Abstract
We performed next-generation sequencing of the 18S rDNA–ITS1–5.8S rDNA region along with traditional Sanger sequencing of rbcL, matK, ndhF, and ITS1–5.8S rDNA–ITS2 to clarify the hybridization pattern in the subtribe Alopecurinae and in the genus Alopecurus in particular. Our data support the hybrid origin of Alopecurus × brachystylus from hybridization between A. geniculatus (sect. Alopecurium) and A. pratensis (sect. Alopecurus). Moreover, in the rDNA of hybrid A. × brachystylus, only A. aequalis-like ribotypes from tetraploid A. geniculatus participated. Surprisingly, we found the traces of introgression of A. arundinaceus-like ribotypes not only in hybrid A. × marssonii (A. geniculatus × A. arundinaceus) but in A. aequalis s. str. as well. A high-polyploid group from the section Alopecurus, A. aggr. alpinus has undoubted hybrid origin: e. g., A. brachystachyus has rDNA from the sect. Alopecurium. Alopecurus alpinus, with its allies, is clearly distinct from other members of the sect. Alopecurus (especially by maternal line) and thus we can re-establish a previous opinion about the separate group to which A. alpinus belongs. Species from the section Colobachne (presumably Alpine grasses from Ancient Mediterranean region) probably hybridized with the A. alpinus group. Even A. myosuroides (sect. Pseudophalaris) that could be referred to the separate genus has ribotypes common with the species of the section Alopecurium (A. aequalis, A. geniculatus) in one of the accessions. Additionally, we found that the possible polyphyletic origin of the genus Limnas. Limnas stelleri is very close to Alopecurus magellanicus according to NGS data, while L. malyschevii is more or less distinct from other studied species of the genus Alopecurus.
1. Introduction
The subtribe Alopecurinae belongs to the largest tribe of the grass family, Poeae, and is widespread in the temperate zone of both Hemispheres. It comprises the genera Alopecurus L. and Limnas Trin. according to the most recent data and is closely related to the genera Beckmannia Host and Rhizocephalus Boiss. [1]. The subtribe Alopecurinae is characterized by very dense, spike-shaped panicles of regular cylindrical form [2]. Alopecurus, the main genus of the subtribe, comprises up to 70 species [2]. It is known for drastic differences in the chromosome numbers of the species: from 2n = 14 in Alopecurus aequalis Sobol. to 2n = ca. 100 in A. alpinus Vill. [3]. This polyploid range most probably originated via interspecific hybridization, which is rather common in the Poeae tribe [1,4,5]. Hybrids in the genus Alopecurus have been noted for a long time, and nothospecies have been described beginning from A. × brachystylus Peterm. (A. geniculatus L. × A. pratensis L.).
Recent phylogenetic research of the subtribe Alopecurinae confirmed its separation from the subtribe Phleinae Dumort. that has similar inflorescences and the affinity of Alopecurinae with some genera from other subtribes, in particular Arctophila (Rupr.) Andersson, Dupontia R.Br., Arctagrostis Griseb., and, only according to the nuclear gene sequence data, Arctopoa (Griseb.) Prob. [1,5,6]. Thus, inflorescence traits of the Alopecurinae members can be explained by parallel evolution when similar structures form independently in two lines of taxa originated from the common ancestor. Additionally, the similar inflorescence has the genus Lagurus L. [2] which belongs to wholly different group in the Poeae tribe—Aveninae J.Presl (Aveneae chloroplast group) [1].
Recently, the genus Alopecurus is divided into four sections: Alopecurus, Colobachne (P. Beauv.) Griseb., Alopecurium Dumort., and Pseudophalaris Tzvelev [2,7]. The section Alopecurus differs from other species by their cylindrical to ellipsoidal panicles and absence of palea [2]. This section also can be divided into two groups: tetra- and hexaploid species of A. aggr. pratensis and A. arundinaceus Poir., and high polyploid Siberian and Arcto-Alpine species A. brachystachyus M.Bieb., A. alpinus, A. borealis Trin., A. glaucus Less. with related taxa. The section Colobachne contains mostly Alpine grasses with short, wide-ellipsoidal panicles and mostly present palea. Members of the sect. Alopecurium have an annual or shortly lived perennial habit with geniculate stems and the sect. Pseudophalaris is distinguished by annual habit and glumes with winged keel [2].
Molecular phylogenetic analysis revealed some lines within Alopecurus that did not correspond with taxonomic division of the genus [1,6] and pointed at the intersectional hybridization events. Previous research of Alopecurus mainly covered American and Middle Eastern species [1,6,8,9]. Meanwhile, Asian mountains, for example, Altai Mountain Country as well as Caucasus could provide us many hybrid taxa that have not been analyzed until now (see [10] about the plant speciation centers). New methods of next-generation sequencing (NGS) allow the detection of multiple hybridization events in plants even when their morphology is uniform. Our goal of this work is to trace the hybridization events in the subtribe Alopecurinae (especially in the genus Alopecurus, taking into account hybrid mountain species from Altai-Sayan region) that could shed light on phylogenetic relationships in the subtribe, and possibly its relationships with other taxa of the tribe Poeae. For our analysis, we used the sequences of the region 18S rDNA–ITS1–5.8S rDNA obtained by NGS and the sequences of ITS1–5.8S rDNA–ITS2, matK, rbcL, and ndhF obtained via standard Sanger sequencing.
2. Results
Since hybridization is known to occur frequently in the tribe Poeae, we treated chloroplast and ITS datasets separately. In order to trace possible hybridization events and assess geographical variability, we analyzed multiple samples of the same species for some chloroplast datasets.
Phylogenetic tree built on concatenated rbcL + matK dataset shows two well-supported clades where the studied samples of the genus Alopecurus fall (Figure 1). The first clade (PP = 1, BS = 100) includes section Alopecurium (A. aequalis) and high polyploid members of the section Alopecurus (A. alpinus, A. brachystachyus). This clade is, in turn, sister to Beckmannia syzigachne with low resolution (PP = 0.56, BS = 70). The second clade (PP = 1, BS = 93) contains tetra- and exaploidy species from the sections Alopecurus (A. pratensis + A. arundinaceus), Colobachne (A. glacialis K.Koch, A. ponticus K.Koch), and one sample of A. brachystachyus from Irkutsk Oblast, Eastern Siberia, Russia. A. laguroides Balansa has an uncertain position in this large clade. Alopecurus myosuroides (sect. Pseudophalaris) falls into a separate clade (PP = 1, BS = 100) that forms a polytomy with the previously discussed clade of Alopecurus, and Apera Adans. (PP = 0.78, BS = 70). The second clade, in addition, has weakly supported grouping with the subtribe Phleinae (PP = 0.72, BS = 53). The overall placement of the Alopecurinae species on the tree built by matK sequence data separately is almost the same (Supplementary Figure S2). But in this case intersectional hybrid A. × brachystylus (A. geniculatus × A. pratensis) falls within the clade including section Alopecurium (we have no rbcL data for A. × brachystylus and thus did not include it in the analysis of concatenated sequences). Limnas stelleri Trin. has an uncertain position on the separate rbcL tree (Supplementary Figure S1) but we do not have matK sequences of L. stelleri for a combined data tree. In phylogenetic analysis based on ndhF sequences, we used fewer samples (Figure 2). The Alopecurinae clade groups with Bellardiochloa variegata (Lam.) Kerguélen (PP = 0.99, BS = 87), whereas Beckmannia syzigachne branches separately (PP = 0.99, BS = 87) within the large clade containing the genus Alopecurus, Hookerochloa hookeriana (F.Muell. ex Hook.f.) E.B.Alexeev, Festucella eriopoda (Vickery) E.B.Alexeev, and Cinna latifolia (Trevir. ex Göpp.) Griseb. (PP = 0.99, BS = 87) (Figure 2). The Alopecurus clade (PP = 0.94, BS = 82) divides into sect. Alopecurus (A. alpinus + A. brachystachyus + A. magellanicus Lam.) + sect. Alopecurium subclade (PP = 0.95, BS = 91) and a subclade that contains sect. Alopecurus (A. pratensis + A. arundinaceus) + sect. Colobachne (A. textilis Boiss.) (PP = 0.78, BS = 93) while A. myosuroides has a sister position to all other species.
