Abstract
Background/Objectives: Natural products containing hydroxyanthracene derivatives (HADs) such as Cascara (Rhamnus purshiana), Frangula (Rhamnus frangula), Rhubarb (Rheum palmatum), and Senna (Cassia angustifolia) have long been used for their laxative properties, but also raise safety concerns due to reported genotoxic and carcinogenic potential. Most studies have focused on quantifying HADs, whereas the broader secondary metabolite landscape of these herbal drugs remains underexplored. We aimed to generate an untargeted metabolomic fingerprint of these four species and to explore their chemical diversity using AI-based structural classification. Methods: Four commercial botanical raw materials were extracted with 60% methanol and analysed by UPLC–HRMS/MS in positive and negative ion modes. Features were processed in Compound Discoverer and annotated by accurate mass and MS/MS matching against spectral databases, then assigned to structural classes using a graph neural network classifier. Multivariate analyses (PCA, HCA) were used to compare metabolic patterns across species. Results: In total, 93, 83, 83 and 51 metabolites were annotated in cascara, frangula, rhubarb, and senna, respectively, spanning flavonoids, anthraquinones, phenylpropanoids and other classes. Only four flavonoids were shared by all species, indicating marked biochemical divergence. Several putatively species-enriched features were observed, including pavine in cascara and frangula, vicenin-2 in senna, and piceatannol in rhubarb. Senna displayed the most distinct metabolic profile, whereas cascara and frangula clustered closely. Conclusions: This work provides a chemistry-centred metabolomic fingerprint of four HAD-containing herbal drugs using graph-based neural networks for natural product classification, supporting future studies on the pharmacological potential, bioavailability and safety of their metabolites.
1. Introduction
Natural products play an important role in disease treatment and still represent the core of traditional medicine systems in several countries [1]. Among these are hydroxyanthracenes (HADs), a class of aromatic organic compounds widely present in nature and particularly in many medicinal plants, including Cassia angustifolia (Senna), Rheum palmatum (Rhubarb), Rhamnus frangula (Frangula), and Rhamnus purshiana (Cascara) [2].
HADs are secondary metabolites characterized by a 9,10-dioxoanthracene core, often substituted with one or more hydroxyl groups and occurring either as free aglycones or as glycosides conjugated to sugar moieties [3,4]. More than 700 natural HADs have been reported, with over 200 identified in flowering plants and the remainder in lichens and fungi [2]. IHADs are distributed in different organs (rhizomes, roots, bark, leaves, fruits) and are typically stored as glycosides, which facilitates their accumulation and modulates their bioactivation in the gastrointestinal tract [4]. Representative examples include Emodin, Aloe-emodin, and Rhein, which are among the main anthraquinone constituents of Rhubarb, Senna, Frangula, and Cascara [5,6].
HADs are known for their pharmacological properties, particularly laxative and digestive activities [6,7], and have been used for decades in numerous pharmaceutical formulations and dietary supplements. However, these compounds are not devoid of toxicity [8]. The genotoxicity of HADs derivatives has been evaluated in numerous in vitro and in vivo studies identified from the public literature. In particular, some epidemiological studies showed an increased risk of colorectal cancer [9,10,11,12]. For this reason, the European Food Safety Authority (EFSA) has re-evaluated the safety of the use of medicinal plants containing HADs in food supplements [13,14], concluding that they should be considered genotoxic and carcinogenic until proven otherwise. This assessment highlights the need for a more in-depth investigations of medicinal plants containing HADs for the whole spectrum of secondary metabolites of Senna, Rhubarb, Frangula, and Cascara.
The chemical and pharmacological properties of the plants Rhamnus frangula (Frangula), Rhamnus purshiana (Cascara), Rheum palmatum (Rhubarb), and Cassia angustifolia (Senna) have been the subject of several investigations. Many studies have already focused on HADs, but comparatively less is known about the broader spectrum of secondary metabolites present in these therapeutic plants. For instance, a recent study [15] combined qualitative–quantitative characterization of HADs in commercial preparations of Senna, Rhubarb, Cascara, and Frangula with cytotoxicity assays and shotgun proteomics in an intestinal cell model, comparing the effects of single HAD molecules with those of whole plant extracts. Together with the broader in vitro and in vivo literature and regulatory evaluations [12,13,16], this work highlights the complexity of HAD-containing products. However, a comprehensive, chemistry-centred description of the overall metabolite landscape of these herbal drugs is still lacking.
In the state of the art, the literature reports different studies on Cascara establishing anthraquinone glycosides as the active constituents of the bark [17]. Additional studies have focused on determining the presence of HADs using methods such as liquid chromatography combined with mass spectrometry [18,19]. While HADs in Frangula and Cascara, belonging to family Rhamnaceae, have been closely quantified and studied, other secondary metabolites have not been explored in as much detail [20,21].
Regarding Rhubarb, specific studies have primarily focused on quantifying HADs and phenolic compounds [22,23,24]. Specifically, rhein has been identified as the metabolite responsible for the toxicity of anthraquinones [16].
Conversely, numerous studies on Senna have concentrated on identifying the generated metabolites without being limited to HADs, in particular those with antibacterial properties [25]. The diversity of bioactive compounds has been revealed in different studies, characterizing and quantifying polyphenols and other phytochemicals [26,27,28].
Despite these efforts, no study has yet provided a comprehensive metabolic profiling of these four species. Untargeted metabolomics offers a powerful means to achieve this, enabling the detection and annotation of a wide diversity of metabolites and shedding light on their biological roles and potential health impacts.
In this context, the present work was designed as a complementary chemistry-centred investigation. Rather than re-evaluating bioavailability and safety, which have been specifically addressed in previous studies such as [15], our primary aim is to provide an untargeted UPLC–MS/MS metabolomic fingerprint of Cascara, Senna, Frangula, and Rhubarb and to explore their chemical diversity using AI-based classification approaches.
In addition to experimental metabolomics, recent advances in artificial intelligence have provided new opportunities for the structural classification of natural products. Tools such as NPClassifier [29] have demonstrated the feasibility of applying deep learning to metabolite categorization, although with limitations in capturing structurally diverse or less represented scaffolds. More recently, graph-based neural networks have shown superior performance in modeling molecular topology and enhancing classification accuracy, as described in [30]. By integrating such approaches, our study not only provides an untargeted metabolic profiling of anthraquinone-rich plants but also leverages state-of-the-art computational methods to achieve a higher-resolution view of their chemical diversity.
