Glaucoma-Associated CDR1 Peptide Promotes RGC Survival in Retinal Explants through Molecular Interaction with Acidic Leucine Rich Nuclear Phosphoprotein 32A (ANP32A)
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthetic CDR Peptides
2.2. Retina Organ Culture Model and Immunohistochemical Staining
2.3. Peptide-Based Immunoprecipitation
2.4. Molecular Dynamics (MD) Simulations and Virtual Docking Analysis
2.5. In-Solution Trypsin Digestion
2.6. Mass Spectrometric (MS) Analysis
2.7. Quantitative Analysis of the Proteomic MS Data
2.8. Statistics and Bioinformatics
2.9. Western Blot Analysis
3. Results
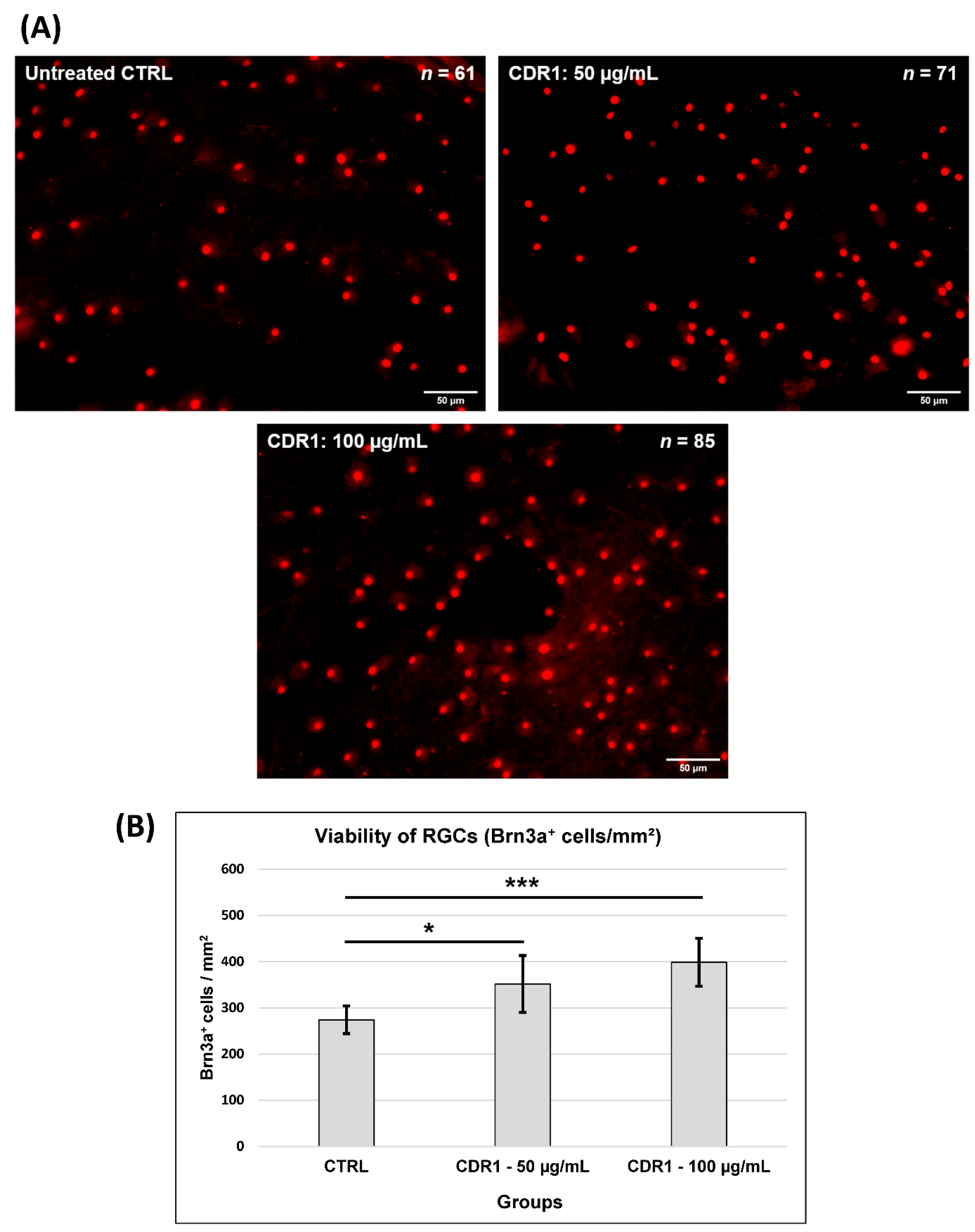
3.1. Effects of Synthetic CDR1 on RGCs Ex Vivo
3.2. Peptide-Based Immunoprecipitation of Retinal Protein Interaction Partners
3.3. MD Simulations and Binding Prediction Analyses of Synthetic CDR1
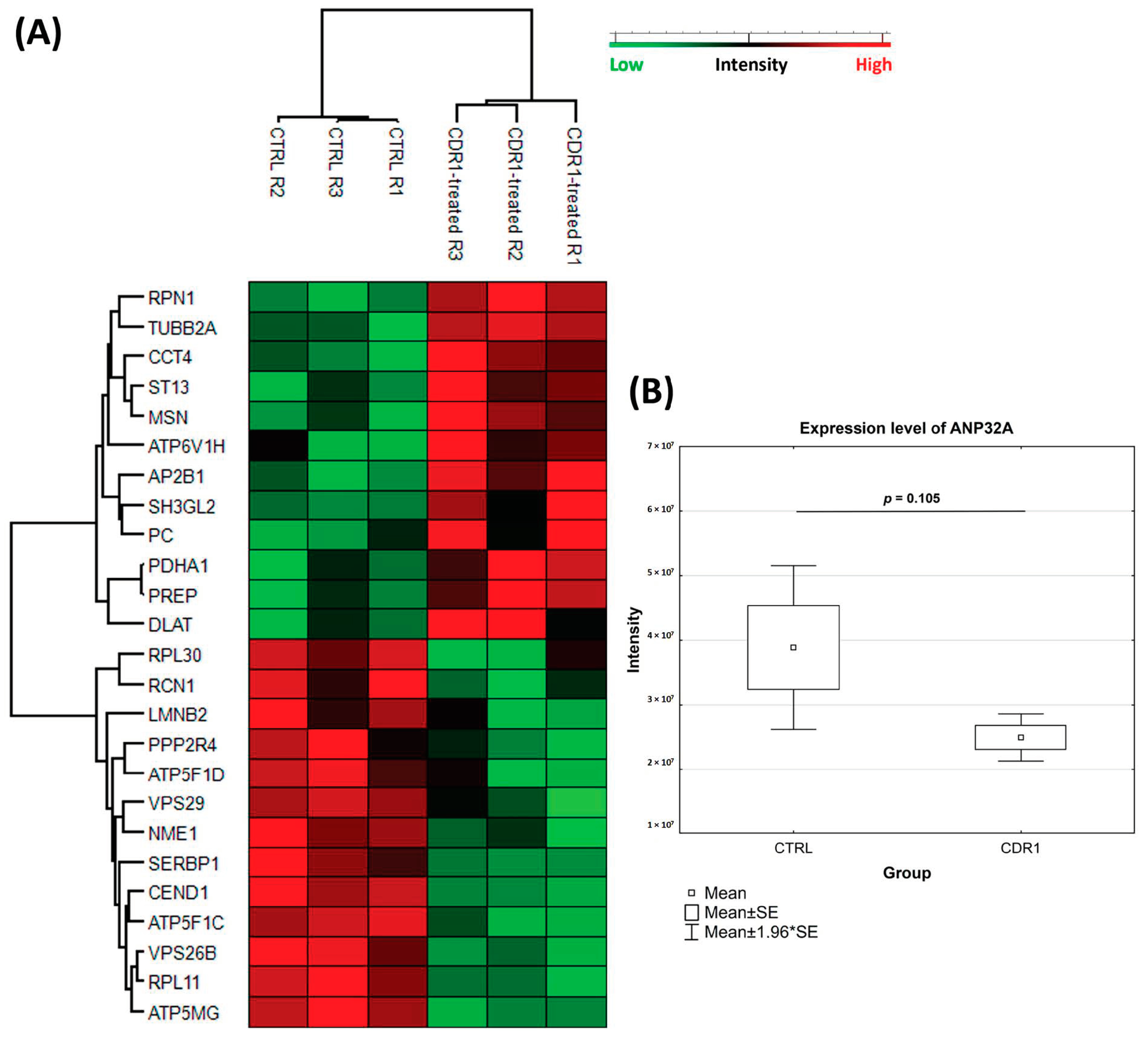
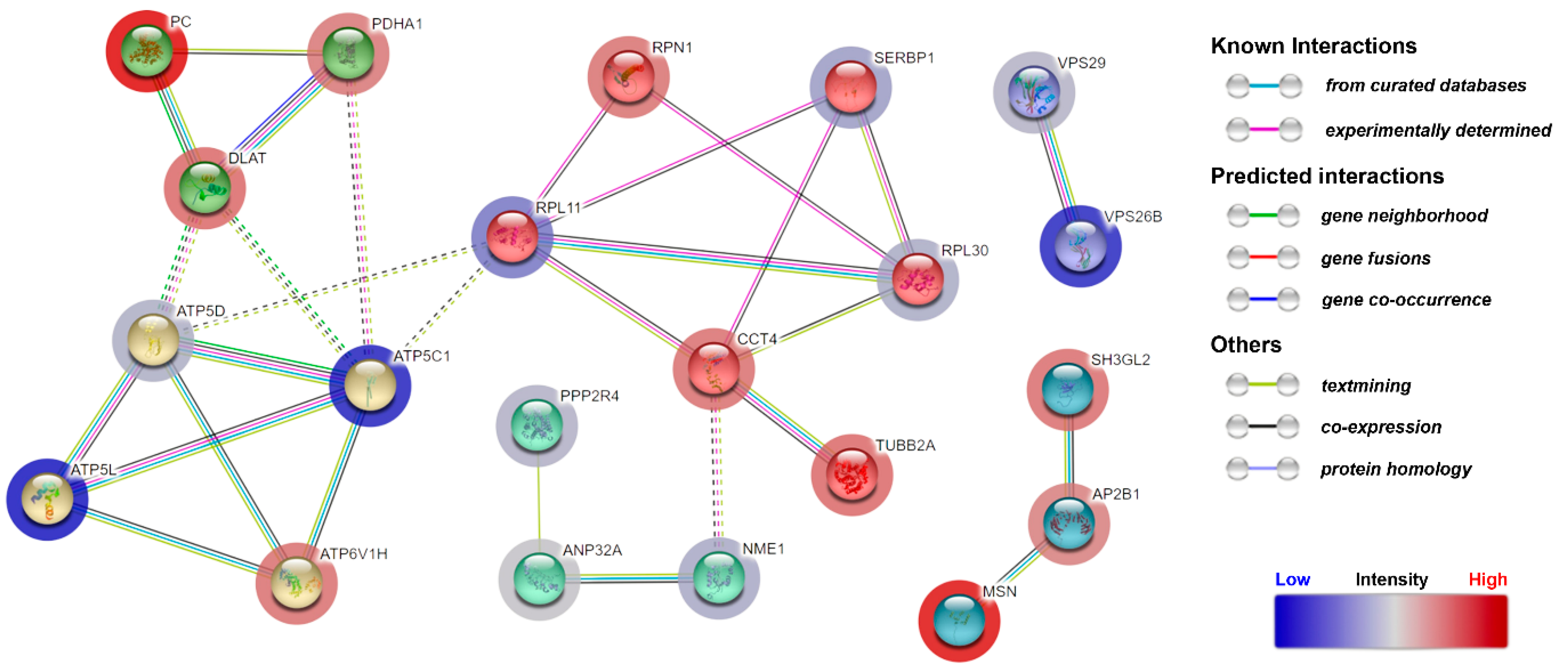
3.4. Quantitative Proteomic Analysis of CDR1-Treated Retinal Explants
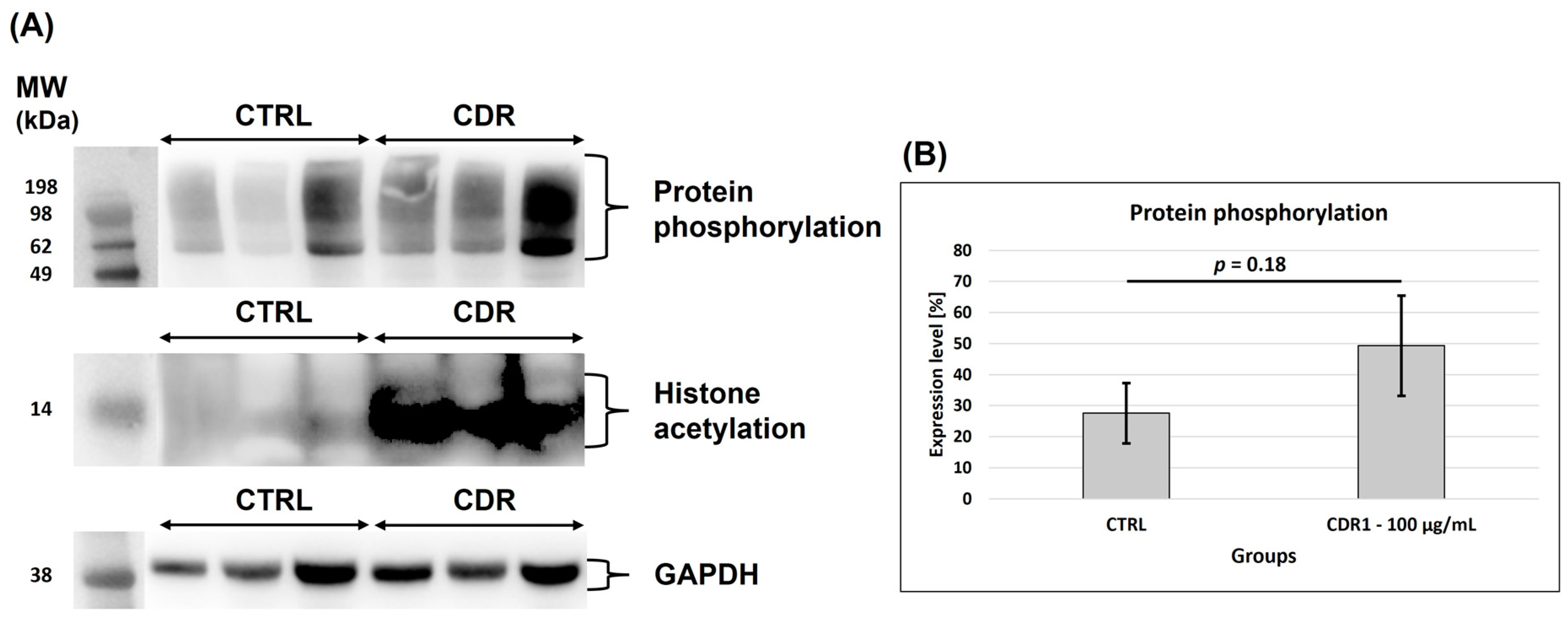
3.5. Western Blot Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tham, Y.-C.; Li, X.; Wong, T.Y.; Quigley, H.A.; Aung, T.; Cheng, C.-Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: A systematic review and meta-analysis. Ophthalmology 2014, 121, 2081–2090. [Google Scholar] [CrossRef]
- Weinreb, R.N.; Aung, T.; Medeiros, F.A. The pathophysiology and treatment of glaucoma: A review. JAMA 2014, 311, 1901–1911. [Google Scholar] [CrossRef]
- Conlon, R.; Saheb, H.; Ahmed, I.I.K. Glaucoma treatment trends: A review. Can. J. Ophthalmol. 2017, 52, 114–124. [Google Scholar] [CrossRef]
- Bettin, P.; Di Matteo, F. Glaucoma: Present challenges and future trends. Ophthalmic Res. 2013, 50, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Gutteridge, I.F. Normal tension glaucoma: Diagnostic features and comparisons with primary open angle glaucoma. Clin. Exp. Optom. 2000, 83, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Melief, C.J.M. Special Review: The future of Immunotherapy. Immunother. Adv. 2021, 1, ltaa005. [Google Scholar] [CrossRef]
- Wahid, B.; Ali, A.; Rafique, S.; Waqar, M.; Wasim, M.; Wahid, K.; Idrees, M. An overview of cancer immunotherapeutic strategies. Immunotherapy 2018, 10, 999–1010. [Google Scholar] [CrossRef]
- Schmelter, C.; Fomo, K.N.; Perumal, N.; Manicam, C.; Bell, K.; Pfeiffer, N.; Grus, F.H. Synthetic Polyclonal-Derived CDR Peptides as an Innovative Strategy in Glaucoma Therapy. J. Clin. Med. 2019, 8, 1222. [Google Scholar] [CrossRef] [PubMed]
- Tonner, H.; Hunn, S.; Auler, N.; Schmelter, C.; Beutgen, V.M.; von Pein, H.D.; Pfeiffer, N.; Grus, F.H. A Monoclonal Anti-HMGB1 Antibody Attenuates Neurodegeneration in an Experimental Animal Model of Glaucoma. IJMS 2022, 23, 4107. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.; Wilding, C.; Funke, S.; Perumal, N.; Beck, S.; Wolters, D.; Holz-Muller, J.; Pfeiffer, N.; Grus, F.H. Neuroprotective effects of antibodies on retinal ganglion cells in an adolescent retina organ culture. J. Neurochem. 2016, 139, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Fomo, K.N.; Schmelter, C.; Atta, J.; Beutgen, V.M.; Schwarz, R.; Perumal, N.; Govind, G.; Speck, T.; Pfeiffer, N.; Grus, F.H. Synthetic antibody-derived immunopeptide provides neuroprotection in glaucoma through molecular interaction with retinal protein histone H3. 1. Front. Med. 2022, 9, 3095. [Google Scholar] [CrossRef] [PubMed]