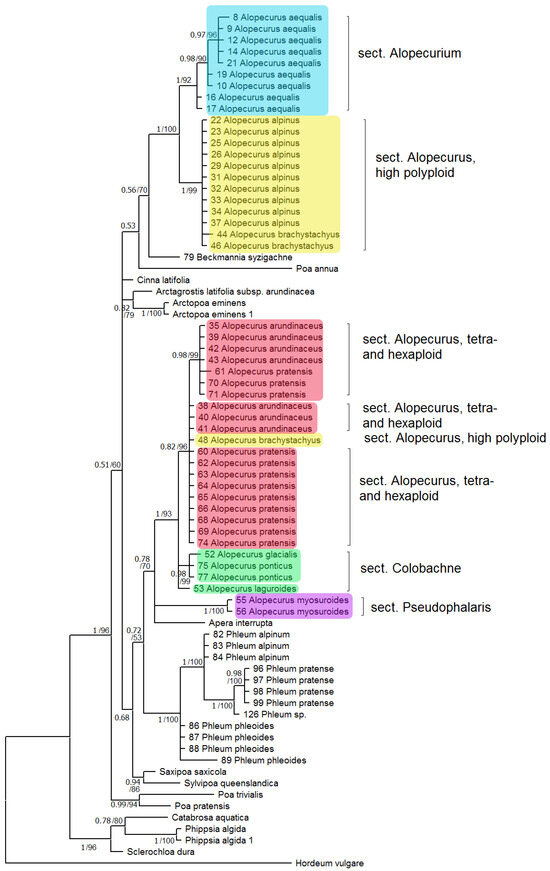
Figure 1.
Phylogenetic tree of the subtribe Alopecurinae and related species according to the rbcL + matK sequence data. The first index on the branch is the posterior probability in Bayesian inference, the second is the bootstrap index obtained by Maximum Likelihood algorithm. When only one index is shown on the branch it is the posterior probability.
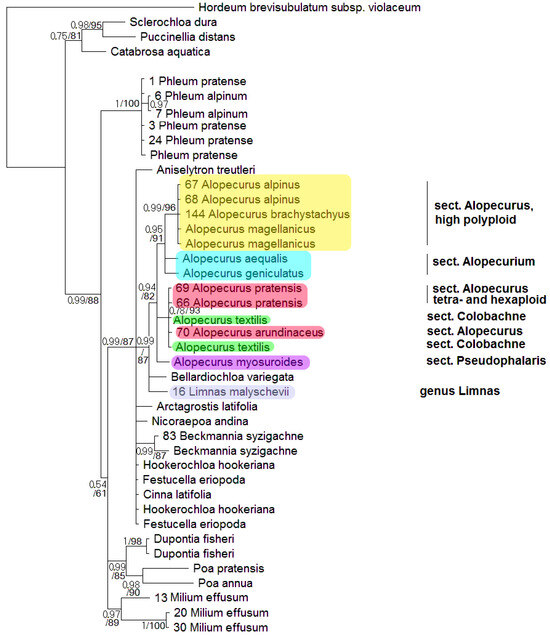
Figure 2.
Phylogenetic tree of the subtribe Alopecurinae and related species according to the ndhF sequence data. The first index on the branch is the posterior probability in Bayesian inference; the second is the bootstrap index obtained by Maximum Likelihood algorithm. When only one index is shown on the branch, it is the posterior probability.
ITS-inferred phylogeny presents a weakly supported clade of Beckmannia + Apera (PP = 0.67, BS = 71), subtribe Alopecurinae, and Cinna L. + Arctopoa (Griseb.) Prob. (PP = 0.63, BS = 55) (Figure 3). This clade forms polytomy with monophyletic Phleinae and Hookerochloa E.B.Alexeev + Saxipoa Soreng, L.J.Gillespie & S.W.L.Jacobs + Sylvipoa Soreng, and L.J.Gillespie & S.W.L.Jacobs + Arctagrostis Griseb. (weakly supported, PP = 0.64). The subclade of Alopecurinae s. str. (without Beckmannia) is supported only according to Bayesian analysis (PP = 0.54, BS unsupported). Rhizocephalus orientalis Boiss. is a sister to all other taxa in the Alopecurinae subclade, though only with weak support in Bayesian inference (PP = 0.54, BS unsupported). Part of the sect. Colobachne (A. brevifolius (G.Westb.) Grossh., A. glacialis, A. textilis) falls into one clade (PP = 1, BS = 99) and occupies a sister position to the clade containing other species of Alopecurus and Limnas + Arctophila (Rupr.) Andersson + Dupontia R.Br. according to Bayesian analysis (PP = 0.70). The clade Limnas + Arctophila + Dupontia is very weakly supported (PP = 0.51, BS = 57). It was discovered as the clade that is the sister to the clade-comprising sect. Alopecurus, A. vaginatus (Willd.) Trin. (sect. Colobachne), the sample of A. myosuroides (sect. Pseudophalaris) from Dagestan Republic, Northern Caucasus, Russia, and sect. Alopecurium (PP = 0.86). In addition, A. brachystachyus (high- polyploid species from the section Alopecurus) is a sister to the clade comprising section Alopecurium and does not group with its possible relative, A. alpinus. One sample of A. myosuroides from Latvia is found in the clade corresponding to the section Alopecurium, according to ITS analysis.
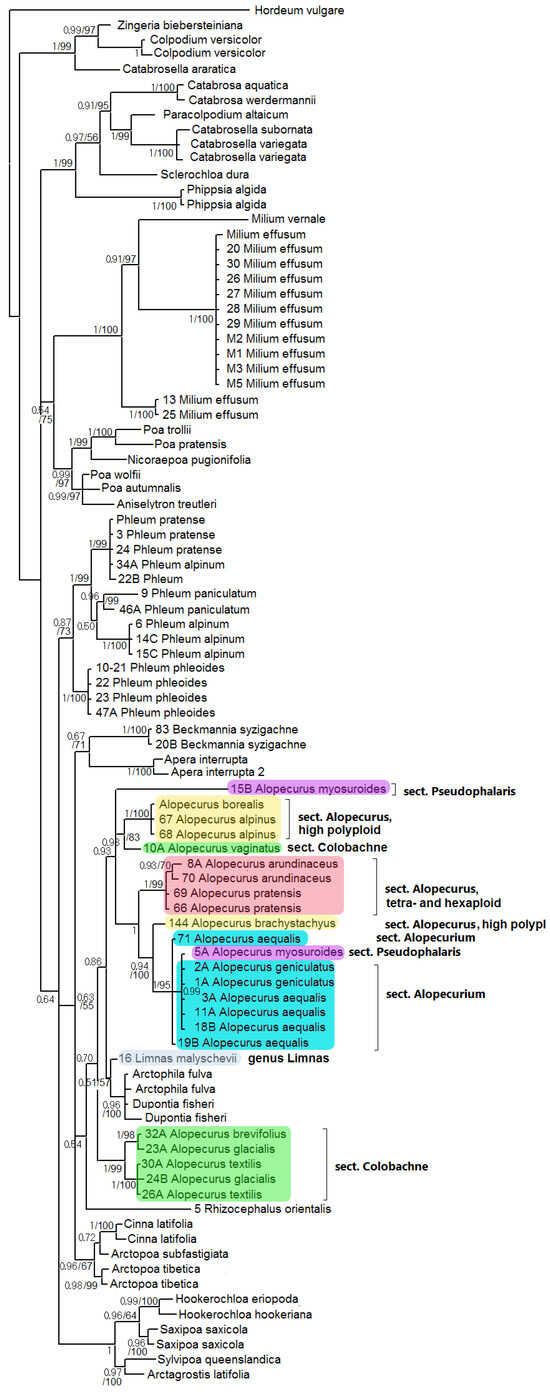
Figure 3.