2. Materials and Methods
The four samples were kindly supplied by different companies. Botanical samples consisted of Rheum palmatum (Rhubarb) (root), Cassia angustifolia (Senna) (leaves), Rhamnus purshiana (Cascara) (bark), and Rhamnus frangula (Frangula) (bark). All materials were obtained as semi-processed dried plant organs in milled form. Samples were stored at 25 °C under controlled humidity in an ISO 9001:2015-certified facility (certificate Q/1765/24) [31], ensuring standardized workflows, traceability, and quality control in line with the storage requirements for semi-processed herbal materials used in food supplement manufacturing. Each powdered plant material (100 mg) was extracted by sonication for 20 min in 10 mL of 60% methanol (Merck Group (Darmstadt, Germany)). This hydroalcoholic mixture was selected on the basis of preliminary tests comparing methanol–water and ethanol–water systems (100%, 80%, 60%), which indicated that 60% methanol provided the best compromise between chromatographic signal intensity and metabolite coverage under the adopted LC–HRMS conditions. After centrifugation at 13,000 rpm for 10 min, the supernatant was collected, filtered through a 0.22 µm membrane, and injected directly into the UPLC–Q Exactive Plus system without dilution. All analyses for each sample were performed in technical triplicate. No biological replicates were included, as the study focused on metabolomic characterization of distinct herbal drugs that are used as sources of anthraquinone-containing ingredients in commercial laxative formulations.
The metabolic profiles of the powdered plant samples were analyzed using an Ultimate 3000 UPLC system (Thermo Fisher Scientific (Waltham, MA, USA)) coupled with a Q-Exactive Plus Hybrid Quadrupole–Orbitrap™ high-resolution mass spectrometer (Thermo Fisher Scientific). Data were acquired in both positive and negative electrospray modes over a scan range of m/z 200–2000. Operating parameters were as follows: spray voltage 3.5 kV (positive mode) and 3.0 kV (negative mode); sheath gas = 20 a.u.; auxiliary gas = 5 a.u.; capillary temperature = 320 °C; and resolution = 35,000. Acquisition was performed in Full MS/dd-MS2 (Top N) mode, selecting and fragmenting precursor ions according to intensity. MS2 spectra were generated using higher-energy collisional dissociation (HCD) at 30 a.u., with a mass accuracy threshold of 5 ppm. Chromatographic separation employed an Acquity UPLC BEH C18 column (2.1 mm × 150 mm, 1.7 µm; Waters (Milford, MA, USA)). The mobile phases were (A) water with 0.1% formic acid and (B) acetonitrile with 0.1% formic acid (Merck Group). A linear gradient was applied starting at 2% B (1 min hold), increasing to 100% B over 50 min, maintained for 2 min, then re-equilibrated to the initial conditions. The flow rate was 0.2 mL , the injection volume was 10 µL, and the column temperature was maintained at 35 °C. Raw LC–MS/MS data were processed using Compound Discoverer 3.3 (Thermo Fisher Scientific). Feature detection and alignment were performed with default settings, except for a retention-time tolerance of 0.2 min and a mass tolerance of 10 ppm. Blank solvent runs were acquired under identical conditions to identify and exclude background peaks originating from the matrix or solvent, thereby enhancing annotation reliability.
2.1. Feature Extraction and Metabolite Annotation
Metabolite features were extracted and processed using Thermo Fisher’s Compound Discoverer (CD) software (v3.3). The workflow included automated feature detection, chromatographic alignment, background subtraction, isotope/adduct grouping, and compound annotation. For each detected feature, CD returned (when available) the compound name, molecular formula, precursor m/z, calculated molecular weight, retention time (RT), maximum peak area, ionization mode (ESI positive or negative), and MS/MS-based annotation obtained through matching against the spectral and structural databases integrated into the platform (mzCloud, mzVault, ChemSpider, Mass List, Metabolika). The full feature tables exported from CD containing these parameters for all detected compounds are provided as Supplementary Materials. In line with the Metabolomics Standards Initiative (MSI), metabolites confirmed with authentic reference standards and MS/MS fragmentation matching (e.g., the main hydroxyanthracene derivatives identified in the different samples) are classified as MSI Level 1, features annotated on the basis of accurate mass and MS/MS spectral similarity to database entries are classified as MSI Level 2, and unannotated or partially characterized features are assigned to MSI Levels 3/4. The MSI confidence level associated with each feature is explicitly indicated in the tables provided in the Supplementary Materials. As such, non–standard-confirmed metabolites discussed in the main text should be regarded as putative annotations (MSI Level 2) pending further validation with authentic standards.
2.2. Metabolite Screening Process
- Database Confirmation: Compounds showing full or partial correspondence in at least one of five reference databases (m/z Cloud, m/z Vault, Metabolika, ChemSpider, or Mass List) were included.
- Mass Accuracy: Deviation within ppm from theoretical m/z.
- RT (Retention Time): Compounds eluting between 5 and 50 min were selected, although the range could be extended (0–120 min) to accommodate specific analytical requirements.
- Peak Area: Features with areas below 1.0 × were excluded in order to minimize low-intensity background signals.
- Availability: Only compounds with corresponding spectra were retained for annotation.
The resulting dataset was cross-checked against the published literature to verify compound identities and contextualize the detected metabolites within known phytochemical profiles.
2.3. Classification of Natural Products
For the structural classification of metabolites, we employed Graph Isomorphism Networks (GINs), following the framework described in [30]. GINs were selected due to their ability to capture molecular graph topology with high fidelity and to improve predictive performance in natural product classification tasks.
Each metabolite was converted from its SMILES notation into a graph representation, where atoms were encoded as nodes with associated features (atom type, degree, hybridization state, formal charge, aromaticity) and bonds were encoded as edges with features describing bond type and conjugation. These molecular graphs were then processed by GINs specifically trained for each classification level (pathway, superclass, and class), as reported in [30]. Technical details of the network architectures and training procedures—including layer composition, activation functions, optimization strategy, and hyperparameters—are provided in Table A1. Model training and validation were performed on curated datasets of annotated natural products using stratified 10-fold cross-validation to ensure robustness. Performance was assessed based on macro-averaged F1 score and accuracy.
The trained models were subsequently applied to the metabolite dataset generated by Compound Discoverer 3.3. The predicted class assignments were merged with experimental annotations, yielding a detailed structural classification of the identified compounds and enabling a more precise comparison of the metabolite composition across the investigated species.
2.4. Statistical Analysis
Intersections among metabolite lists were computed using the web-based tool available at the Bioinformatics & Evolutionary Genomics platform (https://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 15 May 2025)). The application provided both textual and graphical outputs, identifying shared and unique metabolites across the compared datasets.