- Schmelter, C.; Fomo, K.N.; Perumal, N.; Pfeiffer, N.; Grus, F.H. Regulation of the HTRA2 Protease Activity by an Inhibitory Antibody-Derived Peptide Ligand and the Influence on HTRA2-Specific Protein Interaction Networks in Retinal Tissues. Biomedicines 2021, 9, 1013. [Google Scholar] [CrossRef]
- Reilly, P.T.; Yu, Y.; Hamiche, A.; Wang, L. Cracking the ANP32 whips: Important functions, unequal requirement, and hints at disease implications. Bioessays 2014, 36, 1062–1071. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Lu, Q.; Liu, X.; Wang, F.; Ma, X.; Cui, C.; Shi, C.; Li, J.; Zhang, D. The expression and distributions of ANP32A in the developing brain. Biomed Res. Int. 2015, 2015, 207347. [Google Scholar] [CrossRef]
- Chang, J.; Liu, Y.; Zhang, D.-D.; Zhang, D.-J.; Wu, C.-T.; Wang, L.-S.; Cui, C.-P. Hepatopoietin Cn suppresses apoptosis of human hepatocellular carcinoma cells by up-regulating myeloid cell leukemia-1. World J. Gastroenterol. 2010, 16, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Bai, Z.; Wang, J.; Li, X.; Chi, B.; Chen, X. ANP32A modulates cell growth by regulating p38 and Akt activity in colorectal cancer. Oncol. Rep. 2017, 38, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Lu, B.; Han, C.; Qiu, W.; Jin, Q.; Li, D.; Li, Q.; Yang, Q.; Wen, Q.; Opal, P.; et al. ANP32A dysregulation contributes to abnormal megakaryopoiesis in acute megakaryoblastic leukemia. Blood Cancer J. 2017, 7, 661. [Google Scholar] [CrossRef]
- Huang, S.; Huang, Z.; Ma, C.; Luo, L.; Li, Y.-F.; Wu, Y.-L.; Ren, Y.; Feng, C. Acidic leucine-rich nuclear phosphoprotein-32A expression contributes to adverse outcome in acute myeloid leukemia. Ann. Transl. Med. 2020, 8, 345. [Google Scholar] [CrossRef]
- Huyton, T.; Wolberger, C. The crystal structure of the tumor suppressor protein pp32 (Anp32a): Structural insights into Anp32 family of proteins. Protein Sci. 2007, 16, 1308–1315. [Google Scholar] [CrossRef]
- Zamora-Caballero, S.; Šiaučiunaite-Gaubard, L.; Bravo, J. High-resolution crystal structure of the leucine-rich repeat domain of the human tumour suppressor PP32A (ANP32A). Acta Crystallogr. Sect. F 2015, 71, 684–687. [Google Scholar] [CrossRef]
- Yang, X.; Lu, B.; Sun, X.; Han, C.; Fu, C.; Xu, K.; Wang, M.; Li, D.; Chen, Z.; Opal, P.; et al. ANP32A regulates histone H3 acetylation and promotes leukemogenesis. Leukemia 2018, 32, 1587–1597. [Google Scholar] [CrossRef]
