossRef]
- Kiba, T.; Feria-Bourrellier, A.-B.; Lafouge, F.; Lezhneva, L.; Boutet-Mercey, S.; Orsel, M.; Bréhaut, V.; Miller, A.; Daniel-Vedele, F.; Sakakibara, H.; et al. The Arabidopsis Nitrate Transporter NRT2.4 Plays a Double Role in Roots and Shoots of Nitrogen-Starved Plants. Plant Cell 2012, 24, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Bi, L.; Song, G.; Wang, Q.; Jin, G. Species–habitat associations in an old-growth temperate forest in northeastern China. BMC Ecol. 2018, 18, 20. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.-T.; Wei, J.-T.; Li, Y.; Tigabu, M.; Zhao, X.-Y. Genetic Improvement of Pinus koraiensis in China: Current Situation and Future Prospects. Forests 2020, 11, 148. [Google Scholar] [CrossRef]
- Zhang, P.; Lü, X.-T.; Jin, G.; Liu, Z.; Li, M.-H. Leaf nitrogen resorption is more important than litter nitrogen mineralization in mediating the diversity–productivity relationship along a nitrogen-limited temperate forest succession chronosequence. For. Ecosyst. 2023, 10, 100102. [Google Scholar] [CrossRef]
- Cao, J.; Liu, H.; Zhao, B.; Li, Z.; Liang, B.; Shi, L.; Song, Z.; Wu, L.; Wang, Q.; Cressey, E.L.; et al. Nitrogen addition enhances tree radial growth but weakens its recovery from drought impact in a temperate forest in northern China. Sci. Total Environ. 2023, 903, 166884. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Zhu, J.; Lu, D.; Zhu, C.; Gao, P.; Yang, X. Effects of Korean Pine Basal Area in Mixed Broadleaved–Korean Pine Forest Stands on Its Natural Regeneration in Northeast China. For. Sci. 2021, 67, 179–191. [Google Scholar] [CrossRef]
- Xiang, G.; Wu, R.; Zhang, M.; Li, K.; He, X.; Song, Y.; Song, F. Simulated nitrogen deposition alters the ectomycorrhizal fungal community structure in northern Korean pine plantations. Catena 2023, 233, 107525. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, M.; Jin, G. Effects of nitrogen addition on species composition and diversity of early spring herbs in a Korean pine plantation. Ecol. Evol. 2023, 13, e10498. [Google Scholar] [CrossRef]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q. Identification and characterization of the NPF, NRT2 and NRT3 in spinach. Plant Physiol. Biochem. 2021, 158, 297–307. [Google Scholar] [CrossRef]
- Zhang, J.; Han, Z.; Lu, Y.; Zhao, Y.; Wang, Y.; Zhang, J.; Ma, H.; Han, Y.Z. Genome-wide identification, structural and gene expression analysis of the nitrate transporters (NRTs) family in potato (Solanum tuberosum L.). PLoS ONE 2021, 16, e0257383. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yang, D.; Hu, Y.; Xu, J.; Lu, Z. Genome-Wide Identification and Expression Analysis of Nitrate Transporter (NRT) Gene Family in Eucalyptus grandis. Genes 2024, 15, 930. [Google Scholar] [CrossRef]
- Feng, J.; Zhu, C.; Cao, J.; Liu, C.; Zhang, J.; Cao, F.; Zhou, X. Genome-wide identification and expression analysis of the NRT genes in Ginkgo biloba under nitrate treatment reveal the potential roles during calluses browning. BMC Genom. 2023, 24, 633. [Google Scholar] [CrossRef]
- Chiba, Y.; Shimizu, T.; Miyakawa, S.; Kanno, Y.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 2015, 128, 679–686. [Google Scholar] [CrossRef]
- Bai, H.; Euring, D.; Volmer, K.; Janz, D.; Polle, A. The Nitrate Transporter (NRT) Gene Family in Poplar. PLoS ONE 2013, 8, e72126. [Google Scholar] [CrossRef]
- Drechsler, N.; Courty, P.-E.; Brulé, D.; Kunze, R. Identification of arbuscular mycorrhiza-inducible Nitrate Transporter 1/Peptide Transporter Family (NPF) genes in rice. Mycorrhiza 2018, 28, 93–100. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Liu, Y.; Minh, T.N.; Lu, H.; Zhang, P.; Li, W.; Xiao, J.; Ding, X.; Li, Q. Genome-wide identification and expression analyses of nitrate transporter family genes in wild soybean (Glycine soja). J. Appl. Genet. 2020, 61, 489–501. [Google Scholar] [CrossRef]
