Abstract
Gastrointestinal (GI) cancers impose a substantial global health burden, highlighting the necessity for deeper understanding of their intricate pathogenesis and treatment strategies. This review explores the interplay between intratumoral microbiota, tumor metabolism, and major types of GI cancers (including esophageal, gastric, liver, pancreatic, and colorectal cancers), summarizing recent studies and elucidating their clinical implications and future directions. Recent research revealed altered microbial signatures within GI tumors, impacting tumor progression, immune responses, and treatment outcomes. Dysbiosis-induced alterations in tumor metabolism, including glycolysis, fatty acid metabolism, and amino acid metabolism, play critical roles in cancer progression and therapeutic resistance. The integration of molecular mechanisms and potential biomarkers into this understanding further enhances the prognostic significance of intratumoral microbiota composition and therapeutic opportunities targeting microbiota-mediated tumor metabolism. Despite advancements, challenges remain in understanding the dynamic interactions within the tumor microenvironment (TME). Future research directions, including advanced omics technologies and prospective clinical studies, offer promising avenues for precision oncology and personalized treatment interventions in GI cancer. Overall, integrating microbiota-based approaches and molecular biomarkers into GI cancer management holds promise for improving patient outcomes and survival.
1. Introduction
Gastrointestinal (GI) cancers encompass a diverse group of malignancies affecting the digestive system, mainly including the esophagus, stomach, liver, pancreas, colon, and rectum (colorectum) [1,2,3]. These cancers collectively pose a significant global health burden, representing a leading cause of cancer-related morbidity and mortality worldwide. Each type of GI cancer presents distinct challenges in terms of diagnosis, treatment, and prognosis. For instance, colorectal cancer (CRC) is one of the most common GI malignancies, with risk factors including age, family history, dietary habits, and inflammatory bowel diseases (IBDs) [4]. On the other hand, pancreatic cancer (PCA) often presents at an advanced stage, leading to poor prognosis and limited treatment options [5]. Despite advancements in screening, early detection, and treatment modalities such as surgery, chemotherapy, radiation therapy, and targeted therapy, GI cancers remain associated with high mortality rates. Moreover, treatment resistance, disease recurrence, and metastasis pose formidable challenges in the management of these malignancies. Understanding the complex interplay between genetic, environmental, and lifestyle factors in the development and progression of GI cancers is critical for improving patient outcomes.
Intratumoral microbiota/microbiome refers to the diverse community of microorganisms residing within tumor tissues [6,7]. The composition of intratumoral microbiota is incredibly diverse, comprising bacteria, viruses, fungi, and archaea [8]. While the exact sources of these microorganisms are not fully understood, they may originate from the surrounding tissue, bloodstream, gut, or other organs. Additionally, some microbes may be attracted to the unique conditions within tumors, including hypoxia, altered pH levels, and nutrient availability [9]. Research into the role of intratumoral microbiota in cancer revealed both intriguing possibilities and complexities. Some studies suggest that certain microorganisms and their byproducts can promote inflammation, genomic instability, and immune suppression, all of which contribute to tumorigenesis [10,11]. However, there is also evidence to suggest that certain microbes may exert anti-tumor effects by stimulating immune responses against cancer cells [6].
Tumor metabolism, characterized by alterations in cellular energy production and nutrient utilization, plays a crucial role in cancer progression [12,13,14]. Cancer cells exhibit metabolic reprogramming to sustain their rapid proliferation, evade immune surveillance, and adapt to the tumor microenvironment (TME). One hallmark of tumor metabolism is aerobic glycolysis, also known as the Warburg effect, wherein cancer cells preferentially utilize glycolysis for energy production even in the presence of oxygen [15,16,17,18]. This metabolic shift allows cancer cells to generate biomass for proliferation and produce metabolites that support tumor growth and survival. In addition to glycolysis, cancer cells exhibit alterations in other metabolic pathways, including amino acid metabolism, lipid metabolism, and nucleotide biosynthesis [12,13,19]. These metabolic adaptations enable cancer cells to meet the increased demands for building blocks and energy required for sustained growth and proliferation. Furthermore, the TME influences tumor metabolism through factors such as nutrient availability, oxygen tension, and interactions with stromal cells and immune cells [20,21,22,23]. Dysregulated metabolism not only fuels tumor progression, but also contributes to therapeutic resistance and immune evasion. Understanding the metabolic dependencies of cancer cells and the TME is essential for developing targeted therapies that exploit metabolic vulnerabilities in cancer. Moreover, exploring the influence of intratumoral microbiota on tumor metabolism represents a novel avenue for therapeutic intervention in cancer treatment.
The gut microbiota (GM) emerged as a key player in GI health and disease, influencing various aspects of host physiology, including immune function, metabolism, and inflammation [24]. Dysbiosis, or imbalance in the GM composition, was implicated in the pathogenesis of GI disorders, including IBDs and GI cancers [25,26]. Given the close anatomical proximity between the gut and GI tumors, it is plausible that the intratumoral microbiota may influence the development and progression of GI cancers. Moreover, the unique metabolic characteristics of GI tumors, such as altered nutrient utilization and microenvironmental conditions, may create specific niches for microbial colonization and activity within the tumor. Investigating the influence of intratumoral microbiota on tumor metabolism in GI cancers holds significant promise for uncovering novel therapeutic targets and biomarkers. Understanding how microbial communities interact with tumor cells and modulate metabolic pathways may lead to the development of microbiota-based interventions to improve treatment outcomes and patient survival.
Overall, exploring the intricate interplay between intratumoral microbiota, tumor metabolism, and gastrointestinal cancer represents a promising area of research with the potential to advance our understanding of cancer biology and inform the development of personalized therapeutic strategies.
2. Intratumoral Microbiota and GI Cancer
2.1. Origin of Intratumoral Microbiota in GI Cancer
Recent studies identified three main sources of intratumoral microorganisms in GI cancers: mucosal barriers, adjacent normal tissues, and hematogenous spread [7,27]. Mucosal barrier disruption in cancers such as colorectal and pancreatic allows microorganisms from the gut and oral microbiota to invade tumors. Strong evidence suggests that intestinal microbes may be a significant source of intratumoral microbiota [28]. Cancers in the GI system have cavities exposed to the external environment, which favor microbial colonization. Adjacent normal tissues, once thought sterile, can harbor bacteria similar to those found in tumor tissues, aided by the immunosuppressive and hypoxic TME, which fosters microbial colonization. Hematogenous spread involves microorganisms from sites such as the mouth and intestines entering the bloodstream and reaching tumors via damaged blood vessels. For instance, in CRC, Escherichia coli (E. coli) can breach the gut vascular barrier, enter the bloodstream, and colonize the liver, promoting metastasis. Most tumor-associated bacteria and fungi are intracellular, often within cancer or immune cells, suggesting they may be transported as cellular fragments or intact cells, though direct infection through blood vessels cannot be excluded. The TME’s conditions support microbial growth and migration, allowing both “driver” bacteria such as Bacteroides fragilis and Helicobacter pylori (H. pylori), which facilitate carcinogenesis, and “passenger” opportunistic bacteria to thrive [29].
2.2. Recent Studies on the Potential Mechanisms and Biomarker Significance of Intratumoral Microbiota in GI Cancer
2.2.1. Esophageal Cancer
The Cancer Microbiome Atlas (TCMA) provides a pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants [30]. Recent studies leveraging TCMA data revealed distinct microbial signatures between upper (including esophageal and gastric cancer) and lower (including CRC) gastrointestinal tumors, identifying specific bacteria correlated with clinical characteristics of GI cancers [31]. In esophageal carcinoma (ESCA), TCMA analysis unveiled significant alterations in microbial composition between tumor and normal tissues, with notable changes observed in Firmicutes and Proteobacteria. Moreover, a microbial signature consisting of ten microbes was found to correlate with ESCA subtype, tumor stage, and survival status [32]. In esophageal squamous cell carcinoma (ESCC), investigations into the impact of intratumoral microbiota on neoadjuvant chemoimmunotherapy (NACI) response revealed that Streptococcus enrichment correlates with improved treatment outcomes, including increased CD8+ T-cell infiltration and prolonged disease-free survival. Notably, mouse experiments suggest that manipulating the microbiota could enhance the efficacy of immunotherapy. The presence of intratumoral microbiota is linked to the presurgical chemoimmunotherapy response in patients with ESCC. Specifically, the enrichment of Streptococcus signatures correlates with a favorable response to cancer immunotherapy. Additionally, fecal microbiota transplantation (FMT) may alter the composition of tumor-resident microbiota, enhancing the response to immunotherapy [33]. Intratumoral microbiome impacts immune infiltrates in TME and predicts prognosis in ESCC patients [34]. Fusobacterium nucleatum (F. nucleatum) functions as an oncogenic bacterium and can promote ESCC tumor progression. High F. nucleatum burden was associated with poor recurrence-free survival (RFS) in ESCC patients and correlated with reduced response to neoadjuvant chemotherapy, suggesting that targeting this bacterium may improve therapeutic outcomes in ESCC [35].