- Donaghy, P.C.; Cockell, S.J.; Martin-Ruiz, C.; Coxhead, J.; Kane, J.; Erskine, D.; Koss, D.; Taylor, J.-P.; Morris, C.M.; O’Brien, J.T.; et al. Blood mRNA Expression in Alzheimer’s Disease and Dementia with Lewy Bodies. Am. J. Geriatr. Psychiatry 2022, 30, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, I.; Piñol, P.; Corral-Juan, M.; Pandolfo, M.; Matilla-Dueñas, A. A novel function of Ataxin-1 in the modulation of PP2A activity is dysregulated in the spinocerebellar ataxia type 1. Hum. Mol. Genet. 2013, 22, 3425–3437. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Chai, G.-S.; Wang, Z.-H.; Hu, Y.; Sun, D.-S.; Li, X.-G.; Ma, R.-H.; Li, Y.-R.; Ke, D.; Wang, J.-Z.; et al. Knockdown of pp32 Increases Histone Acetylation and Ameliorates Cognitive Deficits. Front. Aging Neurosci. 2017, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.-S.; Feng, Q.; Wang, Z.-H.; Hu, Y.; Sun, D.-S.; Li, X.-G.; Ke, D.; Li, H.-L.; Liu, G.-P.; Wang, J.-Z. Downregulating ANP32A rescues synapse and memory loss via chromatin remodeling in Alzheimer model. Mol. Neurodegener. 2017, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.-S.; Feng, Q.; Ma, R.-H.; Qian, X.-H.; Luo, D.-J.; Wang, Z.-H.; Hu, Y.; Sun, D.-S.; Zhang, J.-F.; Li, X.; et al. Inhibition of Histone Acetylation by ANP32A Induces Memory Deficits. J. Alzheimer’s Dis. 2018, 63, 1537–1546. [Google Scholar] [CrossRef]
- Schmelter, C.; Perumal, N.; Funke, S.; Bell, K.; Pfeiffer, N.; Grus, F.H. Peptides of the variable IgG domain as potential biomarker candidates in primary open-angle glaucoma (POAG). Hum. Mol. Genet. 2017, 26, 4451–4464. [Google Scholar] [CrossRef]
- Schrödinger, L.L. The PyMOL Molecular Graphics System; Version 1.8; LLC: New York, NY, USA, 2015. [Google Scholar]
- Lindahl, E.; Abraham, M.J.; Hess, B.; van der Spoel, D. GROMACS 2021 Manual; Zenodo: Meyrin, Switzerland, 2021. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces; Pullman, B., Ed.; Springer: Dordrecht, The Netherlands, 1981; pp. 331–342. ISBN 978-90-481-8368-5. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Bendixen, E.; Danielsen, M.; Larsen, K.; Bendixen, C. Advances in porcine genomics and proteomics—A toolbox for developing the pig as a model organism for molecular biomedical research. Brief. Funct. Genom. 2010, 9, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Bach, M.; Lehmann, A.; Brunnert, D.; Vanselow, J.T.; Hartung, A.; Bargou, R.C.; Holzgrabe, U.; Schlosser, A.; Chatterjee, M. Ugi Reaction-Derived alpha-Acyl Aminocarboxamides Bind to Phosphatidylinositol 3-Kinase-Related Kinases, Inhibit HSF1-Dependent Heat Shock Response, and Induce Apoptosis in Multiple Myeloma Cells. J. Med. Chem. 2017, 60, 4147–4160. [Google Scholar] [CrossRef] [PubMed]
- Nadal-Nicolás, F.M.; Jiménez-López, M.; Sobrado-Calvo, P.; Nieto-López, L.; Cánovas-Martínez, I.; Salinas-Navarro, M.; Vidal-Sanz, M.; Agudo, M. Brn3a as a marker of retinal ganglion cells: Qualitative and quantitative time course studies in naive and optic nerve-injured retinas. Investig. Ophthalmol. Vis. Sci. 2009, 50, 3860–3868. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Makkinje, A.; Damuni, Z. Molecular identification of I1PP2A, a novel potent heat-stable inhibitor protein of protein phosphatase 2A. Biochemistry 1996, 35, 6998–7002. [Google Scholar] [CrossRef]