The processed data matrix was imported into MetaboAnalyst 6.0 6.0 [32] for multivariate analysis. Exploratory principal component analysis (PCA) was performed to visualize interspecies variation and clustering patterns, while hierarchical cluster analysis (HCA) based on Euclidean distance was used to construct a dendrogram illustrating metabolic relationships among the investigated samples after appropriate data filtering, normalization and scaling. No formal univariate or supervised hypothesis testing (e.g., ANOVA, t-tests, OPLS-DA) was performed; therefore, all comparisons of metabolite abundances are exploratory and descriptive. Consequently, no p-values were calculated and no false discovery rate (FDR) or multiple-testing corrections were applied.
Relative abundances of annotated metabolites were visualized through customized heatmaps generated via a dedicated Python script employing Pandas (v2.1.4), Matplotlib (v3.8), and Seaborn (v0.13).
3. Results
In this work, the dried materials of Cascara, Senna, Frangula, and Rhubarb were investigated to describe their non-volatile profiles through UPLC–MS/MS for the first time. These species are traditionally employed for their laxative properties [33], and their therapeutic relevance has sustained longstanding interest in their chemical composition. As such, a comprehensive characterization of their secondary metabolites is essential both to explain their pharmacological activity and to evaluate potential safety concerns associated with their use.
To complement proteomic findings, an untargeted metabolomic profiling was performed on representative methanolic extracts of Cascara, Frangula, Rhubarb, and Senna. Both positive and negative ionization modes were applied to obtain a comprehensive overview of the metabolite composition. The approach enabled the annotation of 93 metabolites in Cascara, 83 in Frangula, 83 in Rhubarb, and 51 in Senna. Complete compound lists are reported in Appendix A Table A2, Table A5, Table A8 and Table A11).
Given the structural diversity of natural products, a hierarchical classification framework is typically adopted to provide consistent annotation, organizing metabolites into three levels: pathway, superclass, and class [29]. Pathways reflect the major biosynthetic origins, superclasses capture broad chemical categories, and classes resolve scaffold-level diversity within each superclass. This organization enables both global metabolome profiling and detailed analyses of families of biological relevance. To characterize the metabolic diversity of the four species, the identified compounds were classified with the GIN network; results are reported at the superclass level, which offers a good balance between interpretability and comparability with previous studies.
Expert chemists verified the assignments at the superclass level. For Cascara, expert review confirmed 90/93 assignments (96.8% concordance), with only three discordant cases reported in Table A4. For Frangula, 78/83 assignments were confirmed (94.0% concordance). Two compounds were misclassified and three remained unassigned, as detailed in Table A7. For Rhubarb, 75 of 83 assignments (90.0%) were confirmed, with two misclassifications and six unassigned compounds (Table A10). For Senna, 47 of 51 assignments (90.3%) were validated, with three misclassified and two unassigned entries (Table A13).
Per-species classification tables (Appendix A Table A3, Table A6, Table A9 and Table A12) provide the full pathway/superclass/class labels and the associated model confidences (Acc. %).
Overall, automated labels closely matched expert curation, indicating that the approach is suitable for high-throughput profiling while retaining chemical interpretability.
A Venn diagram was generated to visualize the overlap between the four species. Only four metabolites—Phloretin (C15H14O5), Kaempferol (C15H10O6), Hispidulin (C16H12O6), and 4-Heptyloxyphenol (C13H20O2)—were common to all extracts, indicating strong chemical specificity for each botanical source. Cascara and Frangula shared 21 metabolites, ten of which were exclusive to these two Rhamnaceae members, whereas Senna and Rhubarb shared 13 compounds, with five uniquely common to both.
The Venn diagram (Figure 1) further indicated that Cascara and Frangula, belonging to the same family, were similar to each other and showed 21 compounds in common with ten of them characteristic only of these two species. On the other hand, Senna and Rhubarb, seemed to have some metabolites in common, specifically 13 compounds, of which five were exclusive of these two species.
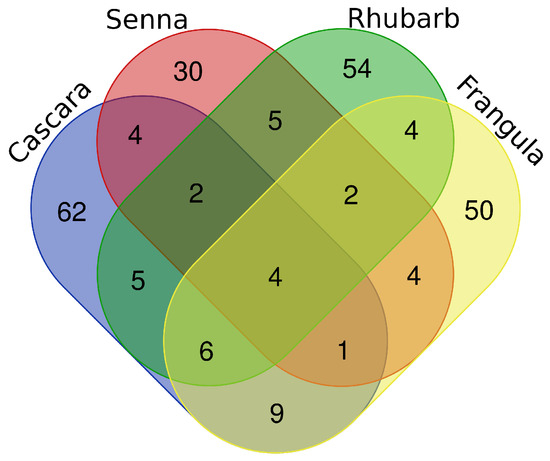
Figure 1.
Four-set Venn diagram showing the overlap of annotated metabolite classes among Cascara, Frangula, Rhubarb, and Senna. Each ellipse represents the set of metabolite classes detected in that plant. A class was considered present when at least one metabolite belonging to that class was annotated by LC–MS/MS (MSI Level ≥ 2) in the corresponding sample. Overlapping regions indicate classes shared by two, three, or all four plants, whereas non-overlapping segments represent plant-specific classes. The diagram highlights both the small core of metabolite classes common to all four species and the substantial proportion of classes unique to each plant.
Below, we report a list of the major metabolite classes identified in each plant along with the most abundant metabolites in each category. Moreover, two pie charts were generated for each plant. The first chart summarizes the distribution of identified metabolite superclasses, highlighting the overall chemical profile of the species. The second chart provides a more detailed view of the most abundant category.
3.1. Distinctive Metabolites in Cascara
- Polycyclic Aromatic Polyketides: This superclass is represented exclusively by anthraquinones and anthrones, including Emodin (C15H10O5), Aloin A (C21H22O9), Aloin B (C21H22O9), and Cascaroside A (C27H32O14).
- Flavonoids: Abundant representatives include Nobiletin (C21H22O8), Isoliquiritigenin (C15H12O4), Primuletin (C15H10O3), Myrciacitrin V (C30H30O13), Naringin (C27H32O14), and Cirsimarin (C23H24O11).
- Isoflavonoids: Detected examples include Genistein (C15H10O5) and Formononetin (C16H12O4).
- Phenylpropanoids (C6–C3): Represented by Chlorogenic acid (C16H18O9) and Caffeic acid (C9H8O4).
- Tyrosine Alkaloids: Exemplified by Pavine (C20H23NO4).
- Benzenoids: Including 4-Ethylcatechol (C8H10O2).
- Monoterpenoids: Including Demethyloleuropein (C24H30O13).
- Phloroglucinols: Represented by 2,4,6-Trimethoxybenzophenone (C16H16O4).
- Aromatic Polyketides: Including 4-Heptyloxyphenol (C13H20O2).
- Trace compounds: Additional minor representatives of coumarins, diterpenoids, and lignans were also detected.