The microbiota of the distal esophagus are affected by acid reflux from the stomach. Acid reflux causes inflammation and mucosal damage, leading to alterations in the microbiome of the distal esophagus. This change allows columnar epithelium to replace the original squamous epithelium, potentially progressing to gastroesophageal reflux disease (GERD), Barrett’s esophagus (BE), and esophageal adenocarcinoma (EAC). In the upper part of the esophagus, the microbiota is influenced by oral resident flora, with Porphyromonas gingivalis (P. gingivalis) playing a role in promoting the development of ESCC [36,37].
2.2.2. Gastric Cancer
H. pylori is a well-known carcinogen associated with gastric cancer (GC), documented extensively in epidemiological and clinical studies [38,39]. H. pylori infection in the stomach primarily contributes to GC through its ability to induce chronic gastritis and subsequent inflammatory cascades. The bacteria stimulate the production of pro-inflammatory cytokines such as IL-1β, TNF-α, and IL-6, creating a pro-tumorigenic environment [40]. Furthermore, chronic inflammation induced by microbial dysbiosis can result in the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS). These reactive molecules can cause oxidative damage to DNA, proteins, and lipids, thereby contributing to mutagenesis and carcinogenesis. The persistent DNA damage and resulting genomic instability foster an environment conducive to malignant transformation and tumor development in the GI tract. Studies investigated the role of microbiome in GC development, recognizing H. pylori, while also identifying additional microbial species enriched in GC samples. Using a robust bioinformatics pipeline, the study found decreased microbial diversity in GC compared to nonmalignant tissue across two large cohorts. Moreover, distinct microbial enrichment patterns were observed among GC molecular subtypes [41]. Altered microbial diversity and enrichment of specific genera were observed in GC intratumoral microbiota, with Methylobacterium being significantly associated with poor prognosis and reduced CD8+ tissue-resident memory T (TRM) cells in the TME. The findings highlight the potential role of Methylobacterium in modulating TGFβ expression and CD8+ TRM cells in GC [42]. A recent study investigated the role of non-H. pylori gastric microbiota in stomach adenocarcinoma (STAD) by analyzing RNA sequencing, clinical, and DNA methylation data from The Cancer Genome Atlas (TCGA) project. The intratumoral microbiome profiles were associated with STAD occurrence, progression, and prognosis, with differential methylation changes observed in genes related to cancer pathways. Bi-directional mediation effects between intratumoral microorganisms and host DNA methylation were identified, highlighting their significance in cancer metastasis and prognosis. Additionally, cell experiments demonstrated that certain microorganisms, such as Staphylococcus saccharolyticus, could influence gastric cell proliferation and invasion. These findings underscore the intricate interplay between the intratumoral microbiome and host epigenetics in STAD progression and TME dynamics [43].
2.2.3. Liver Cancer
As a microbe, hepatitis B virus (HBV) is one well-known risk factor of hepatocellular carcinoma (HCC) [44]. In recent years, the GM was recognized to significantly affect HCC development. However, the comprehensive characterization of the HCC tumor microbiome remained largely elusive. Li et al. employed metagenomic sequencing to characterize the intratumoral microbiota in HBV-related HCC [45]. More recently, Liu et al. found that HBV-related HCC exhibits distinct intratumoral microbiota and immune microenvironment signatures [46]. Using a combination of 16S rRNA fluorescence in situ hybridization (FISH), immunohistochemistry, and sequencing techniques, studies revealed distinct bacterial characteristics and metabolic profiles in HCC tissues compared to adjacent nontumor tissues [47]. Moreover, Huang et al. investigated the intratumoral microbiota associated with HCC progression, revealing increased diversity compared to normal liver tissue. Similarly, findings from another research also reveal higher microbial diversity in HCC tissues compared to adjacent tissues, with increased abundances of certain microorganisms such as Enterobacteriaceae and Fusobacterium, alongside decreased levels of antitumour bacteria such as Pseudomonas. Additionally, alterations in microbial metabolic pathways, particularly enhanced fatty acid and lipid synthesis, were implicated in influencing HCC progression [48]. Predominant phyla included Patescibacteria, Proteobacteria, Bacteroidota, Firmicutes, and Actinobacteriota, with specific taxa such as Streptococcaceae and Lactococcus marking HCC cirrhosis [49]. Additionally, studies demonstrated that the intratumoural microbiome signature can predict the prognosis of HCC [50,51].
Except for liver cancer, a study characterized the intratumor microbiome of intrahepatic cholangiocarcinoma (ICC) tissues. They identified a Gram-positive aerobic bacterium, Staphylococcus capitis, present within the tumors. Additionally, they found a higher abundance of Paraburkholderia fungorum in paracancerous tissues, which exhibited antitumor activity against ICC through modulation of alanine, aspartate, and glutamate metabolism, suggesting a potential therapeutic avenue for ICC treatment [52].
2.2.4. Pancreatic Cancer
Pancreatic cancer (PCA) is an increasingly growing source of cancer-related deaths and is often diagnosed at advanced stages, and thereby resistant to conventional therapies [53]. The investigation of intratumoral microbiota in PCA represents a burgeoning field with profound implications for understanding tumorigenesis, disease progression, and therapeutic interventions [54,55]. Studies elucidated how microbiota and their products can influence the pancreatic TME, modulate the biological behavior of cancer cells, and impact immune system functionality. Intratumoral microbiota, including anaerobic bacteria such as Bacteroides, Lactobacillus, and Peptoniphilus, correlate with immune suppression and poor prognosis in pancreatic ductal adenocarcinoma (PDAC) by creating a hypoxic microenvironment that supports their growth and affects tumor metabolism [56]. While still in its infancy, research showed promising results in treating pancreatic cancer by targeting and modulating the intratumoral microbiota. Table 1 summarizes the recent studies on intratumoral microbiota in PCA research. These emerging evidences not only shed light on the complex interplay between microbiota and PCA, but also offer exciting prospects for developing innovative diagnostic and therapeutic strategies to combat this deadly disease.

Table 1.
Summary of recent studies on intratumoral microbiota in pancreatic cancer.
2.2.5. Colorectal Cancer
Colorectal cancer (CRC) is a multifactorial disease characterized by complex interactions between genetic, environmental, and microbial factors [73,74]. Emerging evidence suggests that the human GM plays a crucial role in CRC development and progression [75]. Specifically, the intratumoral microbiota garnered significant attention due to its potential influence on tumor biology, immune responses, and treatment outcomes. For instance, F. nucleatum is a dominant bacterial species in CRC tissue that is frequently associated with CRC and was implicated in promoting tumorigenesis through various mechanisms. F. nucleatum was reported as being associated with cancer progression and poorer patient prognosis of CRC [76]. Additionally, other bacterial species, such as Bacteroides fragilis, E. coli, and certain members of the Firmicutes phylum, were identified within CRC tumors [6], highlighting the diverse microbial composition within these tissues. Table 2 summarizes the recent studies on intratumoral microbiota in CRC research.
Intratumoral microbiota can contribute to CRC tumorigenesis and progression through various mechanisms. Firstly, certain bacterial species produce genotoxins, such as colibactin produced by some strains of E. coli, which can directly damage DNA. Colibactin induces double-strand breaks in the DNA, leading to mutations and genomic instability, which are critical steps in the initiation and progression of human CRC [77]. Additionally, chronic inflammation is a well-established risk factor for cancer, and intratumoral microbiota can significantly contribute to inflammatory responses within the GI tract. Persistent colonization by pathogenic bacteria can lead to continuous activation of the host immune response, resulting in chronic inflammation. In CRC, bacteria such as enterotoxigenic Bacteroides fragilis produce enterotoxins that trigger inflammation and promote tumorigenesis through the STAT3 signaling pathway [78]. The ongoing inflammation not only induces DNA damage and genomic instability, but also supports a microenvironment that promotes tumor growth and invasion. Moreover, intratumoral microbiota can influence CRC by activating carcinogenic pathways. For example, F. nucleatum can interact with cancer cells and modulate signaling pathways that promote tumorigenesis [79]. F. nucleatum adheres to epithelial cells through FadA adhesin, which activates β-catenin signaling, a pathway often implicated in CRC [80]. Activation of β-catenin signaling leads to increased cell proliferation, invasion, and resistance to apoptosis. Certain microbial metabolites, such as secondary bile acids produced by gut microbiota, can activate nuclear receptors such as farnesoid X receptor (FXR) and constitutive androstane receptor (CAR), influencing cell proliferation and survival pathways [81,82]. Furthermore, intratumoral microbiota can exert immunosuppressive effects that facilitate tumor growth and metastasis in CRC. F. nucleatum promote the recruitment of myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs), which suppress effective immune responses against the tumor [83]. This immunosuppression hampers the host’s capacity to recognize and attack tumor cells, thereby facilitating cancer progression.

Table 2.
Summary of recent studies on intratumoral microbiota in colorectal cancer.
Table 2.