- Chen, S.; Li, B.; Grundke-Iqbal, I.; Iqbal, K. I PP2A 1 affects Tau phosphorylation via association with the catalytic subunit of protein phosphatase 2A. J. Biol. Chem. 2008, 283, 10513–10521. [Google Scholar] [CrossRef]
- Tan, S.; Li, D.; Zhu, X. Cancer immunotherapy: Pros, cons and beyond. Biomed. Pharmacother. 2020, 124, 109821. [Google Scholar] [CrossRef]
- Mortada, I.; Farah, R.; Nabha, S.; Ojcius, D.M.; Fares, Y.; Almawi, W.Y.; Sadier, N.S. Immunotherapies for neurodegenerative diseases. Front. Neurol. 2021, 12, 654739. [Google Scholar] [CrossRef]
- Cabri, W.; Cantelmi, P.; Corbisiero, D.; Fantoni, T.; Ferrazzano, L.; Martelli, G.; Mattellone, A.; Tolomelli, A. Therapeutic Peptides Targeting PPI in Clinical Development: Overview, Mechanism of Action and Perspectives. Front. Mol. Biosci. 2021, 8, 697586. [Google Scholar] [CrossRef]
- Schmelter, C.; Funke, S.; Treml, J.; Beschnitt, A.; Perumal, N.; Manicam, C.; Pfeiffer, N.; Grus, F.H. Comparison of Two Solid-Phase Extraction (SPE) Methods for the Identification and Quantification of Porcine Retinal Protein Markers by LC-MS/MS. Int. J. Mol. Sci. 2018, 19, 3847. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Xie, J.; Bi, Y.; Zhang, H.; Dong, S.; Teng, L.; Lee, R.J.; Yang, Z. Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application. Front. Pharmacol. 2020, 11, 697. [Google Scholar] [CrossRef]
- Seo, S.; Macfarlan, T.; McNamara, P.; Hong, R.; Mukai, Y.; Heo, S.; Chakravarti, D. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J. Biol. Chem. 2002, 277, 14005–14010. [Google Scholar] [CrossRef]
- Wang, M.; Guo, H.; Zhang, X.; Wang, X.; Tao, H.; Zhang, T.; Peng, M.; Zhang, M.; Huang, Z. Small peptide targeting ANP32A as a novel strategy for acute myeloid leukemia therapy. Transl. Oncol. 2022, 15, 101245. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, M.-L.L.; Munier, A.; Mehus, J.G.; Lambeth, D.O. The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000, 32, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Romani, P.; Ignesti, M.; Gargiulo, G.; Hsu, T.; Cavaliere, V. Extracellular NME proteins: A player or a bystander? Lab. Investig. 2018, 98, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Mccorkle, J.R.; Leonard, M.K.; Kraner, S.D.; Blalock, E.M.; Ma, D.; Zimmer, S.G.; Kaetzel, D.M. The metastasis suppressor NME1 regulates expression of genes linked to metastasis and patient outcome in melanoma and breast carcinoma. Cancer Genom. Proteom. 2014, 11, 175–194. [Google Scholar]
- Lodillinsky, C.; Fuhrmann, L.; Irondelle, M.; Pylypenko, O.; Li, X.-Y.; Bonsang-Kitzis, H.; Reyal, F.; Vacher, S.; Calmel, C.; de Wever, O. Metastasis-suppressor NME1 controls the invasive switch of breast cancer by regulating MT1-MMP surface clearance. Oncogene 2021, 40, 4019–4032. [Google Scholar] [CrossRef]
- Anantha, J.; Goulding, S.R.; Tuboly, E.; O’Mahony, A.G.; Moloney, G.M.; Lomansey, G.; McCarthy, C.M.; Collins, L.M.; Sullivan, A.M.; O’Keeffe, G.W. NME1 Protects Against Neurotoxin-, α-Synuclein-and LRRK2-Induced Neurite Degeneration in Cell Models of Parkinson’s Disease. Mol. Neurobiol. 2022, 59, 61–76. [Google Scholar] [CrossRef]