Phylogenetic tree of the subtribe Alopecurinae and related species according to the ITS sequence data. The first index on the branch is the posterior probability in Bayesian inference; the second is the bootstrap index obtained by Maximum Likelihood algorithm. When only one index is shown on the branch, it is the posterior probability.
We analyzed the marker sequences of 18S rDNA–ITS1–5.8S rDNA obtained via NGS to investigate the hidden multiple hybridization. When the sequence was present in more than 10,000 reads per rDNA pool, we calculated the percentage for them and took these sequences as major. In other cases, when the total quantity of reads was below 10,000, we calculated their percentage for the sequences that occurred in 100 and more reads.
The ribotype networks built in TCS 1.21 and visualized in TCSBU are presented in Figure 4, Figure 5, Figure 6, Figure 7 and Figure 8. The first of the networks depicts the origin of hybrid A. × brachystylus (A. geniculatus × A. pratensis) and A. × marssoni (A. geniculatus × A. arundinaceus) with regard to the parental taxa, e. g., diploid A. aequalis (2n = 14) (Figure 4). In addition, we took into analysis A. vlassovii Trin.—a high polyploid (2n = 120) of the sect. Alopecurus, a relative of A. alpinus. We took the main ribotype of A. aequalis as the consensus sequence. Looking at the network, we can distinguish three ribotype subnetworks corresponding to the sections Alopecurium, Alopecurus, and the high-polyploid group (A. vlassovii). Alopecurus aequalis, a diploid species, has four major ribotypes (in this case, upper 100 reads per rDNA pool) (Figure 4). Its main ribotype, Ae1 (473 reads, 22%), is identical to the main ribotype of A. geniculatus (1809 reads, 34%), the main ribotypes of hybrids A. × brachystylus and A. × marssonii Hausskn. (5828 reads, 22%, and 465 reads, 22%, respectively). Ribotype Ae1 also has the minor ribotype fraction of A. vlassovii and A. brachystachyus (below 1%). The second major ribotype of A. aequalis, Ae2 (169 reads, 8%), is common with the second major ribotype of A. × marssonii (164 reads, 8%). Ribotype Ae3 (161 read, 7%) of A. aequalis also corresponds to the second major ribotype of A. × brachystylus (2331 read, 9%) as well as to the third major ribotype of A. × marssonii (158 reads, 7%), and minor ribotype of A. geniculatus (862 reads, 16%). The fourth major ribotype of diploid A. aequalis—Ae4 (140 reads, 6%) is placed between “Alopecurium” and “Alopecurus” subnetworks. This ribotype is shared with the fourth major ribotype of hybrid A. × marssonii (133 reads, 6%) and minor ribotype of A. × brachystylus (150 reads). The second major ribotype of tetraploid A. geniculatus is species-specific (G, 1072 reads, 20%). Surprisingly, the main ribotype of A. brachystachyus (Br, 3485 reads, 47%) also belongs to the “Alopecurium” subnetwork. It is also the second major ribotype of A. vlassovii (2279 reads, 11%). The main ribotype of A. vlassovii is species-specific (Vl1, 5248 reads, 24%) and forms the separate subnetwork along with its derivative, Vl2 (1006 reads, 5%). The second major ribotype of A. pratensis unites with this subnetwork (Pr2, 2763 reads, 11%). The main ribotype of A. pratensis (Pr1, 5561 read, 21%) groups with “Alopecurus” subnetwork and is homologous to the fifth major ribotype of hybrid A. × brachystylus (1087 reads, 4%). The third major ribotype of A. pratensis, Pr3 (1928 reads, 7%), is specific as well as the fourth (Pr4, 1863 reads, 7%) and the sixth (Pr6, 1049 reads, 4%) ones. Unlike this, the fifth major ribotype of A. pratensis (Pr5, 1416 reads, 5%) is identical to the third major ribotype of A. × brachystylus (2095 reads, 8%) and minor ribotype of A. arundinaceus (41 read). Two major ribotypes of A. arundinaceus, Ar1 and Ar2 (2864 reads, 18%, and 2504 reads, 15%, respectively), form a separate cluster within the “Alopecurus” subnetwork. The main ribotype Ar1 is specific, and the second major ribotype is shared with the minor ribotypes of A. aequalis (75 reads) and A. × marssonii (73 reads). The fourth major ribotype of A. × brachystylus (B, 1722 reads, 7%) is species-specific.
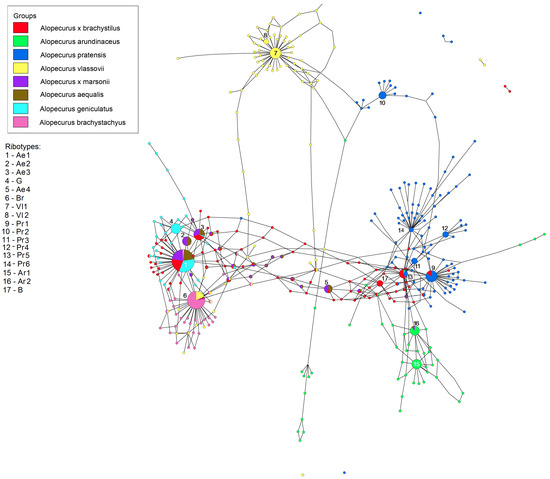
Figure 4.
Ribotype network of hybrid species Alopecurus × brachystylus, A. × marssonii, their putative parental species and high polyploids A. vlassovii and A. brachystachyus. The radius of the circles on the ribotype network is proportional to the percent number of reads for each ribotype. Major ribotypes are larger than others and marked with numbers.
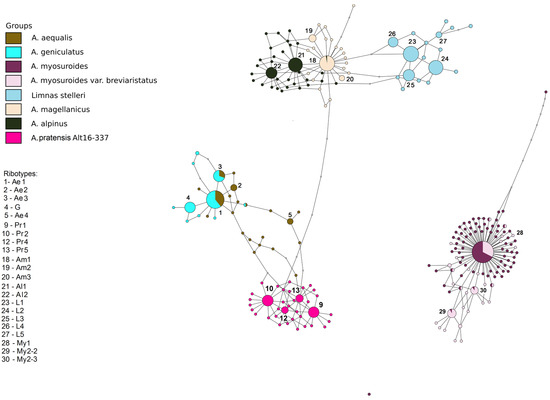
Figure 5.