Summary of recent studies on intratumoral microbiota in colorectal cancer.
| Year of Study | Nature of Study | Participants | Aim and Main Findings | Refs. |
|---|---|---|---|---|
| 2024 | Observational study | CRC patients | This study aimed to elucidate the genetic factors of F. nucleatum facilitating tumor colonization in CRC by analyzing closed genomes of 135 F. nucleatum strains. It identifies a distinct clade, Fna C2, as predominant in CRC tumors, showing increased metabolic potential and colonization of the GI tract, and providing insights into the pathoadaptation of Fna C2 to the CRC tumor niche. | [76] |
| 2023 | Observational study | CRC patients | The study proposes a method to detect bacterial signals in human RNA sequencing data and associates them with clinical and molecular properties of tumors. The analysis reveals correlations between intratumoral microbiome composition and survival, anatomic location, microsatellite instability, consensus molecular subtype, and immune cell infiltration in colon tumors. | [84] |
| 2024 | Prospective-Observational study | CRC patients | This study investigates the tumor microbial profile of young-onset CRC (yoCRC) compared to average-onset CRC (aoCRC), revealing higher microbial diversity and distinct microbial compositions in yoCRC tumors. Akkermansia and Bacteroides are enriched in yoCRC tumors, while aoCRC tumors show more abundances of several other bacteria. | [85] |
| 2023 | Observational study | Patients with locally advanced rectal cancer | This study investigates the tumor-bearing microbiota in patients with locally advanced rectal cancer before neoadjuvant chemoradiation therapy (nCRT) and its association with treatment response. The findings reveal specific microbial biomarkers and functional pathways associated with resistance to nCRT, highlighting the potential role of intratumoral microbiota in modulating treatment outcomes in rectal cancer patients. | [86] |
| 2023 | Observational study | CRC patients (data from the TCGA) | The aim of this study was to unravel the potential remodeling mechanisms of immune cell infiltration and tumorigenesis in CRC by integrating genetic, epigenetic, and intratumor microbial factors. Results reveal the significant influence of intratumor microbes on immune cell infiltration patterns, prognosis, and response to immune checkpoint blockade therapy in CRC. | [87] |
| 2023 | Observational study | Locally advanced rectal cancer (LARC) patients (data from a published European cohort) | This study investigates the intratumoral microbiota in LARC patients and its association with the response to nCRT. It identifies microbial signatures associated with pathological complete response (pCR) and non-pCR groups, highlighting their potential as independent predictive markers for nCRT response and revealing interactions between intratumoral microbes and cancer-associated fibroblasts (CAFs) in mediating treatment response. | [88] |
| 2024 | Observational study | CRC patients | The study aimed to understand how Colibactin-producing E. coli influences tumor heterogeneity, chemoresistance, and patient survival in right-sided CRC tumors. The main findings show that Colibactin-producing E. coli-infected tumors had high glycerophospholipid environments, reduced CD8+ T lymphocyte infiltration, and increased chemoresistance through lipid droplet accumulation and phosphatidylcholine remodeling. | [89] |
| 2021 | Observational study | CRC patients | The study explored the association between the intratumor microbiome and host genetic alterations in CRC patients. Fusobacterium was associated with mutated genes and cell cycle-related pathways, while Campylobacter abundance was linked to mutational signature 3, suggesting a potential role of bacterial-induced DNA damage in CRC. | [90] |
| 2017 | Observational study | Microsatellite instability-high (MSI-H) CRC patients | The study aimed to investigate the clinicopathologic and molecular associations of F. nucleatum in MSI-H CRC patients. High intratumoral F. nucleatum were associated with increased macrophage infiltration and CDKN2A promoter methylation in MSI-H CRC. | [91] |
| 2021 | Observational study | MSI-H CRC patients | In MSI-H CRC, high levels of intratumoral F. nucleatum are associated with larger tumor size and advanced invasion depth. Additionally, F. nucleatum-enriched tumors exhibit decreased density of FoxP3+ T cells and an increased proportion of M2-polarized macrophages in the tumor center. | [92] |
| 2018 | Observational study | CRC patients | The aim of this study was to investigate the association between the amount of Bifidobacteria in CRC tissue and tumor differentiation, specifically the extent of signet ring cells, as well as the immune response to CRC. The main findings reveal that intratumor bifidobacteria were detected in 30% of cases and were associated with the extent of signet ring cells, suggesting a possible role of bifidobacteria in determining distinct tumor characteristics or as an indicator of dysfunctional mucosal barrier in CRC. | [93] |
| 2019 | Observational study | CRC patients | The aim of this study was to investigate the prognostic impact of intratumoral F. nucleatum in CRC patients treated with adjuvant chemotherapy. Intratumoral F. nucleatum load was found to be a potential prognostic factor in stage II/III CRC patients treated with oxaliplatin-based adjuvant chemotherapy, particularly in non-MSI-H/non-sigmoid/non-rectal cancer subsets. | [94] |
3. Role of Intratumoral Microbiota in Modulating GI Tumor Metabolism
3.1. Influence of Microbial Dysbiosis on Tumor Metabolic Reprogramming
Dysbiosis, or the disruption of microbial balance, can lead to alterations in microbial composition and function, influencing tumor metabolism [95]. Dysbiotic conditions within the TME can trigger metabolic reprogramming in cancer cells, promoting tumor growth, invasion, and metastasis [96,97]. This dysregulated metabolism often involves shifts in key metabolic pathways, including glycolysis, fatty acid metabolism, and amino acid metabolism, which support the energetic and biosynthetic demands of rapidly proliferating cancer cells.
3.2. Specific Metabolic Pathways Affected by Intratumoral Microbiota in GI Cancer
3.2.1. Glycolysis
Recently, studies reported the crosstalk between intratumoral bacteria and the tumor. In the immediate TME, marked by vascular hyperplasia, aerobic glycolysis, hypoxia, and immunosuppression, bacterial proliferation becomes favorable. Intratumoral bacteria, integral to this milieu, significantly impacts tumor progression, metastasis, and the efficacy of anti-tumor treatments [98]. The intricate interplay between the intratumoral microbiota and tumor metabolism in GI cancer involves modulation of specific metabolic pathways critical for cancer progression and therapeutic response. One such pathway is glycolysis, the process by which glucose is metabolized to produce energy in the form of ATP. Dysbiosis within the TME can promote glycolytic flux in cancer cells, leading to increased glucose uptake and lactate production, a phenomenon known as the Warburg effect. For instance, the normal esophageal microbiome, which is primarily composed of Gram-positive bacteria, undergoes a shift to a predominantly Gram-negative microbiome during the dysbiosis associated with the EAC cascade. This altered microbiota can then promote the pathogenesis of EAC by activating toll-like receptors (TLRs), inducing cyclooxygenase-2 expression, stimulating inducible nitric oxide synthase, activating the NLRP3 inflammasome, and contributing to the Warburg effect [37]. Additionally, PDAC is known to create a hypoxic microenvironment due to its limited cellularity and dense, desmoplastic stroma [99,100]. Hypoxia in PDAC was shown to enhance the intracellular survival of anaerobic bacteria such as P. gingivalis [101]. Recent research also indicates that intestinal bacteria can intensify the hypoxic conditions, which in turn modulates tissue-resident lymphocytes [102]. Consequently, the tumor stroma and the intratumoral colonization of bacteria may establish the hypoxic environment that supports the growth of anaerobic bacteria in PDAC cases, potentially leading to poor prognosis through immune suppression. Certain bacterial species, such as F. nucleatum, were implicated in promoting glycolysis in CRC cells by upregulating glucose transporters and glycolytic enzymes [103,104]. This metabolic shift towards glycolysis not only provides cancer cells with a sustained energy supply, but also contributes to the acidic TME, fostering tumor growth and immune evasion.
3.2.2. Fatty Acid Metabolism
Another metabolic pathway influenced by intratumoral microbiota is fatty acid metabolism, which plays a crucial role in providing cancer cells with essential lipids for membrane synthesis, energy storage, and signaling [47,105]. Dysbiosis-associated alterations in lipid metabolism were observed in GI tumors, with certain bacterial species implicated in promoting lipogenesis and lipid accumulation within cancer cells [106]. For example, F. nucleatum was shown to induce expression of fatty acid synthase (FASN), a key enzyme involved in de novo lipogenesis in CRC cells [107]. This dysregulated lipid metabolism not only fuels cancer cell proliferation and survival, but also contributes to tumor progression and chemoresistance.
3.2.3. Amino Acid Metabolism
Furthermore, amino acid metabolism represents another metabolic pathway influenced by the intratumoral microbiota in cancer pathogenesis and immunity [27]. Amino acids serve as crucial building blocks for protein synthesis and also play diverse roles in cellular metabolism and signaling pathways. Dysbiosis-induced alterations in amino acid metabolism were linked to tumor progression and therapeutic resistance in GI cancers. For instance, certain bacterial species can modulate the availability of specific amino acids, such as glutamine and arginine, within the TME, thereby impacting cancer cell metabolism and immune cell function [108]. Additionally, dysbiosis-associated alterations in amino acid metabolism can contribute to the generation of immunosuppressive metabolites, such as kynurenine, which suppress anti-tumor immune responses and promote immune evasion in tumors [109,110,111].