- Anantha, J.; Goulding, S.R.; Wyatt, S.L.; Concannon, R.M.; Collins, L.M.; Sullivan, A.M.; O’Keeffe, G.W. STRAP and NME1 mediate the neurite growth-promoting effects of the neurotrophic factor GDF5. iScience 2020, 23, 101457. [Google Scholar] [CrossRef]
- Lieberman, J. Granzyme A activates another way to die. Immunol. Rev. 2010, 235, 93–104. [Google Scholar] [CrossRef]
- Puts, G.S.; Leonard, M.K.; Pamidimukkala, N.V.; Snyder, D.E.; Kaetzel, D.M. Nuclear functions of NME proteins. Lab. Investig. 2018, 98, 211–218. [Google Scholar] [CrossRef]
- Radić, M.; Šoštar, M.; Weber, I.; Ćetković, H.; Slade, N.; Herak Bosnar, M. The subcellular localization and oligomerization preferences of NME1/NME2 upon radiation-induced DNA damage. Int. J. Mol. Sci. 2020, 21, 2363. [Google Scholar] [CrossRef]
- Yu, B.Y.K.; Tossounian, M.-A.; Hristov, S.D.; Lawrence, R.; Arora, P.; Tsuchiya, Y.; Peak-Chew, S.Y.; Filonenko, V.; Oxenford, S.; Angell, R. Regulation of metastasis suppressor NME1 by a key metabolic cofactor coenzyme A. Redox Biol. 2021, 44, 101978. [Google Scholar] [CrossRef]
- Clark, A.R.; Ohlmeyer, M. Protein phosphatase 2A as a therapeutic target in inflammation and neurodegeneration. Pharmacol. Ther. 2019, 201, 181–201. [Google Scholar] [CrossRef]
- O’Connor, C.M.; Perl, A.; Leonard, D.; Sangodkar, J.; Narla, G. Therapeutic targeting of PP2A. Int. J. Biochem. Cell Biol. 2018, 96, 182–193. [Google Scholar] [CrossRef]
- Latarya, G.; Mansour, A.; Epstein, I.; Cotlear, D.; Pikkel, J.; Levartovsky, S.; Yulish, M.; Beit-Yannai, E. Human aqueous humor phosphatase activity in cataract and glaucoma. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1679–1684. [Google Scholar] [CrossRef]
- Chiasseu, M.; Vargas, J.L.C.; Destroismaisons, L.; Velde, C.V.; Leclerc, N.; Di Polo, A. Tau accumulation, altered phosphorylation, and missorting promote neurodegeneration in glaucoma. J. Neurosci. 2016, 36, 5785–5798. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, R.; Kou, X.; Liu, Y.; Qi, C.; Liu, R.; You, W.; Gao, J.; Gao, X. A protein phosphatase 2A deficit in the hippocampal CA1 area impairs memory extinction. Mol. Brain 2019, 12, 51. [Google Scholar] [CrossRef]
- Gaitanou, M.; Segklia, K.; Matsas, R. CEND1, a story with many tales: From regulation of cell cycle progression/exit of neural stem cells to brain structure and function. Stem Cells Int. 2019, 2019, 2054783. [Google Scholar] [CrossRef] [PubMed]
- Politis, P.K.; Makri, G.; Thomaidou, D.; Geissen, M.; Rohrer, H.; Matsas, R. BM88/CEND1 coordinates cell cycle exit and differentiation of neuronal precursors. Proc. Natl. Acad. Sci. USA 2007, 104, 17861–17866. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, A.M.; Sabljic, T.F.; Koeberle, P.D.; Ball, A.K. Downregulation of BM88 after optic nerve injury. Investig. Ophthalmol. Vis. Sci. 2014, 55, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Boehme, N.A.; Hedberg-Buenz, A.; Tatro, N.; Bielecki, M.; Castonguay, W.C.; Scheetz, T.E.; Anderson, M.G.; Dutca, L.M. Axonopathy precedes cell death in ocular damage mediated by blast exposure. Sci. Rep. 2021, 11, 11774. [Google Scholar] [CrossRef] [PubMed]