A clear visualization of the metabolite distribution is shown in Figure 2. In Figure 2a, the pie chart reveals the percentage distribution of major metabolic superclasses in Cascara. The flavonoid class (53%) was clearly the largest, followed by the phenylpropanoids (15%) and anthraquinones (10%). Figure 2b provides a detailed breakdown of the predominant flavonoid class, specifying the contributions of flavones (31%), flavonols (31%), and flavanones (24%).
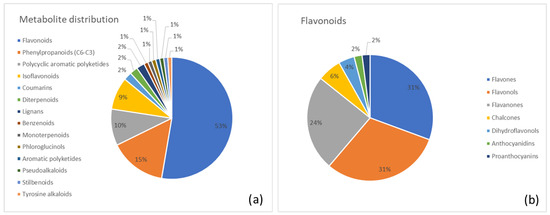
Figure 2.
(a) Distribution of metabolite classes identified in Cascara. Each slice represents the percentage of annotated metabolites (MSI level ≥ 2) assigned to a given class, according to the structural classification pipeline described in the Methods (GIN-based hierarchical classification). (b) Detailed subdivision of the flavonoid superclass in Cascara into the corresponding annotated subclasses. Values are expressed as the percentage of flavonoid metabolites belonging to each subclass.
3.2. Distinctive Metabolites in Frangula
- Polycyclic Aromatic Polyketides: Mainly represented by anthraquinones/anthrones, including Emodin (C15H10O5), Glucofrangulin A (C27H30O14), Glucofrangulin B (C26H28O14), Frangulin A (C21H20O9), and Frangulin B (C20H18O9).
- Flavonoids: Liquiritin (C21H22O9), Isoliquiritigenin (C15H12O4), Kaempferol (C15H10O6).
- Isoflavonoids: Genistein (C15H10O5) and Daidzein (C15H10O4).
- Phenylpropanoids (C6–C3): Caffeic acid (C9H8O4), 3-Caffeoylquinic acid (C16H18O9), and Methyl chlorogenate (C17H20O9).
- Monoterpenoids: Oleuropein (C25H32O13), Demethyloleuropein (C24H30O13), and Loganin (C17H26O10) in trace amounts.
- Stilbenoids: Piceatannol (C14H12O4) and Piceid (C20H22O8) in trace amounts.
- Coumarins: 5,6-O--D-diglucopyranosylangelicin (C23H26O15).
- Aromatic Polyketides: 4-Heptyloxyphenol (C13H20O2).
- Naphthalenes: Nepodin (C13H12O3).
- Fatty Acids and Conjugates: Oleic acid (C18H34O2) detected in trace amounts (observed only in Frangula).
- Unclassified: Four highly prevalent features grouped as “Other” remained unclassified; accurate m/z values were observed but spectral/database evidence was insufficient to assign definitive molecular formulas.
A distinct visualization of metabolite distribution across superclasses is shown in Figure 3. Figure 3a illustrates the percentage distribution in Frangula, highlighting flavonoids (59%) as the most abundant, followed by polycyclic aromatic polyketides (anthraquinones; 12%) and phenylpropanoids (7%). Figure 3b presents a class-level breakdown of the predominant flavonoid group, with contributions from flavonols (35%), flavones (33%), and flavanones (16%).
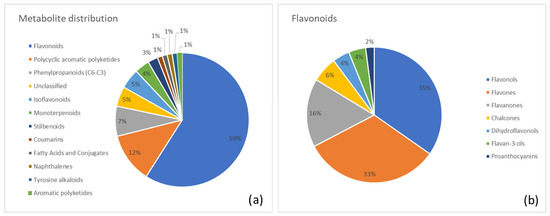
Figure 3.
(a) Distribution of metabolite classes identified in Frangula. Each slice represents the percentage of annotated metabolites (MSI level ≥ 2) assigned to a given class, according to the structural classification pipeline described in the Methods (GIN-based hierarchical classification). (b) Detailed subdivision of the flavonoid superclass in Frangula into the corresponding annotated subclasses. Values are expressed as the percentage of flavonoid metabolites belonging to each subclass.
3.3. Distinctive Metabolites in Rhubarb
Superclass-Level Overview
- Polycyclic Aromatic Polyketides: Mainly represented by anthraquinones such as Emodin (C15H10O5), Rhein (C15H8O6), 1,4-Dihydroxyanthraquinone (C14H8O4), and Rhein-8-glucoside (C21H18O11).
- Flavonoids: Abundant representatives include Catechin (C15H14O6).
- Isoflavonoids: Genistein (C15H10O5) and Daidzein (C15H10O4).
- Stilbenoids: Examples are Piceatannol (C14H12O4), Resveratrol (C14H12O3), and Resveratrol 3-O-glucoside (C20H22O8).
- Phenolic Acids: Protocatechuic aldehyde (C7H6O3), Gallic acid (C7H6O5), and Ellagic acid (C14H6O8).
- Chromanes: Including 5-Acetonyl-7-hydroxy-2-methylchromone (C13H12O4) and Aloesin (C19H22O9).
- Naphthalenes: Exemplified by Torachrysone 8-O--D-glucoside (C20H24O9).
- Aromatic Polyketides: Including 4-Heptyloxyphenol (C13H20O2).
- Trace Compounds: Minor representatives of diterpenoids and additional phenylpropanoids were also detected.
The metabolite composition of Rhubarb is summarized in Figure 4. Figure 4a shows the percentage distribution of major metabolite superclasses, highlighting flavonoids (63%) as the predominant group, followed by polycyclic aromatic polyketides (anthraquinones; 12%) and minor contributions from other superclasses. Figure 4b provides a class-level breakdown of the flavonoid superclass, with flavonols (46%), flavones (23%), and flavan-3-ols (8%) representing the most abundant subclasses.
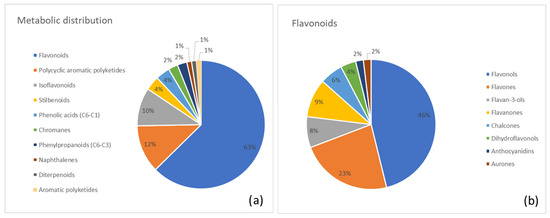
Figure 4.
(a) Distribution of metabolite classes identified in Rhubarb. Each slice represents the percentage of annotated metabolites (MSI level ≥ 2) assigned to a given class, according to the structural classification pipeline described in the Methods (GIN-based hierarchical classification). (b) Detailed subdivision of the flavonoid superclass in Rhubarb into the corresponding annotated subclasses. Values are expressed as the percentage of flavonoid metabolites belonging to each subclass.
3.4. Distinctive Metabolites in Senna
Superclass-Level Overview
- Flavonoids: Abundant representatives include Vicenin 2 (C27H30O15), 2′,2 Bisepigallocatechin digallate (C44H34O22), and Luteolin (C15H10O6).