Overall, the relationship between intratumoral microbiota and tumor metabolism in GI cancers is complex and multifaceted. Intratumoral microbiota can influence tumor metabolism by producing metabolites such as short-chain fatty acids (SCFAs), which alter gene expression and metabolic pathways in cancer cells. In GI cancers, tumor cells often undergo metabolic reprogramming, increasing glycolysis, glutaminolysis, and lipid metabolism. This metabolic environment promotes the growth of specific microbiota, which in turn produce metabolites that further support tumor metabolism and proliferation. Moreover, intratumoral microbiota can affect the TME, influencing the efficacy of metabolic therapies and immunotherapies. Figure 1 summarizes the complex interactions between intratumoral microbiota and tumor metabolism in GI cancers. Additionally, different types of GI cancers exhibit unique patterns of microbiota metabolism interactions, which require further study to understand fully.
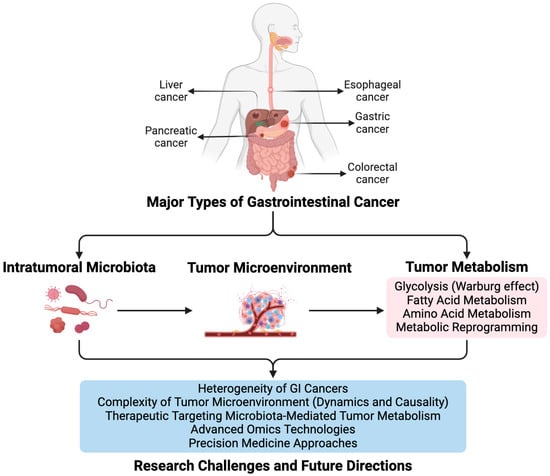
Figure 1.
Summary of Intratumoral Microbiota’s Influence on Tumor Metabolism in Major Types of GI Cancer.
4. Clinical Implications of Intratumoral Microbiota-Mediated Tumor Metabolism in GI Cancer
4.1. Prognostic Significance of Intratumoral Microbiota Composition
The composition of intratumoral microbiota emerged as a potential prognostic marker in GI cancer, offering valuable insights into disease progression and patient outcomes [11,112]. As described above, numerous studies reported associations between specific microbial species or dysbiotic patterns and clinical outcomes in GI cancer patients. For instance, higher abundance of F. nucleatum is consistently associated with poorer prognosis in colorectal CRC, including increased risk of recurrence, metastasis, and reduced overall survival. Similarly, alterations in the composition of intratumoral microbiota, such as decreased microbial diversity or enrichment of pathogenic bacteria, were linked to adverse clinical outcomes in various GI malignancies. Understanding the prognostic significance of intratumoral microbiota composition holds promise for improving risk stratification and informing personalized treatment strategies in GI cancer patients.
4.2. Therapeutic Opportunities Targeting Microbiota-Mediated Tumor Metabolism in GI Cancer
The influence of intratumoral microbiota on tumor metabolism presents novel therapeutic opportunities for the treatment of GI cancer. Targeting microbiota-mediated tumor metabolism represents a promising approach to disrupt cancer progression and enhance therapeutic efficacy in GI malignancies. Several strategies were proposed to modulate intratumoral microbiota and tumor metabolism for therapeutic benefit. These include the use of probiotics, prebiotics, antibiotics, and microbial-based therapies to manipulate the composition and function of intratumoral microbiota. Additionally, targeting specific metabolic pathways dysregulated by microbiota within the TME holds potential for developing innovative treatment modalities. For example, inhibitors targeting key enzymes involved in microbial-induced metabolic reprogramming, such as FASN or glycolytic enzymes, may offer therapeutic benefits in GI cancer. Furthermore, strategies aimed at restoring metabolic homeostasis and immune function within the TME, such as metabolic inhibitors or immunotherapies, hold promise for overcoming microbiota-mediated immune evasion and enhancing antitumor immune responses in GI cancer. To further illustrate these therapeutic and diagnostic methodologies, a schematic pipeline from research to clinical application is provided in Figure 2. This pipeline demonstrates how intratumoral microbiota can be leveraged for therapeutic and diagnostic purposes in GI cancer. Overall, therapeutic targeting of microbiota-mediated tumor metabolism represents a promising avenue for the development of novel treatment strategies in GI cancer, with the potential to improve patient outcomes and survival.
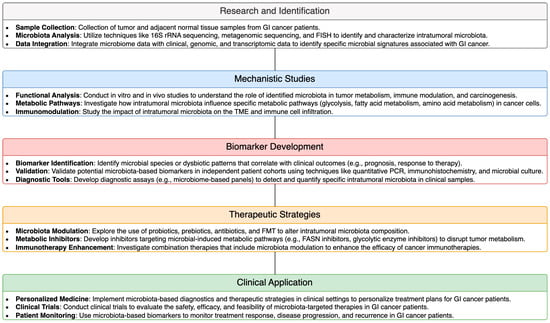
Figure 2.
Schematic Pipeline of Therapeutic and Diagnostic Methodologies Utilizing Intratumoral Microbiota in GI Cancer.
5. Challenges and Future Directions
5.1. Current Limitations in Understanding the Complex Interplay between Intratumoral Microbiota and Tumor Metabolism
Despite significant advancements, several challenges hinder our comprehensive understanding of the intricate relationship between intratumoral microbiota and tumor metabolism in GI cancer. One major limitation is the complexity of the TME, which comprises diverse microbial communities interacting with tumor, stromal, and immune cells [113,114]. Characterizing the spatial distribution and functional activities of intratumoral microbiota within this complex ecosystem remains challenging, requiring advanced techniques and multidisciplinary approaches. Additionally, the dynamic nature of microbial communities and tumor metabolism poses challenges in deciphering causal relationships and temporal dynamics [115,116]. Furthermore, the heterogeneity of GI cancers, both within and between patients, adds another layer of complexity to studying microbiota-mediated tumor metabolism.
5.2. Future Research Directions for Unraveling the Mechanisms and Clinical Applications in GI Cancer
Several promising research directions hold potential for advancing our understanding of intratumoral microbiota-mediated tumor metabolism and translating these insights into clinical applications for GI cancer. Firstly, applying advanced omics technologies, such as metagenomics, metabolomics, and single-cell sequencing, will enable comprehensive profiling of intratumoral microbiota and tumor metabolism at high resolution [117]. Integrative analyses of multi-omics data will provide insights into the functional interactions between microbiota, tumor cells, and the TME. Moreover, mechanistic studies focusing on specific microbial metabolites and metabolic pathways implicated in GI cancer progression will elucidate the underlying molecular mechanisms and identify potential therapeutic targets. Additionally, prospective clinical studies with larger cohorts and longitudinal follow-up are needed to validate the prognostic and predictive value of intratumoral microbiota composition and metabolic signatures in GI cancer. Finally, the development of microbiota-targeted therapeutics and precision medicine approaches tailored to individual patient profiles will improve treatment outcomes and patient survival in GI cancer. Overall, addressing these research priorities will be beneficial for harnessing the potential of intratumoral microbiota in guiding precision oncology strategies and personalized treatment interventions in GI cancer.
6. Conclusions
In conclusion, this review highlighted the significant impact of intratumoral microbiota on tumor metabolism and its potential as a biomarker in five major types of GI cancer, as illustrated in Figure 1. Through the summary of current research, we identified key findings that demonstrate how microbial dysbiosis within the tumor microenvironment (TME) can affect cancer progression, molecular mechanisms, and treatment outcomes. Furthermore, we emphasize the importance of integrating microbiota-based approaches and molecular biomarkers into GI cancer management strategies. By understanding the complex interplay between intratumoral microbiota, tumor metabolism, and molecular mechanisms, clinicians and researchers can develop novel therapeutic interventions and precision medicine strategies tailored to individual patients. Overall, this comprehensive understanding opens new avenues for targeted therapies and personalized treatment modalities, ultimately improving outcomes for patients with GI cancer.