- Segklia, K.; Stamatakis, A.; Stylianopoulou, F.; Lavdas, A.A.; Matsas, R. Increased anxiety-related behavior, impaired cognitive function and cellular alterations in the brain of Cend1-deficient mice. Front. Cell. Neurosci. 2019, 12, 497. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Ojelade, S.A.; Li-Kroeger, D.; Zuo, Z.; Wang, L.; Li, Y.; Gu, J.Y.J.; Tepass, U.; Rodal, A.A.; Bellen, H.J. Retromer subunit, VPS29, regulates synaptic transmission and is required for endolysosomal function in the aging brain. Elife 2020, 9, e51977. [Google Scholar] [CrossRef]
- Muhammad, A.; Flores, I.; Zhang, H.; Yu, R.; Staniszewski, A.; Planel, E.; Herman, M.; Ho, L.; Kreber, R.; Honig, L.S. Retromer deficiency observed in Alzheimer’s disease causes hippocampal dysfunction, neurodegeneration, and Aβ accumulation. Proc. Natl. Acad. Sci. USA 2008, 105, 7327–7332. [Google Scholar] [CrossRef]
- Gray, L.R.; Tompkins, S.C.; Taylor, E.B. Regulation of pyruvate metabolism and human disease. Cell. Mol. Life Sci. 2014, 71, 2577–2604. [Google Scholar] [CrossRef]
- Harder, J.M.; Guymer, C.; Wood, J.P.M.; Daskalaki, E.; Chidlow, G.; Zhang, C.; Balasubramanian, R.; Cardozo, B.H.; Foxworth, N.E.; Deering, K.E. Disturbed glucose and pyruvate metabolism in glaucoma with neuroprotection by pyruvate or rapamycin. Proc. Natl. Acad. Sci. USA 2020, 117, 33619–33627. [Google Scholar] [CrossRef]
- Zilberter, Y.; Gubkina, O.; Ivanov, A.I. A unique array of neuroprotective effects of pyruvate in neuropathology. Front. Neurosci. 2015, 9, 17. [Google Scholar] [CrossRef]
- Sato, K.; Mochida, S.; Tomimoto, D.; Konuma, T.; Kiyota, N.; Tsuda, S.; Shiga, Y.; Omodaka, K.; Nakazawa, T. A pyruvate dehydrogenase kinase inhibitor prevents retinal cell death and improves energy metabolism in rat retinas after ischemia/reperfusion injury. Exp. Eye Res. 2020, 193, 107997. [Google Scholar] [CrossRef]
- Habarou, F.; Brassier, A.; Rio, M.; Chrétien, D.; Monnot, S.; Barbier, V.; Barouki, R.; Bonnefont, J.P.; Boddaert, N.; Chadefaux-Vekemans, B. Pyruvate carboxylase deficiency: An underestimated cause of lactic acidosis. Mol. Genet. Metab. Rep. 2015, 2, 25–31. [Google Scholar] [CrossRef]
- Schousboe, A.; Waagepetersen, H.S.; Sonnewald, U. Astrocytic pyruvate carboxylation: Status after 35 years. J. Neurosci. Res. 2019, 97, 890–896. [Google Scholar] [CrossRef]
- Andersen, J.V.; Christensen, S.K.; Westi, E.W.; Diaz-delCastillo, M.; Tanila, H.; Schousboe, A.; Aldana, B.I.; Waagepetersen, H.S. Deficient astrocyte metabolism impairs glutamine synthesis and neurotransmitter homeostasis in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 2021, 148, 105198. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Patel, A.B. Pyruvate carboxylase and pentose phosphate fluxes are reduced in AβPP-PS1 mouse model of Alzheimer’s disease: A 13C NMR study. J. Alzheimer’s Dis. 2014, 41, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.C.; Sande, P.; Marcos, H.A.; de Zavalía, N.; Sarmiento, M.I.K.; Rosenstein, R.E. Effect of glaucoma on the retinal glutamate/glutamine cycle activity. FASEB J. 2005, 19, 1161–1162. [Google Scholar] [CrossRef]