Ribotype network of the sections Alopecurium, Pseudophalaris, Alopecurus, and the genus Limnas (Limnas stelleri). The radius of the circles on the ribotype network is proportional to the percent number of reads for each ribotype. Major ribotypes are larger than others and marked with numbers.

Figure 6.
Ribotype network depicting the relationships between different samples of Alopecurus pratensis. The radius of the circles on the ribotype network is proportional to the percent number of reads for each ribotype. Major ribotypes are larger than others and marked with numbers.
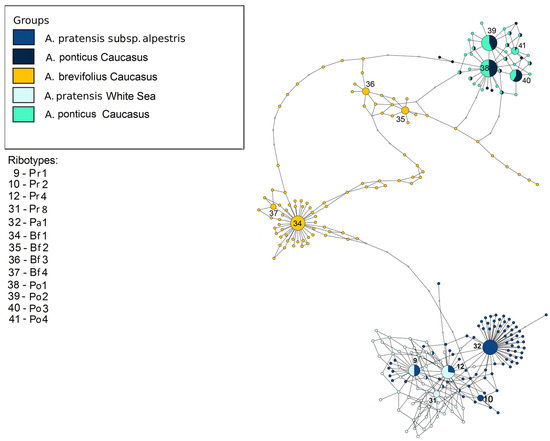
Figure 7.
Ribotype network of the section Alopecurus (A. pratensis) and section Colobachne. The radius of the circles on the ribotype network is proportional to the percent number of reads for each ribotype. Major ribotypes are larger than others and marked with numbers.
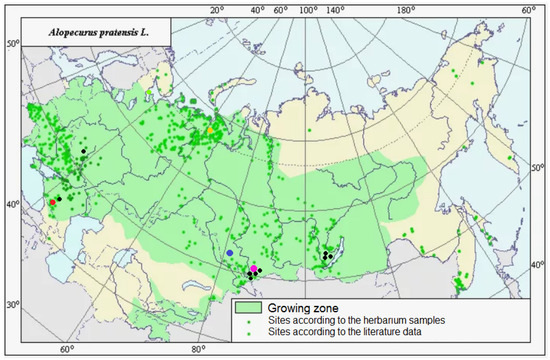
Figure 8.
Area of distribution of Alopecurus pratensis in Russia and adjacent territories and collection points for our herbarium material. Colored rounds depict collection sites of the samples for NGS analysis; black dots—sites of the samples for chloroplast sequence data. Total species area was taken from http://agroatlas.ru, accessed on 5 March 2024.
The second ribotype network concerns the relationships between section Alopecurium, a high-polyploid group of the type section (A. alpinus, A. magellanicus), section Pseudophalaris (A. myosuroides) and the genus Limnas (Figure 5). The studied species of the section Alopecurium do not have the ribotypes of A. alpinus and A. magellanicus in the rDNA pool. The major ribotypes of A. alpinus and A. magellanicus (Al and Am, respectively) are mostly species-specific except for the main ribotype of A. magellanicus (Am1, 2051 read, 40%) that is shared with the minor ribotype of A. alpinus (274 reads). The sample of A. pratensis Alt 16-337 has four major ribotypes forming own subnetwork (Pr1, Pr2, Pr5, Pr4) that are identical to the four ribotypes of the A. pratensis sample from the previous network. Limnas stelleri, a species from the subtribe Alopecurinae, has ribotypes that are close to the Alopecurus magellanicus/A. alpinus subnetwork. Major ribotypes of this species can be called L. The ribotypes of the samples of A. myosuroides have no connection with other samples of Alopecurus in this network, according to NGS data. The main ribotype of A. myosuroides (My1, 10,044 reads, 56%) is identical with that of A. myosuroides var. breviaristatus Asch. & Graebn. (3292 reads, 27%). Other major ribotypes (more than 1000 reads in this case) are present only in A. myosuroides var. breviaristatus (My2-2, 1971 read, 12%, My2-3, 1478 reads, 9%). They are common with minor ribotypes of A. myosuroides.
The third ribotype network shows the phylogenetic structure of A. aggr. pratensis (Figure 6). The main ribotype of A. pratensis s. str. from Altai Krai, Russia (Pr1, 5561 reads, 21%) is shared with the second major ribotype of A. pratensis subsp. alpestris (Wahlenb.) Selander (2889 reads, 11%), the second major ribotype of A. pratensis from Teberda (4775 reads, 22%), and the second major ribotype of A. pratensis from the White Sea (1732 reads, 11%). The second major ribotype of A. pratensis from Altai Krai (Pr2, 2763 reads, 11%) is specific. The third major ribotype of A. pratensis s. str. (Pr3, 1928 reads, 7%) is common with the minor ribotype of Caucasian A. pratensis from Teberda (18 reads). The fourth major ribotype of A. pratensis, Pr4 (1863 reads, 7%) is common with fourth major ribotype of A. pratensis subsp. alpestris (1611, 5%). The fifth major ribotype (Pr5, 1416 reads, 5%) is identical to the main ribotype of A. pratensis from Teberda (5359 reads, 24%), the main ribotype of A. pratensis from the White Sea (3153 reads, 21%), and the third major ribotype of A. pratensis subsp. alpestris (1903 reads, 7%) while the sixth major ribotype, Pr6 (1049 reads, 4%) is shared with minor ribotype of A. pratensis from the White Sea (49 reads). The third major ribotype of A. pratensis from Teberda (Pr7, 3539 reads, 16%) is specific as well as the third major ribotype of A. pratensis from the White Sea (Pr8, 1041 read, 7%), and the main ribotype of A. pratensis subsp. alpestris (Pa1, 9301, 37%) is common with minor fraction of A. pratensis (547 reads, below 1%).
The next picture describes the relationships between the studied samples of the sections Alopecurus (A. pratensis s. l.) and Colobachne (Figure 7). ITS1 sequences of A. pratensis s. str. samples from the White Sea and A. pratensis subsp. alpestris obtained by NGS group separately from those of A. ponticus. The main ribotype of A. pratensis from the White Sea is the same as the main ribotype of A. pratensis subsp. alpestris (Pr1). The main ribotype of A. pratensis subsp. alpestris (Pa1, 9301, 37%) is related to the ribotypes of A. pratensis but is specific on this scheme. The fourth major ribotype of A. pratensis subsp. alpestris (1611, 5%) belongs to the group Pr and is identical to the ribotype Pr4 (see above). The ribotype structure of the species of the sect. Colobachne is diverse. A. brevifolius sample from Teberda (T89) forms the peculiar subnetwork that is only distantly related to the sect. Alopecurus. Its ribotypes, Bf1–Bf4 are specific. Ribotypes of two samples of A. ponticus from Teberda (T57, T59) are almost identical except for the fourth major ribotype of A. ponticus T59 (Po4) that is shared with the minor ribotype of A. ponticus T57.
3. Discussion
When a species has substantially discordant positions between the cpDNA and nrDNA phylogenies, there is a possibility that the species may be a hybrid [11,12,13,14]. Moreover, intraspecific genome polymorphism is widely used in research for interspecific and, in some cases, intraspecific phylogenetic reconstructions, especially in cases when introgression is involved [15,16,17,18]. For example, some genera of the tribe Triticeae Dumort. (Poaceae) appeared to be of intergeneric origin themselves [19,20,21]. Molecular phylogenetic analysis of different gene sets that allowed to trace possible hybridization events proposed generic names based on the genome combinations that the species has [22]. Other works presented new data of multiple polyploid origin of many Hordeum L. species (sometimes auto- and allopolyploid species were in the same aggregate) [21].