Author Contributions
Conceptualization, C.L.; writing—original draft preparation, X.B. and J.W.; writing—review and editing, C.L.; funding acquisition, J.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Open Funds for Shaanxi Provincial Key Laboratory of Infection and Immune Diseases (No. 2023-KFMS-1).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
GI: gastrointestinal; CRC: colorectal cancer; IBDs: inflammatory bowel diseases; PCA: pancreatic cancer; TME: tumor microenvironment; GM: gut microbiota; E. coli: Escherichia coli; H. pylori: Helicobacter pylori; TCMA: The Cancer Microbiome Atlas; ESCA: esophageal carcinoma; ESCC: esophageal squamous cell carcinoma; NACI: neoadjuvant chemoimmunotherapy; FMT: fecal microbiota transplantation; F. nucleatum: Fusobacterium nucleatum; RFS: recurrence-free survival; GERD: gastroesophageal reflux disease; BE: Barrett’s esophagus; EAC: esophageal adenocarcinoma; P. gingivalis: Porphyromonas gingivalis; GC: gastric cancer; ROS: reactive oxygen species; RNS: reactive nitrogen species; TRM: tissue-resident memory T; STAD: stomach adenocarcinoma; TCGA: The Cancer Genome Atlas; HBV: hepatitis B virus; HCC: hepatocellular carcinoma; FISH: fluorescence in situ hybridization; ICC: intrahepatic cholangiocarcinoma; PDAC: pancreatic ductal adenocarcinoma; LTS: long-term survival; ILC2: innate lymphoid cells 2; NE: neutrophil elastase; IPMNs: intraductal papillary mucinous neoplasms; FXR: farnesoid X receptor; CAR: constitutive androstane receptor; MDSCs: myeloid-derived suppressor cells; Tregs: regulatory T cells; nCRT: neoadjuvant chemoradiation therapy; LARC: locally advanced rectal cancer; CAFs: cancer-associated fibroblasts; MSI-H: microsatellite instability-high; TLRs: toll-like receptors; FASN: fatty acid synthase; and SCFAs: short-chain fatty acids.
References
- Arnold, M.; Abnet, C.C.; Neale, R.E.; Vignat, J.; Giovannucci, E.L.; McGlynn, K.A.; Bray, F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology 2020, 159, 335–349.e15. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.H.; Guan, C.A.; Bujang, M.A.; Lai, W.H.; Voon, P.J.; Sim, E.U.H. Identification of Phenomic Data in the Pathogenesis of Cancers of the Gastrointestinal (GI) Tract in the UK Biobank. Sci. Rep. 2024, 14, 1997. [Google Scholar] [CrossRef] [PubMed]
- Jardim, S.R.; de Souza, L.M.P.; de Souza, H.S.P. The Rise of Gastrointestinal Cancers as a Global Phenomenon: Unhealthy Behavior or Progress? Int. J. Environ. Res. Public Health 2023, 20, 3640. [Google Scholar] [CrossRef] [PubMed]
- Marabotto, E.; Kayali, S.; Buccilli, S.; Levo, F.; Bodini, G.; Giannini, E.G.; Savarino, V.; Savarino, E.V. Colorectal Cancer in Inflammatory Bowel Diseases: Epidemiology and Prevention: A Review. Cancers 2022, 14, 4254. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic Cancer: A Review of Clinical Diagnosis, Epidemiology, Treatment and Outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, P.; Mei, W.; Zeng, C. Intratumoral Microbiota: Implications for Cancer Onset, Progression, and Therapy. Front. Immunol. 2023, 14, 1301506. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, A.; Wang, Y.; Zhang, Y. Intratumoral Microbiota: Roles in Cancer Initiation, Development and Therapeutic Efficacy. Signal Transduct. Target. Ther. 2023, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, J.; Cai, W.; Huang, Y.; Liu, X.; Ma, Z.; Tang, Z.; Bian, X.; Zheng, J.; Jiang, J.; et al. The Emerging Tumor Microbe Microenvironment: From Delineation to Multidisciplinary Approach-Based Interventions. Acta Pharm. Sin. B 2024, 14, 1560–1591. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, K.; Taylor, C.T. The Impact of Hypoxia on Bacterial Infection. FEBS J. 2015, 282, 2260–2266. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Yu, B.; Rao, B.; Sun, Y.; Yu, J.; Wang, D.; Cui, G.; Ren, Z. The Effect of the Intratumoral Microbiome on Tumor Occurrence, Progression, Prognosis and Treatment. Front. Immunol. 2022, 13, 1051987. [Google Scholar] [CrossRef]
- Xuan, M.; Gu, X.; Liu, Y.; Yang, L.; Li, Y.; Huang, D.; Li, J.; Xue, C. Intratumoral Microorganisms in Tumors of the Digestive System. Cell Commun. Signal 2024, 22, 69. [Google Scholar] [CrossRef]
- Hammoudi, N.; Ahmed, K.B.; Garcia-Prieto, C.; Huang, P. Metabolic Alterations in Cancer Cells and Therapeutic Implications. Chin. J. Cancer 2011, 30, 508–525. [Google Scholar] [CrossRef]
- Yang, J.; Shay, C.; Saba, N.F.; Teng, Y. Cancer Metabolism and Carcinogenesis. Exp. Hematol. Oncol. 2024, 13, 10. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Zhu, J.; Thompson, C.B. The Hallmarks of Cancer Metabolism: Still Emerging. Cell Metab. 2022, 34, 355–377. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Zhou, D.; Duan, Z.; Li, Z.; Ge, F.; Wei, R.; Kong, L. The Significance of Glycolysis in Tumor Progression and Its Relationship with the Tumor Microenvironment. Front. Pharmacol. 2022, 13, 1091779. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gillies, R.J. Why Do Cancers Have High Aerobic Glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef]
- Elia, I.; Haigis, M.C. Metabolites and the Tumour Microenvironment: From Cellular Mechanisms to Systemic Metabolism. Nat. Metab. 2021, 3, 21–32. [Google Scholar] [CrossRef]
- Zhao, Y.; Shen, M.; Wu, L.; Yang, H.; Yao, Y.; Yang, Q.; Du, J.; Liu, L.; Li, Y.; Bai, Y. Stromal Cells in the Tumor Microenvironment: Accomplices of Tumor Progression? Cell Death Dis. 2023, 14, 587. [Google Scholar] [CrossRef]
- Lobel, G.P.; Jiang, Y.; Simon, M.C. Tumor Microenvironmental Nutrients, Cellular Responses, and Cancer. Cell Chem. Biol. 2023, 30, 1015–1032. [Google Scholar] [CrossRef]
- Kaymak, I.; Williams, K.S.; Cantor, J.R.; Jones, R.G. Immunometabolic Interplay in the Tumor Microenvironment. Cancer Cell 2021, 39, 28–37. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the Human Gut Microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Singh, R.; Zogg, H.; Wei, L.; Bartlett, A.; Ghoshal, U.C.; Rajender, S.; Ro, S. Gut Microbial Dysbiosis in the Pathogenesis of Gastrointestinal Dysmotility and Metabolic Disorders. J. Neurogastroenterol. Motil. 2021, 27, 19–34. [Google Scholar] [CrossRef]
- Agagunduz, D.; Cocozza, E.; Cemali, O.; Bayazit, A.D.; Nani, M.F.; Cerqua, I.; Morgillo, F.; Saygili, S.K.; Berni Canani, R.; Amero, P.; et al. Understanding the Role of the Gut Microbiome in Gastrointestinal Cancer: A Review. Front. Pharmacol. 2023, 14, 1130562. [Google Scholar] [CrossRef]
- Wang, M.; Yu, F.; Li, P. Intratumor Microbiota in Cancer Pathogenesis and Immunity: From Mechanisms of Action to Therapeutic Opportunities. Front. Immunol. 2023, 14, 1269054. [Google Scholar] [CrossRef]
- Xie, Y.; Xie, F.; Zhou, X.; Zhang, L.; Yang, B.; Huang, J.; Wang, F.; Yan, H.; Zeng, L.; Zhang, L.; et al. Microbiota in Tumors: From Understanding to Application. Adv. Sci. 2022, 9, e2200470. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A Bacterial Driver-Passenger Model for Colorectal Cancer: Beyond the Usual Suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Dohlman, A.B.; Arguijo Mendoza, D.; Ding, S.; Gao, M.; Dressman, H.; Iliev, I.D.; Lipkin, S.M.; Shen, X. The Cancer Microbiome Atlas: A Pan-Cancer Comparative Analysis to Distinguish Tissue-Resident Microbiota from Contaminants. Cell Host Microbe 2021, 29, 281–298.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Li, Z.; Gao, X.; Huang, D. Global Analysis of Microbiota Signatures in Four Major Types of Gastrointestinal Cancer. Front. Oncol. 2021, 11, 685641. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, H.; Gao, X.; Wang, J. The Intratumor Microbiota Signatures Associate with Subtype, Tumor Stage, and Survival Status of Esophageal Carcinoma. Front. Oncol. 2021, 11, 754788. [Google Scholar] [CrossRef]