- García-Bermúdez, M.Y.; Freude, K.K.; Mouhammad, Z.A.; van Wijngaarden, P.; Martin, K.K.; Kolko, M. Glial cells in glaucoma: Friends, foes, and potential therapeutic targets. Front. Neurol. 2021, 12, 169. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, Q.; Ma, M.; Chen, L.-J.; Yu, J.; Xiong, K.; Wu, J. Involvement of moesin phosphorylation in ischemia/reperfusion induced inner blood-retinal barrier dysfunction. Int. J. Ophthalmol. 2020, 13, 545. [Google Scholar] [CrossRef]
- Al-Humimat, G.; Marashdeh, I.; Daradkeh, D.; Kooner, K. Investigational Rho kinase inhibitors for the treatment of glaucoma. J. Exp. Pharmacol. 2021, 13, 197. [Google Scholar] [CrossRef]
- Lin, C.-W.; Sherman, B.; Moore, L.A.; Laethem, C.L.; Lu, D.-W.; Pattabiraman, P.P.; Rao, P.V.; deLong, M.A.; Kopczynski, C.C. Discovery and preclinical development of netarsudil, a novel ocular hypotensive agent for the treatment of glaucoma. J. Ocul. Pharmacol. Ther. 2018, 34, 40–51. [Google Scholar] [CrossRef]


| No. | Protein ID | Protein Name | Gene Name | Score | Mol. Weight [kDa] | Peptides | Fold Change | p-Value | CDR1 vs. CTRL |
|---|---|---|---|---|---|---|---|---|---|
| 1 | P11498 | Pyruvate carboxylase, mitochondrial | PC | 46.55 | 129.6 | 3 | 2.99 | <0.05 | ↑ |
| 2 | P26038 | Moesin | MSN | 48.79 | 67.8 | 3 | 2.88 | <0.05 | ↑ |
| 3 | P50502 | Hsc70-interacting protein | ST13 | 33.33 | 41.3 | 4 | 1.55 | <0.05 | ↑ |
| 4 | Q13885 | Tubulin beta-2A chain | TUBB2A | 65.75 | 49.9 | 21 | 1.52 | <0.01 | ↑ |
| 5 | P10515 | Dihydrolipoyllysine-residue acetyltransferase component of PDH, mitochondrial | DLAT | 9.34 | 69.0 | 3 | 1.51 | <0.05 | ↑ |
| 6 | Q99962 | Endophilin-A1 | SH3GL2 | 55.17 | 40.0 | 7 | 1.45 | <0.05 | ↑ |
| 7 | Q9UI12 | V-type proton ATPase subunit H | ATP6V1H | 128.60 | 55.9 | 8 | 1.43 | <0.05 | ↑ |
| 8 | P50991 | T-complex protein 1 subunit delta | CCT4 | 50.63 | 57.9 | 7 | 1.41 | <0.01 | ↑ |
| 9 | Q9GMB0 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1 | RPN1 | 23.80 | 68.7 | 8 | 1.39 | <0.001 | ↑ |
| 10 | P23687 | Prolyl endopeptidase | PREP | 137.82 | 80.8 | 11 | 1.37 | <0.05 | ↑ |
| 11 | P29804 | Pyruvate dehydrogenase E1 component subunit α, mitochondrial | PDHA1 | 29.28 | 43.1 | 7 | 1.31 | <0.05 | ↑ |
| 12 | P63010 | AP-2 complex subunit beta | AP2B1 | 65.96 | 104.6 | 11 | 1.16 | <0.01 | ↑ |
| No. | Protein ID | Protein Name | Gene Name | Score | Mol. Weight [kDa] | Peptides | Fold Change | p-Value | CDR1 vs. CTRL |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Q9UBQ0 | Vacuolar protein sorting-associated protein 29 | VPS29 | 38.05 | 20.5 | 2 | 1.33 | <0.05 | ↓ |
| 2 | Q15257 | Serine/threonine-protein phosphatase 2A activator | PPP2R4 | 23.28 | 40.7 | 2 | 1.56 | <0.05 | ↓ |