- Isoflavonoids: Demethylwedelolactone (C15H8O7) and Irilone (C16H6O3).
- Polycyclic Aromatic Polyketides: Represented by anthraquinones distinctive of Senna, including Rhein (C15H8O6), Rhein-8-glucoside (C21H18O11), Sennoside A (C42H38O20), and Sennoside B (C42H38O20).
- Phenylpropanoids (C6–C3): Examples include Guaethol (C8H10O2) and Eugenol (C10H12O2).
- Coumarins: Exemplified by 11-O-Galloylbergenin (C21H20O13).
- Benzenoids: Including Creosol (C8H10O2).
- Aromatic Polyketides: Including 4-Heptyloxyphenol (C13H20O2), also observed in the other species.
- Trace Compounds: Minor representatives of diterpenoids, lignans, and naphthalenes were also detected.
A clear representation of the metabolite distribution in Senna is provided in Figure 5. Figure 5a shows the percentage distribution of major metabolite superclasses, with flavonoids (41%) as the most abundant group, followed by polycyclic aromatic polyketides (anthraquinones; 21%) and phenylpropanoid (12%). Figure 5b presents a class-level breakdown of the flavonoid superclass, highlighting the relative contributions of flavones (29%), flavonols (28%), and proanthocyanins (14%).
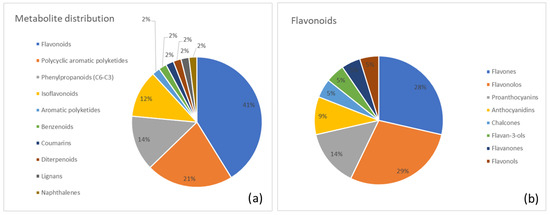
Figure 5.
(a) Distribution of metabolite classes identified in Senna. Each slice represents the percentage of annotated metabolites (MSI level ≥ 2) assigned to a given class, according to the structural classification pipeline described in the Methods (GIN-based hierarchical classification). (b) Detailed subdivision of the flavonoid superclass in Senna into the corresponding annotated subclasses. Values are expressed as the percentage of flavonoid metabolites belonging to each subclass.
Taken together, the hierarchical classification and species-specific metabolite distributions provide a comprehensive overview of the chemical diversity across the four plants. The distribution of major compound superclasses is summarized in Table 1.

Table 1.
Distribution of major compound superclasses across Cascara, Frangula, Rhubarb, and Senna.
Principal component analysis (PCA) was applied to the normalized metabolite dataset in order to explore the interspecies variability. The first two principal components accounted for 87.1% (PC1) and 10.9% (PC2) of total variance, respectively. The PCA score plot (Figure 6a) reveals a clear separation of Senna from the other species, indicating a distinct metabolomic signature.
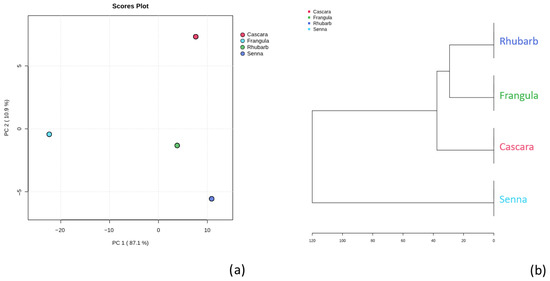
Figure 6.
(a) Principal component analysis (PCA) score plot of the metabolomic profiles of Cascara, Frangula, Rhubarb, and Senna. Data were autoscaled (z-score normalization). PC1 explains 87.1% of the total variance and PC2 explains 10.9%. Each point represents a technical replicate, and samples are colour-coded by species (Cascara: red; Frangula: green; Senna: light blue; Rhubarb: blue). Cascara clusters to the right along PC1 and Senna to the left, while Frangula and Rhubarb occupy the lower-right quadrant, reflecting interspecies differences in overall metabolite composition. (b) Hierarchical cluster analysis (HCA) dendrogram of the same samples based on Euclidean distance. Colours correspond to the PCA groups. Rhubarb and Frangula cluster first and then merge with Cascara, while Senna forms the most distant branch. The horizontal axis represents dissimilarity (0–120).
Hierarchical cluster analysis (HCA) based on the Euclidean distance (Figure 6b) produced comparable results: Frangula and Rhubarb clustered closely, followed by Cascara, whereas Senna formed an independent branch, confirming its unique chemical composition.
A heatmap representation (Figure 7) illustrates the distribution of metabolites across classes. Two main clusters can be observed: the first is dominated by flavonoids common to all plants, while the second is subdivided into two subclusters: (i) phenylpropanoids, anthraquinones, and anthrones, suggesting shared biosynthetic pathways; and (ii) additional metabolic groups such as aromatic polyketides, chromanes, stilbenoids, and coumarins, reflecting high chemical diversity and species-specific biosynthetic specialization.
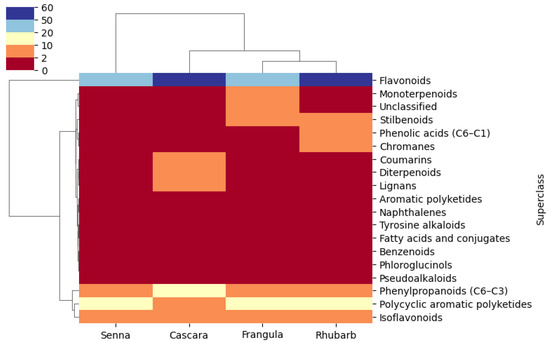
Figure 7.
Heatmap showing the distribution of 19 metabolite superclasses across the analysed samples. Each row corresponds to a distinct superclass and each column to a single LC–MS/MS analysis (technical replicate), grouped by plant species. The colour scale ranges from red (low relative abundance, value = 0) to blue (high relative abundance, maximum value = 60; not further normalized). Unsupervised hierarchical clustering was applied to both rows and columns using the Euclidean distance and average linkage; the corresponding dendrograms are displayed alongside the heatmap. The plot highlights a clear predominance of flavonoids, which are the only superclass showing consistently high abundance across all samples, whereas the remaining superclasses display moderate to low levels, reflected by intermediate shades between red and blue.
This compositional diversity contributes to the distinct biochemical identity observed for each plant species.
4. Discussion
A chemical fingerprint profile can comprehensively reflect the types of chemical components contained in medicinal plants and their products, which can then be used to describe and evaluate their quality as a whole [34]. To this end, a method was developed by combining the high separation performance of HPLC applied to complex samples with the high selectivity and sensitivity of MS, allowing for a comprehensive evaluation of the medicinal plants under investigation. In fact, by contributing to a better understanding of the distribution and variability of these compounds within species, plant metabolomics provides a formidable resource for exploring the richness and complexity of metabolites found in plants.