- Wu, H.; Leng, X.; Liu, Q.; Mao, T.; Jiang, T.; Liu, Y.; Li, F.; Cao, C.; Fan, J.; Chen, L.; et al. Intratumoral Microbiota Composition Regulates Chemoimmunotherapy Response in Esophageal Squamous Cell Carcinoma. Cancer Res. 2023, 83, 3131–3144. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, S.; Ma, X.; Zhan, J.; Pan, C.; Zhang, H.; Xie, X.; Wen, J.; Xie, X. Intratumoral Microbiome Impacts Immune Infiltrates in Tumor Microenvironment and Predicts Prognosis in Esophageal Squamous Cell Carcinoma Patients. Front. Cell Infect. Microbiol. 2023, 13, 1165790. [Google Scholar] [CrossRef]
- Yamamura, K.; Izumi, D.; Kandimalla, R.; Sonohara, F.; Baba, Y.; Yoshida, N.; Kodera, Y.; Baba, H.; Goel, A. Intratumoral Fusobacterium nucleatum Levels Predict Therapeutic Response to Neoadjuvant Chemotherapy in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res. 2019, 25, 6170–6179. [Google Scholar] [CrossRef]
- Li, Y.; Wei, B.; Xue, X.; Li, H.; Li, J. Microbiome Changes in Esophageal Cancer: Implications for Pathogenesis and Prognosis. Cancer Biol. Med. 2023, 21, 163–174. [Google Scholar] [CrossRef]
- Gillespie, M.R.; Rai, V.; Agrawal, S.; Nandipati, K.C. The Role of Microbiota in the Pathogenesis of Esophageal Adenocarcinoma. Biology 2021, 10, 697. [Google Scholar] [CrossRef]
- Khatoon, J.; Rai, R.P.; Prasad, K.N. Role of Helicobacter pylori in Gastric Cancer: Updates. World J. Gastrointest. Oncol. 2016, 8, 147–158. [Google Scholar] [CrossRef]
- Maleki Kakelar, H.; Barzegari, A.; Dehghani, J.; Hanifian, S.; Saeedi, N.; Barar, J.; Omidi, Y. Pathogenicity of Helicobacter pylori in Cancer Development and Impacts of Vaccination. Gastric Cancer 2019, 22, 23–36. [Google Scholar] [CrossRef]
- Xi, Y.; Zhang, X.L.; Luo, Q.X.; Gan, H.N.; Liu, Y.S.; Shao, S.H.; Mao, X.H. Helicobacter pylori Regulates Stomach Diseases by Activating Cell Pathways and DNA Methylation of Host Cells. Front. Cell Dev. Biol. 2023, 11, 1187638. [Google Scholar] [CrossRef]
- Abate, M.; Vos, E.; Gonen, M.; Janjigian, Y.Y.; Schattner, M.; Laszkowska, M.; Tang, L.; Maron, S.B.; Coit, D.G.; Vardhana, S.; et al. A Novel Microbiome Signature in Gastric Cancer: A Two Independent Cohort Retrospective Analysis. Ann. Surg. 2022, 276, 605–615. [Google Scholar] [CrossRef]
- Peng, R.; Liu, S.; You, W.; Huang, Y.; Hu, C.; Gao, Y.; Jia, X.; Li, G.; Xu, Z.; Chen, Y. Gastric Microbiome Alterations Are Associated with Decreased CD8+ Tissue-Resident Memory T Cells in the Tumor Microenvironment of Gastric Cancer. Cancer Immunol. Res. 2022, 10, 1224–1240. [Google Scholar] [CrossRef]
- Yue, K.; Sheng, D.; Xue, X.; Zhao, L.; Zhao, G.; Jin, C.; Zhang, L. Bidirectional Mediation Effects between Intratumoral Microbiome and Host DNA Methylation Changes Contribute to Stomach Adenocarcinoma. Microbiol. Spectr. 2023, 11, e0090423. [Google Scholar] [CrossRef]
- Ji, J.; Ji, F.; Bayarsaikhan, E. Intratumoral Microbiota in HCC: A New Kid on the Block? Hepatology 2023, 78, 1012–1014. [Google Scholar] [CrossRef]
- Li, S.; Xia, H.; Wang, Z.; Zhang, X.; Song, T.; Li, J.; Xu, L.; Zhang, N.; Fan, S.; Li, Q.; et al. Intratumoral Microbial Heterogeneity Affected Tumor Immune Microenvironment and Determined Clinical Outcome of HBV-Related HCC. Hepatology 2023, 78, 1079–1091. [Google Scholar] [CrossRef]
- Liu, Y.; Kim, E.S.; Guo, H. Hepatitis B Virus-Related Hepatocellular Carcinoma Exhibits Distinct Intratumoral Microbiota and Immune Microenvironment Signatures. J. Med. Virol. 2024, 96, e29485. [Google Scholar] [CrossRef]
- Xue, C.; Gu, X.; Shi, Q.; Ma, X.; Jia, J.; Su, Y.; Bao, Z.; Lu, J.; Li, L. The Interaction between Intratumoral Bacteria and Metabolic Distortion in Hepatocellular Carcinoma. J. Transl. Med. 2024, 22, 237. [Google Scholar] [CrossRef]
- He, Y.; Zhang, Q.; Yu, X.; Zhang, S.; Guo, W. Overview of Microbial Profiles in Human Hepatocellular Carcinoma and Adjacent Nontumor Tissues. J. Transl. Med. 2023, 21, 68. [Google Scholar] [CrossRef]
- Huang, J.H.; Wang, J.; Chai, X.Q.; Li, Z.C.; Jiang, Y.H.; Li, J.; Liu, X.; Fan, J.; Cai, J.B.; Liu, F. The Intratumoral Bacterial Metataxonomic Signature of Hepatocellular Carcinoma. Microbiol. Spectr. 2022, 10, e0098322. [Google Scholar] [CrossRef]
- Sun, L.; Ke, X.; Guan, A.; Jin, B.; Qu, J.; Wang, Y.; Xu, X.; Li, C.; Sun, H.; Xu, H.; et al. Intratumoural Microbiome Can Predict the Prognosis of Hepatocellular Carcinoma after Surgery. Clin. Transl. Med. 2023, 13, e1331. [Google Scholar] [CrossRef]
- Song, Y.; Xiang, Z.; Lu, Z.; Su, R.; Shu, W.; Sui, M.; Wei, X.; Xu, X. Identification of a Brand Intratumor Microbiome Signature for Predicting Prognosis of Hepatocellular Carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 11319–11332. [Google Scholar] [CrossRef]
- Chai, X.; Wang, J.; Li, H.; Gao, C.; Li, S.; Wei, C.; Huang, J.; Tian, Y.; Yuan, J.; Lu, J.; et al. Intratumor Microbiome Features Reveal Antitumor Potentials of Intrahepatic Cholangiocarcinoma. Gut Microbes 2023, 15, 2156255. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, W.; Zhang, Z.; Sha, G.; Wang, D.; Tang, D. Intratumoral Microbiota: A New Force in Diagnosing and Treating Pancreatic Cancer. Cancer Lett. 2023, 554, 216031. [Google Scholar] [CrossRef]
- Amara, S.; Yang, L.V.; Tiriveedhi, V.; Muzaffar, M. Complex Role of Microbiome in Pancreatic Tumorigenesis: Potential Therapeutic Implications. Cells 2022, 11, 1900. [Google Scholar] [CrossRef]
- Panebianco, C.; Ciardiello, D.; Villani, A.; Maiorano, B.A.; Latiano, T.P.; Maiello, E.; Perri, F.; Pazienza, V. Insights into the Role of Gut and Intratumor Microbiota in Pancreatic Ductal Adenocarcinoma as New Key Players in Preventive, Diagnostic and Therapeutic Perspective. Semin. Cancer Biol. 2022, 86, 997–1007. [Google Scholar] [CrossRef]
- Abe, S.; Masuda, A.; Matsumoto, T.; Inoue, J.; Toyama, H.; Sakai, A.; Kobayashi, T.; Tanaka, T.; Tsujimae, M.; Yamakawa, K.; et al. Impact of Intratumoral Microbiome on Tumor Immunity and Prognosis in Human Pancreatic Ductal Adenocarcinoma. J. Gastroenterol. 2024, 59, 250–262. [Google Scholar] [CrossRef]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef]
- Alam, A.; Levanduski, E.; Denz, P.; Villavicencio, H.S.; Bhatta, M.; Alhorebi, L.; Zhang, Y.; Gomez, E.C.; Morreale, B.; Senchanthisai, S.; et al. Fungal Mycobiome Drives IL-33 Secretion and Type 2 Immunity in Pancreatic Cancer. Cancer Cell 2022, 40, 153–167.e11. [Google Scholar] [CrossRef]
- Tan, Q.; Ma, X.; Yang, B.; Liu, Y.; Xie, Y.; Wang, X.; Yuan, W.; Ma, J. Periodontitis Pathogen Porphyromonas gingivalis Promotes Pancreatic Tumorigenesis via Neutrophil Elastase from Tumor-Associated Neutrophils. Gut Microbes 2022, 14, 2073785. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Z.; Shen, X.; Xu, J.; Weng, Y.; Wang, W.; Xue, J. Clostridium butyricum and Its Metabolite Butyrate Promote Ferroptosis Susceptibility in Pancreatic Ductal Adenocarcinoma. Cell Oncol. 2023, 46, 1645–1658. [Google Scholar] [CrossRef] [PubMed]