| 3 | P30049 | ATP synthase subunit delta, mitochondrial | ATP5F1D | 60.95 | 17.5 | 2 | 1.58 | <0.05 | ↓ |
| 4 | Q03252 | Lamin-B2 | LMNB2 | 40.46 | 69.9 | 3 | 1.67 | <0.05 | ↓ |
| 5 | Q15293 | Reticulocalbin-1 | RCN1 | 66.83 | 38.9 | 3 | 1.68 | <0.05 | ↓ |
| 6 | P62888 | 60S ribosomal protein L30 | RPL30 | 15.89 | 12.8 | 3 | 1.89 | <0.05 | ↓ |
| 7 | P15531 | Nucleoside diphosphate kinase A | NME1 | 108.02 | 17.1 | 6 | 1.99 | <0.05 | ↓ |
| 8 | Q8NC51 | Plasminogen activator inhibitor 1 RNA-binding protein | SERBP1 | 65.36 | 45.0 | 3 | 2.90 | <0.05 | ↓ |
| 9 | Q29205 | 60S ribosomal protein L11 | RPL11 | 35.69 | 20.2 | 2 | 5.16 | <0.01 | ↓ |
| 10 | Q4G0F5 | Vacuolar protein sorting-associated protein 26B | VPS26B | 10.83 | 39.2 | 3 | 9.97 | <0.01 | ↓ |
| 11 | P36542 | ATP synthase subunit gamma, mitochondrial | ATP5F1C | 67.00 | 33.0 | 2 | 10.30 | <0.01 | ↓ |
| 12 | O75964 | ATP synthase subunit g, mitochondrial | ATP5MG | 37.14 | 11.4 | 2 | 11.08 | <0.001 | ↓ |
| 13 | Q29026 | Cell cycle exit and neuronal differentiation protein 1 | CEND1 | 66.37 | 14.0 | 2 | 11.81 | <0.001 | ↓ |
| Signaling Pathways | −log (p-Value) | Molecules |
|---|---|---|
| Acetyl-CoA Biosynthesis I | 4.62 | DLAT, PDHA1 |
| Mitochondrial Dysfunction | 4.47 | ATP5F1C, ATP5F1D, ATP5MG, PDHA1 |
| Granzyme A Signaling | 3.71 | ANP32A, NME1 |
| Oxidative Phosphorylation | 3.62 | ATP5F1C, ATP5F1D, ATP5MG |
| Mechanisms of Viral Exit from Host Cells | 3.04 | LMNB2, SH3GL2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmelter, C.; Fomo, K.N.; Brueck, A.; Perumal, N.; Markowitsch, S.D.; Govind, G.; Speck, T.; Pfeiffer, N.; Grus, F.H. Glaucoma-Associated CDR1 Peptide Promotes RGC Survival in Retinal Explants through Molecular Interaction with Acidic Leucine Rich Nuclear Phosphoprotein 32A (ANP32A). Biomolecules 2023, 13, 1161. https://doi.org/10.3390/biom13071161
Schmelter C, Fomo KN, Brueck A, Perumal N, Markowitsch SD, Govind G, Speck T, Pfeiffer N, Grus FH. Glaucoma-Associated CDR1 Peptide Promotes RGC Survival in Retinal Explants through Molecular Interaction with Acidic Leucine Rich Nuclear Phosphoprotein 32A (ANP32A). Biomolecules. 2023; 13(7):1161. https://doi.org/10.3390/biom13071161
Chicago/Turabian StyleSchmelter, Carsten, Kristian Nzogang Fomo, Alina Brueck, Natarajan Perumal, Sascha D. Markowitsch, Gokul Govind, Thomas Speck, Norbert Pfeiffer, and Franz H. Grus. 2023. "Glaucoma-Associated CDR1 Peptide Promotes RGC Survival in Retinal Explants through Molecular Interaction with Acidic Leucine Rich Nuclear Phosphoprotein 32A (ANP32A)" Biomolecules 13, no. 7: 1161. https://doi.org/10.3390/biom13071161
APA StyleSchmelter, C., Fomo, K. N., Brueck, A., Perumal, N., Markowitsch, S. D., Govind, G., Speck, T., Pfeiffer, N., & Grus, F. H. (2023). Glaucoma-Associated CDR1 Peptide Promotes RGC Survival in Retinal Explants through Molecular Interaction with Acidic Leucine Rich Nuclear Phosphoprotein 32A (ANP32A). Biomolecules, 13(7), 1161. https://doi.org/10.3390/biom13071161