The genus Alopecurus belongs to the subtribe Alopecurinae (tribe Poeae s. l.), which is known for widespread hybridization. Molecular phylogenetic studies of the genus showed polyphyletic placement of the previously described sections Alopecurus and Colobachne [7,23]. Some replacements were also made on the species level that can have, in fact, different explanations [7,23]. Researching the ribotype composition of Alopecurus and some taxa from allied genera, we found many ribotypes due to the active hybridization and high polyploidy in the subtribe Alopecurinae.
In the genus Alopecurus, four nothospecies were described that are usually sterile and rather rare [2]. We analyzed two of them, Alopecurus × brachystylus and A. × marssonii. A. × brachystylus is an intersectional hybrid according to taxonomists [24,25]. One of the putative parent species, the Euro–Mediterranean–South Asian A. geniculatus, belongs to the section Alopecurium, the members of which are distinguished by their characteristic narrow cylindrical panicles and geniculate stems. The second parent taxon, the Euro–Siberian–Central Asian A. pratensis, belongs to the type section, which includes species with predominantly wide cylindrical or ellipsoidal panicles and straight stems. Parental species, growing in the same habitats, can hybridize, usually forming sterile offspring [26,27]. A. × brachystylus was not previously studied by molecular phylogenetic methods. Our data confirm this statement but add some details. On the maternal side, A. × brachystylus probably originated from A. geniculatus (Figure 5). Our NGS analysis shows the presence of A. pratensis ribotypes and ribotypes that are common with diploid A. aequalis (also belonging to the sect. Alopecurium). Tetraploid A. geniculatus (2n = 28), in its turn, could originate from diploid A. aequalis and an unknown diploid progenitor (Figure 4). The second possible interpretation is that A. geniculatus has the second ribotype that passed the stages of post-hybridization transformation. Thus, the hybrid genome of A. × brachystylus has only A. aequalis ribotypes from an A. geniculatus parent. A. × marssonii is a hybrid between A. geniculatus and Predominantly Euro–Siberian–Caucasian–Central Asian meadow–coastal weakly halophilic species A. arundinaceus. Ribotype structure of A. × marssonii indicates its origin from A. geniculatus (hybrid with A. aequalis as parental species). A. arundinaceus-related ribotypes occurred only in a minor fraction of A. × marssonii. But, surprisingly, a minor ribotype fraction that is common with A. arundinaceus was also found in A. aequalis ribotypes (Figure 4). This unexpected fact can point at the cases of introgression that took place in in the fairly distant past. All trees built on the sequence data, obtained by the Sanger method, clearly show distinction of the sect. Alopecurium.
South Siberian mountain species A. vlassovii, which belongs to the high polyploid group of the sect. Alopecurus (A. aggr. borealis), has poly- and aneuploid chromosome numbers, 2n = ca. 98–130, ca. 120, and ca. 150, without predominance of any one of them [28,29,30]. According to the NGS data, A. vlassovii has two subgenomes that correspond to the A. borealis group and A. brachystachyus (being close to the sect. Alopecurium) (Figure 4). High polyploidy of A. vlassovii is clearly a result of multiple hybridization and introgression within the group and probably between the different sections. A. vlassovii can be a member of a peculiar introgressive–hybridization complex of species [31] belonging to the affinity group of A. alpinus. A. vlassovii is possible hybrid of A. alpinus s. l. and A. brachystachyus. The latter species, according to rbcL and matK sequence analysis, has different maternal taxa. The majority of A. brachystachyus are relatives of A. alpinus s. l. and other high-polyploid mountain species but some samples occupy an uncertain position (Figure 1). ITS data obtained by the Sanger method, along with the NGS data, show the relationship between A. brachystachyus and A. aequalis + A. geniculatus.
R. Soreng et al. [32] synonymized the Antarctic species Alopecurus magellanicus, Holarctic mountain-tundra species A. alpinus, and A. borealis, North Pacific mountain-meadow A. stejnegeri Vasey and endemic to the Ural Mountains, weakly halophilic A. glaucus. In our opinion, we cannot accept such a broad interpretation of A. magellanicus, since these undoubtedly closely related species nevertheless differ in both morphological and molecular genetic characteristics. A comparison of herbarium material of these species with specimens of A. magellanicus (South Georgia and the South Sandwich Islands) did not reveal absolute similarity between them. In contrast, A. magellanicus has its own species-specific characters not found in other closely related species. For example, A. magellanicus has larger spikelets (4–5 mm long) than A. alpinus, less abundant pubescence of the glumes, more developed awns, and the presence of a membranous border at the tips of the lemmas. Subantarctic A. magellanicus and Arctic A. alpinus have their own specific ribotypes (Figure 5). Only a minor ribotype fraction of A. alpinus is shared with A. magellanicus. A. magellanicus is a close relative of A. alpinus and A. brachystachyus according to ndhF sequence analysis. A. alpinus, as an ancestor of A. magellanicus, could spread across Cordilleras, this being the classic example of interpolar disjunction (see also [33]).
Alopecurus myosuroides, belonging to the sect. Pseudophalaris, stands apart from other species of the genus. The main distinction of A. myosuroides is its annual life form and the winged keel on its glumes. We see that A. myosuroides also stands separately according to the molecular phylogenetic data (Figure 1, Figure 2, Figure 3, Figure 4 and Figure 6). Morphological separateness along with distinction of the sequences can point at possible generic specificity of A. myosuroides. But one sample of A. myosuroides from Latvia falls into a clade with A. aequalis and A. geniculatus (sect. Alopecurium). This unusual position can reflect the ancient homoploid hybridization.
Another interesting fact is the position of Limnas. The genus Limnas is a member of subtribe Alopecurinae and is distinguished by more or less loose panicles and leathery-membranous glumes [2]. There are two endemic species of Limnas in Russia: Eastern Siberian Limnas stelleri and Eastern Siberian/Far Eastern Limnas malyschevii. It is interesting that Limnas stelleri turned out to be more or less closely related with North American Alopecurus alpinus and Subantarctic A. magellanicus (Figure 5). The close relationship of Limnas and Alopecurus species was previously identified by Soreng [23], using ITS data obtained by the Sanger method (Figure 3). The second species, L. malyschevii, is weakly related to the clade Dupontia + Arctophila by ITS data and is sister to Alopecurus clade by ndhF sequences (Figure 2). Such unusual placement of two Limnas species could be due to the intergeneric reticulation or even polyphyletic origin of the genus.