The GIN-based hierarchical classification proved robust across the dataset. Close inspection of the few discordant cases revealed chemically plausible failure modes that are typical in large-scale automated annotation. These included ambiguities between structurally similar scaffolds, misclassification driven by partial structural features (e.g., side-chain length, oxidation state, or ring substitution patterns), and limited representation of certain compound families in the training data. Such discrepancies were readily resolved by expert review and did not alter the overall conclusions at the pathway or superclass level.
Confidence values were highly informative: assignments above 99% were almost invariably confirmed, while discordant cases showed lower or imbalanced scores across hierarchy levels. This supports a pragmatic workflow in which automated predictions are retained as defaults and selectively curated when confidence flags emerge. Such an approach balances throughput with accuracy, preserves reproducibility, and minimizes the risk of propagating annotation errors. Studies integrating untargeted metabolomics with transparent uncertainty estimates and expert validation remain relatively uncommon, underscoring current limitations in evaluating the efficacy and safety of plant-derived products [35].
Using this framework, we identified 93 compounds in Cascara, 83 in Rhubarb, 83 in Frangula, and 51 in Senna.
Among the identified metabolites (MSI level 2 unless otherwise specified), several species-enriched features emerged in our dataset. In particular, Pavine, a tyrosine alkaloid, was detected in Cascara and Frangula preparations; to the best of our knowledge, this represents its first report in these herbal drugs. Likewise, Vicenin 2 was observed in Senna, whereas Piceatannol showed comparatively higher abundances in Rhubarb. These assignments are based on high-resolution MS and MS/MS database matching, and as such should be regarded as putative annotations pending confirmation with authentic standards. In all four plants, flavonoids represented the dominant superclass, confirming their central role in the phytochemical composition of these species, which is in agreement with previous studies [36,37,38,39].
With regard to HADs, each plant was characterized by compounds typical of the species itself, confirming what has previously been stated in several studies [18,19,20,23]. Despite the complexity of the analysed samples, their natural variability, and their origin from different plant parts (e.g., leaves and bark), it was possible to confirm consistent, species-specific trends in HAD composition [15]. In Frangula, the trend was defined by a predominance of frangulins and glucofrangulins A and B, followed by emodin. In Cascara, the profile was marked by cascaroside A, aloin A, and aloin B, with emodin and traces of aloe-emodin. In Rhubarb, rhein and rhein-8-glucoside dominated together with emodin, while Senna was characterized by abundant sennidins (A and B), sennosides (A and B), and rhein derivatives. These profiles illustrate not only the expected complexity of HAD distribution but also the taxonomic consistency across species despite differences in the analysed plant parts.
From a pharmacological perspective, the observed chemical diversity is expected to influence both bioavailability and safety. For instance, glycosylated hydroxyanthracene derivatives generally require metabolic activation in the gut before absorption, whereas aglycones and low-molecular-weight phenolics are typically more readily absorbed but may also display different toxicity profiles [13]. Likewise, flavonoids and other polyphenols can modulate intestinal permeability, metabolism, and oxidative stress, potentially affecting the overall response to these preparations [40]. However, the present study was not designed to directly evaluate bioavailability or safety, and no pharmacokinetic or toxicological measurements were performed. Therefore, our data should be interpreted as a comprehensive chemical framework that can inform future functional studies, rather than as a direct assessment of clinical efficacy or risk.
The data highlight the significant differences in the composition of compound classes among the plants. Cascara and Frangula, both of which belong to the Rhamnaceae family, exhibit notable similarities, especially in the abundance of flavonoids and anthraquinones. Rhubarb, on the other hand, is characterized by a higher presence of phenolic acids and chromanes. Senna stands out with a distinct distribution, particularly in terms of coumarins and specific monoterpenoids. This comparative view emphasizes the metabolic diversity among the species and highlights both family-specific and species-specific chemical signatures.
An additional key observation is the limited number of metabolites shared among the four species. The Venn diagram analysis vividly illustrates that only four metabolites are common across all plants. This scarcity of shared compounds indicates a high degree of specificity in the secondary metabolites present in each species. This observation prompted us to further explore inter-species relationships through multivariate approaches.
Our statistical analysis, including principal component analysis (PCA) and hierarchical cluster analysis (HCA), provided a deeper understanding of the metabolic relationships among the plant samples. Senna emerged as distinct, reinforcing its unique metabolic profile compared to the other varieties. Moreover, the heatmap analysis highlighted two main clusters, revealing the prevalence of flavonoids across all plants and indicating potential biosynthetic connections in the pathways of phenylpropanoids and anthraquinones. The complex interrelationships among the various metabolites that make up the bigger subcluster highlight the intricacy and interconnectivity of these plants’ metabolic profiles.
5. Conclusions
This study provides a comprehensive metabolomic characterization of four medicinal plants widely used for their laxative properties: Cascara (Rhamnus purshiana), Senna (Cassia angustifolia), Rhubarb (Rheum palmatum), and Frangula (Rhamnus frangula). By combining untargeted LC–MS/MS with bioinformatic approaches, we generated a high-resolution chemical fingerprint that extends beyond the well-known hydroxyanthracene derivatives (HADs).
The analysis revealed species-specific metabolic signatures, including the first report of Pavine in the Rhamnaceae family, and highlighted the predominance of flavonoids, anthraquinones, phenylpropanoids, and other bioactive classes. Strikingly, only four flavonoids were shared across all species, underscoring the remarkable biochemical diversity and taxonomic specificity of these plants. Our approach proved effective for fingerprinting complex botanical matrices and offers a robust framework for the discovery of distinctive metabolites with potential biological relevance. Accurate structural classification not only deepens our understanding of plant metabolic diversity but also provides a chemical basis for evaluating the safety and efficacy of phytotherapeutic preparations. Future studies combining metabolomic profiling with bioactivity assays and biosynthetic pathway analysis will be crucial in translating these findings into pharmacological applications in order to assess the therapeutic potential of these plants.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/metabo15120779/s1, Table S1: Excel file reporting, for each detected feature in Cascara, the following fields: Correspondent name, molecular formula, annotation mass error (DeltaMass, ppm), molecular weight, m/z, retention time (min), maximum peak area, MSI level, MS2 information, and reference ion; Table S2: Excel file reporting, for each detected feature in Frangula, the same set of fields; Table S3: Excel file reporting, for each detected feature in Rhubarb, the same set of fields; Table S4: Excel file reporting, for each detected feature in Senna, the same set of fields.