- Pfisterer, N.; Ammer-Herrmenau, C.; Antweiler, K.; Kuffer, S.; Ellenrieder, V.; Neesse, A. Dynamics of Intestinal and Intratumoral Microbiome Signatures in Genetically Engineered Mice and Human Pancreatic Ductal Adenocarcinoma. Pancreatology 2023, 23, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Chakladar, J.; Kuo, S.Z.; Castaneda, G.; Li, W.T.; Gnanasekar, A.; Yu, M.A.; Chang, E.Y.; Wang, X.Q.; Ongkeko, W.M. The Pancreatic Microbiome Is Associated with Carcinogenesis and Worse Prognosis in Males and Smokers. Cancers 2020, 12, 2672. [Google Scholar] [CrossRef] [PubMed]
- Nalluri, H.; Jensen, E.; Staley, C. Role of Biliary Stent and Neoadjuvant Chemotherapy in the Pancreatic Tumor Microbiome. BMC Microbiol. 2021, 21, 280. [Google Scholar] [CrossRef] [PubMed]
- Merali, N.; Chouari, T.; Terroire, J.; Jessel, M.D.; Liu, D.S.K.; Smith, J.H.; Wooldridge, T.; Dhillon, T.; Jimenez, J.I.; Krell, J.; et al. Bile Microbiome Signatures Associated with Pancreatic Ductal Adenocarcinoma Compared to Benign Disease: A UK Pilot Study. Int. J. Mol. Sci. 2023, 24, 16888. [Google Scholar] [CrossRef] [PubMed]
- Kohi, S.; Macgregor-Das, A.; Dbouk, M.; Yoshida, T.; Chuidian, M.; Abe, T.; Borges, M.; Lennon, A.M.; Shin, E.J.; Canto, M.I.; et al. Alterations in the Duodenal Fluid Microbiome of Patients With Pancreatic Cancer. Clin. Gastroenterol. Hepatol. 2022, 20, e196–e227. [Google Scholar] [CrossRef] [PubMed]
- Hozaka, Y.; Oi, H.; Satake, S.; Uchino, Y.; Goto, Y.; Idichi, T.; Tanoue, K.; Yamasaki, Y.; Kawasaki, Y.; Mataki, Y.; et al. Are Intratumoral Microbiota Involved in the Progression of Intraductal Papillary Mucinous Neoplasms of the Pancreas? Surgery 2023, 173, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.S.; Yang, C.Y.; Yeh, C.C.; Lin, R.T.; Chen, C.C.; Bai, L.Y.; Hung, M.C.; Lin, C.C.; Wu, C.Y.; Lin, J.T. Endoscopic Ultrasound-Guided Fine-Needle Biopsy as a Tool for Studying the Intra-Tumoral Microbiome in Pancreatic Ductal Adenocarcinoma: A Pilot Study. Sci. Rep. 2022, 12, 107. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhu, N.; Zheng, X.; Liu, Y.; Lu, H.; Yin, X.; Hao, H.; Tan, Y.; Wang, D.; Hu, H.; et al. Intratumor Microbiome Analysis Identifies Positive Association Between Megasphaera and Survival of Chinese Patients with Pancreatic Ductal Adenocarcinomas. Front. Immunol. 2022, 13, 785422. [Google Scholar] [CrossRef]
- Udayasuryan, B.; Ahmad, R.N.; Nguyen, T.T.D.; Umana, A.; Monet Roberts, L.; Sobol, P.; Jones, S.D.; Munson, J.M.; Slade, D.J.; Verbridge, S.S. Fusobacterium nucleatum Induces Proliferation and Migration in Pancreatic Cancer Cells through Host Autocrine and Paracrine Signaling. Sci. Signal 2022, 15, eabn4948. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, J.; Li, H.; Huang, B.; Zhang, B.; Song, B.; Bao, C.; Liu, Y.; Wang, Z. Integrated Multi-Omics Identified the Novel Intratumor Microbiome-Derived Subtypes and Signature to Predict the Outcome, Tumor Microenvironment Heterogeneity, and Immunotherapy Response for Pancreatic Cancer Patients. Front. Pharmacol. 2023, 14, 1244752. [Google Scholar] [CrossRef] [PubMed]
- Kinskey, J.C.; Huda, T.I.; Gozlan, E.C.; Quach, J.U.; Arturo, J.F.; Chobrutskiy, A.; Chobrutskiy, B.I.; Blanck, G. The Presence of Intratumoral Porphyromonas gingivalis Correlates with a Previously Defined Pancreatic Adenocarcinoma, Immune Cell Expression Phenotype and with Tumor Resident, Adaptive Immune Receptor Features. Carcinogenesis 2023, 44, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.L.; Wang, Y.; Zeng, Z.; Deng, B.; Zhu, B.S.; Cao, T.; Li, Y.K.; Xiao, J.; Han, Q.; Wu, Q. Colorectal Cancer (CRC) as a Multifactorial Disease and Its Causal Correlations with Multiple Signaling Pathways. Biosci. Rep. 2020, 40, BSR20200265. [Google Scholar] [CrossRef]
- Heavey, P.M.; McKenna, D.; Rowland, I.R. Colorectal Cancer and the Relationship between Genes and the Environment. Nutr. Cancer 2004, 48, 124–141. [Google Scholar] [CrossRef]
- Li, J.; Zhu, Y.; Yang, L.; Wang, Z. Effect of Gut Microbiota in the Colorectal Cancer and Potential Target Therapy. Discov. Oncol. 2022, 13, 51. [Google Scholar] [CrossRef]
- Zepeda-Rivera, M.; Minot, S.S.; Bouzek, H.; Wu, H.; Blanco-Miguez, A.; Manghi, P.; Jones, D.S.; LaCourse, K.D.; Wu, Y.; McMahon, E.F.; et al. A Distinct Fusobacterium nucleatum Clade Dominates the Colorectal Cancer Niche. Nature 2024, 628, 424–432. [Google Scholar] [CrossRef]
- Leake, I. Genotoxins from Gut Bacteria. Nat. Biotechnol. 2022, 40, 1765. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.T.; Kantilal, H.K.; Davamani, F. The Mechanism of Bacteroides fragilis Toxin Contributes to Colon Cancer Formation. Malays. J. Med. Sci. 2020, 27, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.M.; Liu, H.L. Fusobacterium nucleatum and Colorectal Cancer: A Review. World J. Gastrointest. Oncol. 2018, 10, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Liu, X.; Liu, Z.; Pan, C.; Zhang, X.; Zhao, Z.; Sun, H. Fusobacterium nucleatum in Tumors: From Tumorigenesis to Tumor Metastasis and Tumor Resistance. Cancer Biol. Ther. 2024, 25, 2306676. [Google Scholar] [CrossRef]
- Xiang, D.; Yang, J.; Liu, L.; Yu, H.; Gong, X.; Liu, D. The Regulation of Tissue-Specific Farnesoid X Receptor on Genes and Diseases Involved in Bile Acid Homeostasis. Biomed. Pharmacother. 2023, 168, 115606. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Qian, L. Research on Gut Microbiota-Derived Secondary Bile Acids in Cancer Progression. Integr. Cancer Ther. 2022, 21, 15347354221114100. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, Y.; Mima, K.; Ishimoto, T.; Ogata, Y.; Imai, K.; Miyamoto, Y.; Akiyama, T.; Daitoku, N.; Hiyoshi, Y.; Iwatsuki, M.; et al. Relationship between Fusobacterium nucleatum and Antitumor Immunity in Colorectal Cancer Liver Metastasis. Cancer Sci. 2021, 112, 4470–4477. [Google Scholar] [CrossRef] [PubMed]
- Sambruni, G.; Macandog, A.D.; Wirbel, J.; Cagnina, D.; Catozzi, C.; Dallavilla, T.; Borgo, F.; Fazio, N.; Fumagalli-Romario, U.; Petz, W.L.; et al. Location and Condition Based Reconstruction of Colon Cancer Microbiome from Human RNA Sequencing Data. Genome Med. 2023, 15, 32. [Google Scholar] [CrossRef] [PubMed]
- Barot, S.V.; Sangwan, N.; Nair, K.G.; Schmit, S.L.; Xiang, S.; Kamath, S.; Liska, D.; Khorana, A.A. Distinct Intratumoral Microbiome of Young-Onset and Average-Onset Colorectal Cancer. eBioMedicine 2024, 100, 104980. [Google Scholar] [CrossRef]
- Huang, X.; Chen, C.; Xie, W.; Zhou, C.; Tian, X.; Zhang, Z.; Wang, Q.; Chang, H.; Xiao, W.; Zhang, R.; et al. Metagenomic Analysis of Intratumoral Microbiome Linking to Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2023, 117, 1255–1269. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, X.; Zhang, H.; Zhang, H.; Yi, Z.; Zhang, Q.; Liu, Q.; Liu, X. Multi-Omics Analysis Reveals Intratumor Microbes as Immunomodulators in Colorectal Cancer. Microbiol. Spectr. 2023, 11, e0503822. [Google Scholar] [CrossRef]
- Sun, L.; Qu, J.; Ke, X.; Zhang, Y.; Xu, H.; Lv, N.; Leng, J.; Zhang, Y.; Guan, A.; Feng, Y.; et al. Interaction between Intratumoral Microbiota and Tumor Mediates the Response of Neoadjuvant Therapy for Rectal Cancer. Front. Microbiol. 2023, 14, 1229888. [Google Scholar] [CrossRef]