- Fan, X.; Tang, Z.; Tan, Y.; Zhang, Y.; Luo, B.; Yang, M.; Lian, X.; Shen, Q.; Miller, A.J.; Xu, G. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. USA 2016, 113, 7118–7123. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Qiao, D.; Zhang, Q.; Li, Y.; Xu, H.; Wei, L.; Gu, Y.; Cao, Y. Cloning and expression study of a putative high-affinity nitrate transporter gene from Dunaliella salina. J. Appl. Phycol. 2004, 16, 395–400. [Google Scholar] [CrossRef]
- Yin, X.-M.; Luo, W.; Wang, S.-W.; Shen, Q.-R.; Long, X.-H. Effect of Nitrogen Starvation on the Responses of Two Rice Cultivars to Nitrate Uptake and Utilization. Pedosphere 2014, 24, 690–698. [Google Scholar] [CrossRef]
- Gao, Y.; Qi, S.; Wang, Y. Nitrate signaling and use efficiency in crops. Plant Commun. 2022, 3, 100353. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.E.; Serezino, L.H.D.; Alves, M.K.; Tagliaferro, A.L.; Vitti, M.; Creste, S.; Riaño-Pachón, D.M.; dos Santos, R.V.; Figueira, A. Root nitrate uptake in sugarcane (Saccharum spp.) is modulated by transcriptional and presumably posttranscriptional regulation of the NRT2.1/NRT3.1 transport system. Mol. Genet. Genom. 2022, 297, 1403–1421. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wu, J.; Yang, S.; Schiefelbein, J.; Gan, Y. Nitrate regulation of lateral root and root hair development in plants. J. Exp. Bot. 2020, 71, 4405–4414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jennings, A.; Barlow, P.W.; Forde, B.G. Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 1999, 96, 6529–6534. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yordanov, Y.S.; Georgieva, T.; Li, X.; Busov, V. Nitrogen deprivation promotes Populus root growth through global transcriptome reprogramming and activation of hierarchical genetic networks. New Phytol. 2013, 200, 483–497. [Google Scholar] [CrossRef]
- Lv, X.; Zhang, Y.; Hu, L.; Zhang, Y.; Zhang, B.; Xia, H.; Du, W.; Fan, S.; Kong, L. Low-Nitrogen Stress Stimulates Lateral Root Initiation and Nitrogen Assimilation in Wheat: Roles of Phytohormone Signaling. J. Plant Growth Regul. 2021, 40, 436–450. [Google Scholar] [CrossRef]
- Ötvös, K.; Marconi, M.; Vega, A.; O’Brien, J.; Johnson, A.; Abualia, R.; Antonielli, L.; Montesinos, J.C.; Zhang, Y.; Tan, S.; et al. Modulation of plant root growth by nitrogen source-defined regulation of polar auxin transport. EMBO J. 2021, 40, e106862. [Google Scholar] [CrossRef]
- Huang, N.-C.; Liu, K.-H.; Lo, H.-J.; Tsay, Y.-F. Cloning and Functional Characterization of an Arabidopsis Nitrate Transporter Gene That Encodes a Constitutive Component of Low-Affinity Uptake. Plant Cell 1999, 11, 1381–1392. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-H.; Kuo, H.-F.; Canivenc, G.v.; Lin, C.-S.; Lepetit, M.; Hsu, P.-K.; Tillard, P.; Lin, H.-L.; Wang, Y.-Y.; Tsai, C.-B.; et al. Mutation of the Arabidopsis NRT1.5 Nitrate Transporter Causes Defective Root-to-Shoot Nitrate Transport. Plant Cell 2008, 20, 2514–2528. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Song, Y.; Zhu, J.; Yan, Q.; Wang, G. Korean pine seed: Linking changes in dormancy to germination in the 2 years following dispersal. For. Int. J. For. Res. 2018, 91, 98–109. [Google Scholar] [CrossRef]
- Ren, H.; Gao, G.; Ma, Y.; Li, Z.; Wang, S.; Gu, J. Shift of root nitrogen-acquisition strategy with tree age is mediated by root functional traits along the collaboration gradient of the root economics space. Tree Physiol. 2023, 43, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).