Author Contributions
Conceptualization, A.L.P., P.N., M.C., V.C., L.T., and L.S.; methodology, A.L.P., P.N., and L.S.; software, A.L.P. and F.C.; formal analysis, V.C. and L.T.; writing—original draft, A.L.P. and P.N.; writing—review and editing, V.C., L.T., L.S., and M.B.; supervision, L.S. and M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All processed metabolomic data supporting this study are provided in the Supplementary Materials as Excel tables exported from Compound Discoverer, including for each detected feature the precursor m/z, retention time, peak area, ionization mode, database-derived annotation, and MSI confidence level. The full implementation of the graph-based neural network used for metabolite classification (model architectures, trained weights and analysis scripts) is openly available in the associated GitHub repository: https://github.com/bcorrad/ginestra25 (accessed on 20 November 2025).
Acknowledgments
We would like to thank FEI—Federazione Erboristi Italiani (Italian Herbalists Federation) and Linneus Consulting for their support.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| HADs | Hydroxyanthracene Derivatives |
| UPLC–MS/MS | Ultra-High-Performance Liquid Chromatography–Tandem Mass Spectrometry |
| HCA | Hierarchical Cluster Analysis |
| GIN | Graph Isomorphism Network |
Appendix A
Appendix A reports the model architecture and the complete set of tables for each species.
Appendix A.1. Model Architecture
The GIN (Graph Isomorphism Network) architectures reported in Table A1 were used for hierarchical metabolite classification at the pathway, superclass, and class levels. Each model processes molecular graphs using successive GIN convolutional layers followed by batch normalization and fully connected layers. Output dimensions correspond to the number of categories at each hierarchy level (7, 70, and 653, respectively).

Table A1.
Layer-by-layer GIN architectures for pathway, superclass, and class classification tasks.
Table A1.
Layer-by-layer GIN architectures for pathway, superclass, and class classification tasks.
| Model | Pathway | Superclass | Class | |||
|---|---|---|---|---|---|---|
| Layer | Params | Layer | Params | Layer | Params | |
| GINConv | (23, 512) | GINConv | (23, 512) | GINConv | (23, 512) | |
| BatchNorm1d | (512) | BatchNorm1d | (512) | BatchNorm1d | (512) | |
| GINConv ×3 | (512, 512) | GINConv ×3 | (512, 512) | GINConv ×2 | (512, 512) | |
| BatchNorm1d | (512) | BatchNorm1d | (512) | BatchNorm1d | (512) | |
| GIN | Linear | (2560, 1024) | Linear | (2560, 1024) | Linear | (2048, 1024) |
| BatchNorm1d | (1024) | BatchNorm1d | (1024) | BatchNorm1d | (1024) | |
| Output layer | (1024, 7) | Output layer | (1024, 70) | Output layer | (1024, 653) | |
Appendix A.2. Cascara
For Cascara, Table A2 lists all identified metabolites with validated superclass assignments, while Table A3 provides the hierarchical classification with model-derived confidence scores. Table A4 highlights the few discordant cases, which are discussed further below. For full-precision m/z values (reported to four decimal places) together with the corresponding retention times (RT) and mass errors (, ppm), readers are referred to Supplementary Table S1.

Table A2.
Cascara—Identified metabolites with validated Superclass. Molecular weights are rounded to two decimals.
Table A2.
Cascara—Identified metabolites with validated Superclass. Molecular weights are rounded to two decimals.
| Component | Formula | Molecular Weight | Superclass |
|---|---|---|---|
| Pavine | C20H23NO4 | 341.16 | Tyrosine alkaloids |
| Emodin | C15H10O5 | 270.05 | Polycyclic aromatic polyketides |
| Aloin A | C21H22O9 | 418.13 | Polycyclic aromatic polyketides |
| Nobiletin | C21H22O8 | 402.13 | Flavonoids |
| Aloin B | C21H22O9 | 418.13 | Polycyclic aromatic polyketides |
| Isoliquiritigenin | C15H12O4 | 256.07 | Flavonoids |
| 2-Ethyl-9,10-anthraquinone | C16H12O2 | 236.08 | Polycyclic aromatic polyketides |
| Primuletin | C15H10O3 | 238.06 | Flavonoids |
| Cascaroside A | C27H32O14 | 580.18 | Polycyclic aromatic polyketides |
| myrciacitrin V | C30H30O13 | 598.17 | Flavonoids |
| Naringin | C27H32O14 | 580.18 | Flavonoids |
| Cascaroside C/Cascaroside D | C27H32O13 | 564.19 | Polycyclic aromatic polyketides |
| Cirsimarin | C23H24O11 | 476.13 | Flavonoids |
| myrciacitrin IV | C32H32O13 | 624.19 | Flavonoids |
| Demethyloleuropein | C24H30O13 | 526.17 | Monoterpenoids |
| Flavone | C15H10O2 | 222.07 | Flavonoids |
| 1,8-Dihydroxy-9(10H)-anthracenone | C14H10O3 | 226.06 | Polycyclic aromatic polyketides |
| Kaempferol | C15H10O6 | 286.05 | Flavonoids |
| Chrysoeriol 7-O-glucoside | C22H22O11 | 462.12 | Flavonoids |
| Narirutin -O-glucoside | C33H42O19 | 742.23 | Flavonoids |
| Narirutin | C27H32O14 | 580.18 | Flavonoids |
| Chlorogenic acid | C16H18O9 | 354.09 | Phenylpropanoids (C6–C3) |
| Caffeic acid | C9H8O4 | 180.04 | Phenylpropanoids (C6–C3) |
| Aloe emodin | C15H10O5 | 254.06 | Polycyclic aromatic polyketides |
| 3-Feruloylquinic acid | C17H20O9 | 368.11 | Phenylpropanoids (C6–C3) |
| Genistein | C15H10O5 | 270.05 | Isoflavonoids |
| Rhoifolin -O-glucoside | C33H40O19 | 740.22 | Flavonoids |