- de Oliveira Alves, N.; Dalmasso, G.; Nikitina, D.; Vaysse, A.; Ruez, R.; Ledoux, L.; Pedron, T.; Bergsten, E.; Boulard, O.; Autier, L.; et al. The Colibactin-Producing Escherichia coli Alters the Tumor Microenvironment to Immunosuppressive Lipid Overload Facilitating Colorectal Cancer Progression and Chemoresistance. Gut Microbes 2024, 16, 2320291. [Google Scholar] [CrossRef]
- Okuda, S.; Shimada, Y.; Tajima, Y.; Yuza, K.; Hirose, Y.; Ichikawa, H.; Nagahashi, M.; Sakata, J.; Ling, Y.; Miura, N.; et al. Profiling of Host Genetic Alterations and Intra-Tumor Microbiomes in Colorectal Cancer. Comput. Struct. Biotechnol. J. 2021, 19, 3330–3338. [Google Scholar] [CrossRef]
- Park, H.E.; Kim, J.H.; Cho, N.Y.; Lee, H.S.; Kang, G.H. Intratumoral Fusobacterium nucleatum Abundance Correlates with Macrophage Infiltration and CDKN2A Methylation in Microsatellite-Unstable Colorectal Carcinoma. Virchows Arch. 2017, 471, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.A.; Yoo, S.Y.; Oh, H.J.; Jeong, S.; Cho, N.Y.; Kang, G.H.; Kim, J.H. Differential Immune Microenvironmental Features of Microsatellite-Unstable Colorectal Cancers According to Fusobacterium nucleatum Status. Cancer Immunol. Immunother. 2021, 70, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Kosumi, K.; Hamada, T.; Koh, H.; Borowsky, J.; Bullman, S.; Twombly, T.S.; Nevo, D.; Masugi, Y.; Liu, L.; da Silva, A.; et al. The Amount of Bifidobacterium Genus in Colorectal Carcinoma Tissue in Relation to Tumor Characteristics and Clinical Outcome. Am. J. Pathol. 2018, 188, 2839–2852. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.J.; Kim, J.H.; Bae, J.M.; Kim, H.J.; Cho, N.Y.; Kang, G.H. Prognostic Impact of Fusobacterium nucleatum Depends on Combined Tumor Location and Microsatellite Instability Status in Stage II/III Colorectal Cancers Treated with Adjuvant Chemotherapy. J. Pathol. Transl. Med. 2019, 53, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Proffitt, C.; Bidkhori, G.; Moyes, D.; Shoaie, S. Disease, Drugs and Dysbiosis: Understanding Microbial Signatures in Metabolic Disease and Medical Interventions. Microorganisms 2020, 8, 1381. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Li, R.; Chen, Z.; Li, G.; Liu, B.; Guo, S.; Yue, Q.; Yang, S.; Xie, L.; Zhang, Y.; et al. The Role of the Symbiotic Microecosystem in Cancer: Gut Microbiota, Metabolome, and Host Immunome. Front. Immunol. 2023, 14, 1235827. [Google Scholar] [CrossRef]
- Liu, J.; Luo, F.; Wen, L.; Zhao, Z.; Sun, H. Current Understanding of Microbiomes in Cancer Metastasis. Cancers 2023, 15, 1893. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Mao, Y.; Wang, L. The Crosstalk of Intratumor Bacteria and the Tumor. Front. Cell Infect. Microbiol. 2023, 13, 1273254. [Google Scholar] [CrossRef]
- Ye, L.Y.; Zhang, Q.; Bai, X.L.; Pankaj, P.; Hu, Q.D.; Liang, T.B. Hypoxia-Inducible Factor 1alpha Expression and Its Clinical Significance in Pancreatic Cancer: A Meta-Analysis. Pancreatology 2014, 14, 391–397. [Google Scholar] [CrossRef]
- Tan, Z.; Xu, J.; Zhang, B.; Shi, S.; Yu, X.; Liang, C. Hypoxia: A Barricade to Conquer the Pancreatic Cancer. Cell Mol. Life Sci. 2020, 77, 3077–3083. [Google Scholar] [CrossRef]
- Gnanasekaran, J.; Binder Gallimidi, A.; Saba, E.; Pandi, K.; Eli Berchoer, L.; Hermano, E.; Angabo, S.; Makkawi, H.A.; Khashan, A.; Daoud, A.; et al. Intracellular Porphyromonas gingivalis Promotes the Tumorigenic Behavior of Pancreatic Carcinoma Cells. Cancers 2020, 12, 2331. [Google Scholar] [CrossRef] [PubMed]
- Harada, Y.; Sujino, T.; Miyamoto, K.; Nomura, E.; Yoshimatsu, Y.; Tanemoto, S.; Umeda, S.; Ono, K.; Mikami, Y.; Nakamoto, N.; et al. Intracellular Metabolic Adaptation of Intraepithelial CD4(+)CD8alphaalpha(+) T Lymphocytes. iScience 2022, 25, 104021. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Guo, F.; Lu, S.Y.; Shen, C.; Ma, D.; Zhang, X.; Xie, Y.; Yan, T.; Yu, T.; Sun, T.; et al. F. Nucleatum Targets LncRNA ENO1-IT1 to Promote Glycolysis and Oncogenesis in Colorectal Cancer. Gut 2021, 70, 2123–2137. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Wang, H.; Tao, Y.; Luo, K.; Ye, J.; Ran, S.; Guan, Z.; Wang, Y.; Hu, H.; Huang, R. Fusobacterium nucleatum and Colorectal Cancer: From Phenomenon to Mechanism. Front. Cell Infect. Microbiol. 2022, 12, 1020583. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, Q.; Lu, W. Intratumoral Microbiome and Gastrointestinal Cancers. Front. Oncol. 2022, 12, 1047015. [Google Scholar] [CrossRef]
- Sheflin, A.M.; Whitney, A.K.; Weir, T.L. Cancer-Promoting Effects of Microbial Dysbiosis. Curr. Oncol. Rep. 2014, 16, 406. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Du, J.; Chao, S.; Li, S.; Cai, H.; Zhang, H.; Chen, G.; Liu, P.; Bu, P. Fusobacterium nucleatum Promotes Colorectal Cancer Cell to Acquire Stem Cell-Like Features by Manipulating Lipid Droplet-Mediated Numb Degradation. Adv. Sci. 2022, 9, e2105222. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Cai, J.; Hou, W.; Xu, K.; Wu, X.; Song, Y.; Bai, C.; Mo, Y.Y.; Zhang, Z. Microbiome and Spatially Resolved Metabolomics Analysis Reveal the Anticancer Role of Gut Akkermansia Muciniphila by Crosstalk with Intratumoral Microbiota and Reprogramming Tumoral Metabolism in Mice. Gut Microbes 2023, 15, 2166700. [Google Scholar] [CrossRef]
- Pacheco, J.H.L.; Elizondo, G. Interplay between Estrogen, Kynurenine, and AHR Pathways: An Immunosuppressive Axis with Therapeutic Potential for Breast Cancer Treatment. Biochem. Pharmacol. 2023, 217, 115804. [Google Scholar] [CrossRef]
- Dehhaghi, M.; Kazemi Shariat Panahi, H.; Heng, B.; Guillemin, G.J. The Gut Microbiota, Kynurenine Pathway, and Immune System Interaction in the Development of Brain Cancer. Front. Cell Dev. Biol. 2020, 8, 562812. [Google Scholar] [CrossRef]
- Canavese, M.; Wijesundara, D.; Maddern, G.J.; Grubor-Bauk, B.; Hauben, E. Hepatitis C Virus Drives the Pathogenesis of Hepatocellular Carcinoma: From Immune Evasion to Carcinogenesis. Clin. Transl. Immunol. 2016, 5, e101. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Chu, Q.; Zheng, Q.; Yuan, X.; Su, Y.; Bao, Z.; Lu, J.; Li, L. Current Understanding of the Intratumoral Microbiome in Various Tumors. Cell Rep. Med. 2023, 4, 100884. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor Microenvironment Complexity and Therapeutic Implications at a Glance. Cell Commun. Signal 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Xue, X.; Bukhari, I.; Qiu, C.; Li, Y.; Zheng, P.; Mi, Y. Gut Microbiota and Its Therapeutic Implications in Tumor Microenvironment Interactions. Front. Microbiol. 2024, 15, 1287077. [Google Scholar] [CrossRef]
- Antoniewicz, M.R. A Guide to Deciphering Microbial Interactions and Metabolic Fluxes in Microbiome Communities. Curr. Opin. Biotechnol. 2020, 64, 230–237. [Google Scholar] [CrossRef]
- Sieow, B.F.; Nurminen, T.J.; Ling, H.; Chang, M.W. Meta-Omics- and Metabolic Modeling-Assisted Deciphering of Human Microbiota Metabolism. Biotechnol. J. 2019, 14, e1800445. [Google Scholar] [CrossRef] [PubMed]
- Arikan, M.; Muth, T. Integrated Multi-Omics Analyses of Microbial Communities: A Review of the Current State and Future Directions. Mol. Omi. 2023, 19, 607–623. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).