Molecular phylogenetic data often help in distinguishing different species in one large geographically heterogeneous complex. For example, analysis of chloroplast and nuclear (ITS) genes revealed a different position of Malaysian samples of Spiranthes sinensis (Pers.) Ames (Orchidaceae Juss.) compared with other East Asian samples [34]. They were considered as crypto-hybrids [34]. In grasses (Poaceae), even samples of Hyalopoa pontica (Balansa) Tzvelev from different but closely located gorges of Greater Caucasus (Teberda, Carachay-Cherkessia Republic) can fall into different clades [35]. Alopecurus pratensis has almost a worldwide area, occurring in many non-tropical areas. It is tetraploid with 2n = 28 [2]. It forms a group of related taxa: A. pratensis s. str., A. pratensis subsp. alpestris, A. arundinaceus, and A. brachystachyus. A. pratensis subsp. alpestris is an Arctic or hypoarctic plant with glaucous tinge of ligules and glaucous leaves. Our NGS data confirm that A. pratensis subsp. alpestris can be can be distinguished as a separate species because it has specific main ribotype (Figure 6). In addition, A. pratensis from Northern Caucasus (Teberda) and A. pratensis from the White Sea coast have peculiar major ribotypes but not main one. It can be related with the geographical variability of A. pratensis s. l. (Figure 8). In addition, geographical variability was detected by chloroplast sequence data (rbcL and matK). Samples from Stavropol Krai and Altai Republic differ from those from Irkutsk Oblast and other samples from Altai Republic (Figure 1). It is possible that Alopecurus pratensis is heterogeneous by the maternal line. A. brachystachyus, on the contrary, has different affinity according to the chloroplast sequence data; it is closer to the Arctic and Sub-Arctic high polyploid species but also can have an A. pratensis-like maternal genome (Figure 1, see above).
Species from the section Colobachne differ from the other Alopecurus members by wide ellipsoid or shortly cylindrical panicles and by predominant presence of palea [2,24]. The center of their diversity is Ancient Mediterranean and Caucasus region ([36], Figure 9).
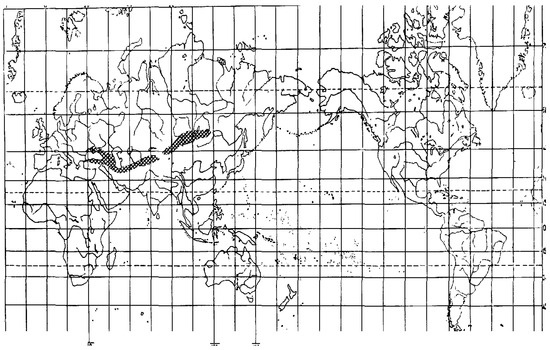
Figure 9.
Area of distribution of the Vaginatae group (=section Colobachne) [36].
They form polyploid range from 2n = 14 [30] to 2n = 56 [37]. We need to note that A. brachystachyus (sect. Alopecurus) was previously treated as the species of the Vaginatae group (=sect. Colobachne) as well [36]. Molecular phylogenetic analysis clearly confirms distinction of this section from high-polyploid relatives of A. alpinus and from A. brachystachyus and comparative unity of the section Colobachne on the maternal side (rbcL and matK, Figure 1 and Figure 2). At the same time, the nrDNA of the Colobachne species indicates probable hybridizations: A. vaginatus falls into a clade with A. alpinus and allied taxa (Figure 3) and is distant from other members of the sect. Colobachne. Additionally, the section Colobachne (excl. A. vaginatus) is rather distant form all other Alopecurus members based on ITS sequences (Figure 3). According to NGS data, studied species of the section Colobachne are fairly distinct from each other and do not form common ribotypes (Figure 7). We can assume that species of the sect. Colobachne probably arose from intersectional hybridization but are rather stabile themselves.
The position of two genera, Beckmannia and Rhizocephalus, remains controversial. Earlier, these genera were placed near the genus Alopecurus in the tribe Phleeae [24,38]. These genera were placed in the special subtribe Beckmanniinae. Our data present Beckmannia as the genus allied to the subtribe Alopecurinae that is closer to the latter on the chloroplast trees (Figure 1 and Figure 2). Rhizocephalus orientalis occupies an uncertain position being sister to Alopecurinae only by Bayesian method according to the ITS data (PP = 0.54) (Figure 3).
Thus, our data prove developed hybridization in the subtribe Alopecurinae, not only in the sterile nothospecies. The degree of introgressive hybridization processes varies depending on the sections and groups; e.g., some Alopecurus species do not hybridize with closely related taxa according to NGS data but have plastid DNA from different sections. We tend to re-establish the high polyploid group of Alopecurus previously named as the group Alpinae [36] that now is placed within the sect. Alopecurus. The genus Limnas is probably polyphyletic.
4. Materials and Methods
For our molecular phylogenetic analysis, we took species from all sections of the genus Alopecurus, paying special attention to the potential hybrid taxa. We included also species from other genera of the subtribe Alopecurinae s. l.: Beckmannia, Limnas, Rhizocephalus as well. We analyzed chloroplast sequences of the region matK of 53 samples of 13 species, rbcL in 61 samples of 12 species, ndhF of 19 samples of 11 species, and 27 ITS sequences of 16 species (all obtained by Sanger method). Then, we concatenated rbcL and matK sequences of the studied species when there were both sequences of the same species. The sequences of rbcL + matK regions were presented for 48 samples of 10 species. We paid more attention to the hybrid species and polyploid species that can have some differences along their habitat. Information about the species studied via Sanger method is given in Table 1. For NGS analysis, we used 23 species of the subtribe Alopecurinae. The list of studied species is presented in Table 2.

Table 1.
Sequences of the species of subtribe Alopecurinae and related subtribes obtained in the present study by the Sanger method and their numbers in GenBank.

Table 2.
Summary of the Alopecurinae species used in the present NGS study and their major ribotypes.
In the case of 10,000 reads per rDNA pool, we assumed the ribotypes more than 1000 reads per rDNA pool to be major. When the total quantity of reads was below 10,000, we took the sequences of ribotypes that occurred in 100 or more reads as menomic DNA, which was extracted from leaf material with the aid of a Qiagen Plant Mini Kit (Qiagen Inc., Hilden, Germany), according to the instruction manual. The fragments were amplified and sequenced at the Center for Shared Use “Genomic Technologies, Proteomics, and Cell Biology” of the All-Russian Research Institute of Agricultural Microbiology on an Illumina Platform MiSeq. PCR was performed in 15 μL of the reaction mixture containing 0.5–1 unit of activity of Q5® High-Fidelity DNA Polymerase (NEB, Ipswich, MA, USA), 5 pM of forward and reverse primers, 10 ng of DNA template, and 2 nM of each dNTP (Life Technologies, ThermoScientific, Waltham, MA, USA). The fragments were amplified under the following conditions: initial denaturation at 94 °C for 1 min, followed by 25 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and a final elongation 72 °C for 5 min using ITS 1P [39] and ITS 2 [40] primers. PCR products were then purified according to the Illumina recommended method using AMPureXP (Beckman Coulter, Indianapolis, IN, USA). The libraries for sequencing were prepared according to the manufacturer’s MiSeq Reagent Kit Preparation Guide (Illumina) (http://web.uri.edu/gsc/files/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 6 September 2022)). They were sequenced on an Illumina MiSeq instrument (Illumina, San Diego, CA, USA) using a MiSeq® ReagentKit v3 (600 cycles) with double-sided reading (2 × 300 n) following the manufacturer’s instructions. The sequences were trimmed with Trimmomatic [41], included in Unipro Ugene [42] as follows: PE reads, sliding window trimming with size 4, quality threshold 12, and minimal read length 130. Further, paired marker sequences were combined, dereplicated, and sorted into the ribotypes by vsearch 2.7.1 [43]. The resulting sequences represent ribotypes with certain frequency in the whole genome pool. These sequences were analyzed by TCS 1.21 [44]. The network built by algorithms of statistic parsimony was visualized and processed in TCSBU [45].
Forward and reverse sequences of ITS and chloroplast regions obtained by Sanger method were observed in Chromas Lite 2.1 (https://technelysium.com.au/wp/chromas/, accessed on 1 October 2021) and concatenated in MEGA XI [46]. The sequencing was performed according to the standard protocol provided with a BigDyeTM Terminator Kit ver. 3.1 set of reagents on the sequencer ABI PRIZM 3100 sequencer at the Center for the collective use of scientific equipment “Cellular and molecular technologies for the study of plants and fungi” of the Komarov Botanical Institute, St. Petersburg. They were aligned by Muscle [47] implemented in MEGA XI [46]. Evolutionary models were computed using jModelTest 2.1.10 [48]. For the ITS dataset, we obtained GTR + I + G, for matK TPM3uf + G, for ndhF, the evolutionary model was TVM + I + G, and for rbcL we computed TVM + G. Bayesian inference was performed using Mr. Bayes 3.2.2 [49] as follows: 3–5 million of a generation, sampling trees every 100 generations, and the first 25% trees were discarded as burn-in. ML analysis was conducted by iqtree 1.6.12 (http://www.iqtree.org/, accessed 5 March 2023) under the fast bootstrap option, 1000 generations. The resulting trees combined Bayesian and ML data. The first index was posterior probability, and the second was a bootstrap index.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13070919/s1, Table S1. rbcL sequences used in our analysis. Table S2. matK sequences used in our analysis. Table S3. ndhF sequences used in our analysis. Table S4. ITS sequences used in our analysis. Table S5. Primary structure of the major ribotypes of the genus Alopecurus and Limnas stelleri obtained by NGS. Numbers show the position in the alignment of the major ribotypes. D is a deletion. Table S6. Aligned ribotype sequences of the hybrid species Alopecurus × brachystylus, A. × marssonii, their putative parental species and high polyploids A. vlassovii and A. brachystachyus. Table S7. Aligned ribotype sequences of species of the sections Alopecurium, Pseudophalaris, Alopecurus, and the genus Limnas (Limnas stelleri). Table S8. Aligned ribotype sequences of different samples of Alopecurus pratensis (including A. pratensis subsp. alpestris). Table S9. Aligned ribotype sequences of A. pratensis sample (sect. Alopecurus) and species of the section Colobachne. Figure S1. Phylogenetic tree of the subtribe Alopecurinae and related species according to the rbcL sequence data. The first index on the branch is the posterior probability in Bayesian inference, the second is the bootstrap index obtained by Maximum Likelihood algorithm. When only one index is shown on the branch it is the posterior probability. Figure S2. Phylogenetic tree of the subtribe Alopecurinae and related species according to the matK sequence data. The first index on the branch is the posterior probability in Bayesian inference, the second is the bootstrap index obtained by Maximum Likelihood algorithm. When only one index is shown on the branch it is the posterior probability.
Author Contributions
A.A.G. and N.N.N. carried out the experiments. A.A.G., N.N.N., E.O.P. and A.V.R. analyzed the data. I.G.L. provided seed material and revised a manuscript. A.A.G., N.N.N. and A.V.R. wrote the manuscript. V.S.S. thoroughly corrected and edited the text. S.A.C. carried out bioinformatics data processing. All authors have read and agreed to the published version of the manuscript.
Funding
Our research was supported by the Russian Science Foundation grant No. 22-24-01085.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
The authors are grateful to A.G. Pinaev and all researchers of the Center for Shared Use “Genomic Technologies, Proteomics and Cell Biology” of the All-Russian Research Institute of Agricultural Microbiology for next-generation sequencing, to E.E. Krapivskaya for sequencing of the samples by Sanger method, to E.M. Machs for invaluable help in data processing. The work was conducted using the equipment of the core facility center “Center Bio-Bank” of St. Petersburg State University.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Soreng, R.J.; Peterson, P.M.; Zuloaga, F.O.; Romaschenko, K.; Clark, L.G.; Teisher, J.K.; Gillespie, L.J.; Barberá, P.; Welker, C.A.D.; Kellogg, E.A.; et al. A worldwide phylogenetic classification of the Poaceae (Gramineae) III: An update. J. Syst. Evol. 2022, 60, 476–521. [Google Scholar] [CrossRef]
- Tzvelev, N.N.; Probatova, N.S. Grasses of Russia; KMK Scientific Press: Moscow, Russia, 2019; p. 646. (In Russian) [Google Scholar]
- Krogulevich, R.E. Role of polyploidy in the formation of flora of Putorana. In Flora of Putorana. Materials for Understanding the Characteristics of the Composition and Genesis of Mountain Subarctic Floras; Malyshev, L.I., Ed.; Nauka: Novosibirsk, Russia, 1976; pp. 217–235. (In Russian) [Google Scholar]
- Tzvelev, N.N. Problems of Theoretical Morphology and the Evolution of Higher Plants; KMK: Moscow and St.-Petersburg, Russia, 2005; p. 407. (In Russian) [Google Scholar]
- Tkach, N.; Schneider, J.; Döring, E.; Wölk, A.; Hochbach, A.; Nissen, J.; Winterfeld, G.; Meyer, S.; Gabriel, J.; Hoffmann, M.H.; et al. Phylogenetic lineages and the role of hybridization as driving force of evolution in grass supertribe Poodae. Taxon 2020, 69, 234–277. [Google Scholar] [CrossRef]
- Soreng, R.J.; Peterson, P.M.; Romaschenko, K.; Davidse, G.; Judziewicz, E.J.; Zuloaga, F.O.; Filgueiras, T.S.; Morrone, O. A worldwide phylogenetic classification of the Poaceae (Gramineae). J. Syst. Evol. 2015, 53, 117–137. [Google Scholar] [CrossRef]
- Cabi, E.; Soreng, R.J.; Gillespie, L.J.; Boudko, E. Alopecurus goekyigitiana (Poaceae, subtribe Alopecurinae sensu stricto), a new species from Turkey based on morphological and molecular investigation. Turk. J. Bot. 2017, 41, 189–199. [Google Scholar] [CrossRef]
- Gillespie, L.J.; Soreng, R.J.; Bull, R.D.; Jacobs, S.W.L.; Refulio-Rodriguez, N.F. Phylogenetic relationships in subtribe Poinae (Poaceae, Poeae) based on nuclear ITS and plastid trnT-trnL-trnF sequences. Botany 2008, 86, 938–967. [Google Scholar] [CrossRef]
- Rhaif, Q.A.; Shaheed, K.A.; Al-Musawi, B.H. Molecular study for two species of Alopecurus L. (Poaceae) in Iraq. Ann. For. Res. 2022, 65, 8525–8534. [Google Scholar]
- Hartley, W. Studies on the origin, evolution and distribution of the Gramineae. IV. The genus Poa L. Austr. J. Bot. 1961, 9, 152–161. [Google Scholar] [CrossRef]
- Soltis, D.E.; Kuzoff, H.K. Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxifragaceae). Evolution 1995, 49, 727–742. [Google Scholar] [CrossRef]
- Kellogg, E.A. When Genes Tell Different Stories: The Diploid Genera of Triticeae (Gramineae). Syst. Bot. 1996, 21, 321–347. [Google Scholar] [CrossRef]
- Patterson, J.P.; Larson, S.R.; Johnson, P.G. Genome relationships in polyploid Poa pratensis and other Poa species inferred from phylogenetic analysis of nuclear and chloroplast DNA sequences. Genome 2005, 48, 76–87. [Google Scholar] [CrossRef]
- Stull, G.W.; Pham, K.K.; Soltis, P.S.; Soltis, D.E. Deep reticulation: The long legacy of hybridization in vascular plant evolution. The Plant J. 2023, 114, 743–766. [Google Scholar] [CrossRef]
- Baldwin, B.G.; Sanderson, M.J.; Porter, J.M.; Wojciechowski, M.F.; Campbell, C.S.; Donoghue, M.J. The ITS Region of Nuclear Ribosomal DNA: A Valuable Source of Evidence on Angiosperm Phylogeny. Ann. Mis. Bot. Gard. 1995, 82, 247–277. [Google Scholar] [CrossRef]
- Mahelka, V.; Krak, K.; Kopecký, D.; Fehrer, J.; Šafář, J.; Bartoš, J.; Hobza, R.; Blavet, N.; Blattner, F.R. Multiple horizontal transfers of nuclear ribosomal genes between phylogenetically distinct grass lineages. Biol. Sci. 2017, 114, 1726–1731. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.D.P.N.; Riina, R.; Valduga, E.; Caruzo, M.B.R. A new species of Croton (Euphorbiaceae) endemic to the Brazilian Pampa and its phylogenetic affinities. Plant Syst. Evol. 2022, 308, 14. [Google Scholar] [CrossRef]
- Suyama, Y.; Hirota, S.K.; Matsuo, A.; Tsunamoto, Y.; Mitsuyuki, C.; Shimura, A.; Okano, K. Complementary combination of multiplex high-throughput DNA sequencing for molecular phylogeny. Ecol. Res. 2022, 37, 171–181. [Google Scholar] [CrossRef]
- Dewey, D.R. The genomic system of classification as a guite to intergeneric hybridization with the perennial Triticeae II. In Gene Manipulation in Plant Improvement; Gustafson, J.P.N.Y., Ed.; Plenum Publ.: New York, NY, USA, 1984; pp. 209–279. [Google Scholar]
- Mason-Gamer, R.J. Phylogeny of a genomically diverse group of Elymus (Poaceae) allopolyploids reveals multiple levels of reticulation. PLoS ONE 2013, 8, e78449. [Google Scholar] [CrossRef]
- Brassac, J.; Blattner, F.R. Species-Level Phylogeny and Polyploid Relationships in Hordeum (Poaceae) Inferred by Next-Generation Sequencing and In Silico Cloning of Multiple Nuclear Loci. Syst. Biol. 2015, 64, 792–808. [Google Scholar] [CrossRef]
- Tan, L.; Huang, Q.-X.; Song, Y.; Wu, D.-D.; Cheng, Y.-R.; Zhang, C.-B.; Sha, L.-N.; Fan, X.; Kang, H.-Y.; Wang, Y.; et al. Biosystematics studies on Elymus breviaristatus and Elymus sinosubmuticus (Poaceae: Triticeae). BMC Plant Biol. 2022, 22, 57. [Google Scholar] [CrossRef]
- Soreng, R.J.; Gillespie, L.J.; Boudko, E.A.; Cabi, E. Biogeography, timing, and life-history traits in the PPAM clade: Coleanthinae (syn. Puccinelliinae), Poinae, Alopecurinae superclade, Miliinae, and Avenulinae and Phleinae (Poaceae, Pooideae, Poeae). J. Syst. Evol. 2022, 60, 591–620. [Google Scholar] [CrossRef]
- Tzvelev, N.N. Grasses of the USSR; Nauka: Moscow, Russia, 1976; p. 788. (In Russian) [Google Scholar]
- Stace, C.A.; Preston, C.D.; Pearman, D.A. Hybrid Flora of the British Isles; BSBI Publications: Durham, UK, 2015; p. 501. [Google Scholar]
- Sieber, V.K.; Murray, B.G. The cytology of the genus Alopecurus (Gramineae). Bot. J. Lin. Soc. 1979, 79, 343–355. [Google Scholar] [CrossRef]
- Sieber, V.K.; Murray, B.G. Spontaneous polyploids in marginal populations of Alopecurus bulbosus Gouan (Poaceae). Bot. J. Lin. Soc. 1980, 81, 293–300. [Google Scholar] [CrossRef]
- Bolkhovskikh, Z.V.; Grif, V.G.; Zakharyeva, O.I.; Matveeva, T.S. Chromosomal Numbers of Flowering Plants; Nauka: Leningrad, Russia, 1969; p. 926. (In Russian) [Google Scholar]
- Sokolovskaya, A.P.; Probatova, N.S. Chromosome numbers of some species of Alopecurus L. of the USSR flora. Vestn. Len. Univ. Ser. 16 Biol. [Leningrad Univ. Biol. Sci. Bull.] 1974, 21, 62–67. (In Russian) [Google Scholar]
- Agapova, N.D.; Arkharova, K.B.; Vakhtina, E.A.; Zemskova, E.A.; Tarvis, L.V. Chromosome Numbers in Flowering Plants of the Flora of the USSR: Moraceae—Zygophyllaceae; Nauka: St. Petersburg, Russia, 1993; p. 430. (In Russian) [Google Scholar]
- Kamelin, R.V. The peculiarities of angiosperm speciation. In Trudy Zoologicheskogo Instituta RAN; Works of Zoological Institute of RAS. Appl. 1; Works of Zoological Institute of RAS: Saint Petersburg, Russia, 2009; pp. 141–149. [Google Scholar]
- Soreng, R.J.; Peterson, P.M.; Davidse, G.; Judziewicz, E.J.; Zuloaga, F.O.; Filgueiras, T.S.; Morrone, O. Catalogue of New World Grasses (Poaceae): IV. Subfamily Pooideae. Contr. US Nat. Herb. 2003, 48, 1–730. [Google Scholar]
- Soreng, R.J. Chloroplast-DNA phylogenetics and biogeography in a reticulating group: Study in Poa (Poaceae). Am. J. Bot. 1990, 77, 1383–1400. [Google Scholar] [CrossRef]
- Surveswaran, S.; Kumar, P.; Sun, M. Spiranthes himalayensis (Orchidaceae, Orchidoideae) a new species from Asia. PhytoKeys 2017, 89, 115–128. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nosov, N.N.; Tikhomirov, V.N.; Machs, E.M.; Rodionov, A.V. On polyphyly of the former section Ochlopoa and the hybridogenic section Acroleucae (Poa, Poaceae): Insights from molecular phylogenetic analyses. Nord. J. Bot. 2019, 37, e02015. [Google Scholar] [CrossRef]
- Strelkova, O.S. Polyploidy and Geographo-systematic Groups in the Genus Alopecurus L. Cytologia 1938, 8, 468–480. [Google Scholar] [CrossRef][Green Version]
- Rodionov, A.V.; Punina, E.O.; Dobroradova, M.A.; Tyupa, N.B.; Nosov, N.N. Chromosome numbers of some grasses (Poaceae): Aveneae, Poeae, Phalarideae, Phleeae, Bromeae, Triticeae. Bot. Zhurn. 2006, 91, 615–627. (In Russian) [Google Scholar]
- Tzvelev, N.N. The system of Grasses (Poaceae) and their evolution. Bot. Rev. 1989, 55, 141–203. [Google Scholar] [CrossRef]
- Ridgway, K.P.; Duck, J.M.; Young, J.P.W. Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnL (UAA) intron. BMC Ecol. 2003, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; the UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef] [PubMed]
- Múrias dos Santos, A.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. tcsBU: A tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).