| Procyanidin dimer B1 | C30H26O12 | 578.14 | Flavonoids |
| Formononetin | C16H12O4 | 268.07 | Isoflavonoids |
| Luteolin | C15H10O6 | 286.05 | Flavonoids |
| Prunin | C21H22O10 | 434.12 | Flavonoids |
| Kaempferol 3-O-glucosyl-rhamnosyl-galactoside | C33H40O20 | 756.21 | Flavonoids |
| Nepetin | C16H12O7 | 316.06 | Flavonoids |
| 3-Caffeoylquinic acid | C16H18O9 | 354.10 | Phenylpropanoids (C6–C3) |
| -O-Acetylglycitin | C24H24O11 | 488.13 | Isoflavonoids |
| 2,4,6-Trimethoxybenzophenone | C16H16O4 | 272.11 | Phloroglucinols |
| Daidzin | C21H20O9 | 416.11 | Isoflavonoids |
| Geraldone | C16H12O5 | 284.07 | Flavonoids |
| 6-Methoxyflavonol | C16H12O4 | 268.07 | Flavonoids |
| 1-Sinapoyl-2-feruloylgentiobiose | C33H40O18 | 724.22 | Phenylpropanoids (C6–C3) |
| 3-Methoxynobiletin | C22H24O9 | 432.14 | Flavonoids |
| 1,2-Diferuloylgentiobiose | C32H38O17 | 694.21 | Phenylpropanoids (C6–C3) |
| Quercetin | C15H10O7 | 302.04 | Flavonoids |
| Phloretin | C15H14O5 | 274.08 | Flavonoids |
| Galangin | C15H10O5 | 270.05 | Flavonoids |
| Coumestrol | C15H8O5 | 268.04 | Isoflavonoids |
| Apocynin | C9H10O3 | 166.06 | Phenylpropanoids (C6–C3) |
| Phloridzin | C21H24O10 | 436.14 | Flavonoids |
| Sinensetin | C20H20O7 | 372.12 | Flavonoids |
| Eriodictyol | C15H12O6 | 288.06 | Flavonoids |
| Tectoridin | C22H22O11 | 462.12 | Isoflavonoids |
| Chrysin | C15H10O4 | 254.06 | Flavonoids |
| Taxifolin | C15H12O7 | 304.06 | Flavonoids |
| Vanillin | C8H8O3 | 152.05 | Phenylpropanoids (C6–C3) |
| Cyanidin 3-O-glucosyl-rutinoside | C33H41O20 | 757.22 | Flavonoids |
| Sinapaldehyde | C11H12O4 | 208.07 | Phenylpropanoids (C6–C3) |
| 3-hydroxyflavanone | C15H12O3 | 240.08 | Flavonoids |
| Eugenol | C10H12O2 | 164.08 | Phenylpropanoids (C6–C3) |
| p-Coumaric acid ethyl ester | C11H12O3 | 192.08 | Phenylpropanoids (C6–C3) |
| Carnosol | C20H26O4 | 330.18 | Diterpenoids |
| isoquercetin | C21H20O12 | 464.10 | Flavonoids |
| 3-p-Coumaroylquinic acid | C16H18O8 | 338.10 | Phenylpropanoids (C6–C3) |
| Morin | C15H10O7 | 302.04 | Flavonoids |
| p-Coumaroyl glycolic acid | C11H10O5 | 222.05 | Phenylpropanoids (C6–C3) |
| Homoeriodictyol | C16H14O6 | 302.08 | Flavonoids |
| 4-Vinylguaiacol | C9H10O2 | 150.07 | Phenylpropanoids (C6–C3) |
| Apigenin 7-O-(-malonyl-apiosyl-glucoside) | C29H30O17 | 650.15 | Flavonoids |
| Kaempferitrin | C27H30O14 | 578.16 | Flavonoids |
| olmelin | C16H12O5 | 284.07 | Isoflavonoids |
| Coumarin | C9H6O2 | 146.04 | Coumarins |
| Hesperetin | C16H14O6 | 302.08 | Flavonoids |
| m-Coumaric acid | C9H8O3 | 164.05 | Phenylpropanoids (C6–C3) |
| 3,4,7,8-TETRAHYDROXYFLAVONE | C15H10O6 | 286.05 | Monoterpenoids |
| -O-Malonyldaidzin | C24H22O12 | 502.11 | Isoflavonoids |
| Hispidulin | C16H12O6 | 300.06 | Flavonoids |
| Apigenin | C15H10O5 | 270.05 | Flavonoids |
| Syringaresinol | C22H26O8 | 418.16 | Lignans |
| Secoisolariciresinol | C20H26O6 | 362.17 | Lignans |
| Kalambroside B | C30H34O17 | 666.18 | Flavonoids |
| Naringenin 7-O-glucoside | C21H22O10 | 434.12 | Flavonoids |
| Scopoletin | C10H8O4 | 192.04 | Coumarins |
| Kaempferol 3-O--rutinoside | C27H30O15 | 594.16 | Flavonoids |
| Acetyl eugenol | C12H14O3 | 206.09 | Phenylpropanoids (C6–C3) |
| Didymin/Poncirin | C28H34O14 | 594.20 | Flavonoids |
| 1,2-Disinapoylgentiobiose | C34H42O19 | 754.23 | Phenylpropanoids (C6–C3) |
| Carnosic acid | C20H28O4 | 332.20 | Diterpenoids |
| Dihydroquercetin 3-O-rhamnoside | C21H22O11 | 450.12 | Flavonoids |
| Isorhamnetin 3-O-glucoside 7-O-rhamnoside | C28H32O16 | 624.17 | Flavonoids |
| Quercetin 3-O-xylosyl-rutinoside | C32H38O20 | 742.20 | Flavonoids |
| 2-O-Rhamnosylvitexin | C27H30O14 | 578.16 | Flavonoids |
| 4-Ethylcatechol | C8H10O2 | 138.07 | Benzenoids |
| 9,10-Dihydroxyanthracene | C14H10O2 | 210.07 | Polycyclic aromatic polyketides |
| 4-Heptyloxyphenol | C13H20O2 | 208.15 | Aromatic polyketides |

Table A3.
Cascara—Hierarchical classification of identified metabolites with model accuracy expressed as percentage (Acc. %).
Table A3.
Cascara—Hierarchical classification of identified metabolites with model accuracy expressed as percentage (Acc. %).
| Component | Pathway | Acc.% | Superclass | Acc.% | Class | Acc.% |
|---|---|---|---|---|---|---|
| Pavine | Alkaloids | 99.99 | Tyrosine alkaloids | 100.00 | Isoquinoline alkaloids | 88.90 |
| Emodin | Polyketides | 99.90 | Polycyclic aromatic polyketides | 100.00 | Anthraquinones and anthrones | 99.90 |
| Aloin A | Polyketides | 99.80 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Nobiletin | Shikimates and Phenylpropanoids | 99.97 | Flavonoids | 100.00 | Flavones | 99.98 |
| Aloin B | Polyketides | 99.80 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Isoliquiritigenin | Shikimates and Phenylpropanoids | 99.99 | Flavonoids | 100.00 | Chalcones | 100.00 |
| 2-Ethyl-9,10-anthraquinone | Polyketides | 100.00 | Polycyclic aromatic polyketides | 100.00 | Anthraquinones and anthrones | 100.00 |
| Primuletin | Shikimates and Phenylpropanoids | 98.60 | Flavonoids | 100.00 | Flavones | 100.00 |
| Cascaroside A | Polyketides | 99.70 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.70 |
| myrciacitrin V | Shikimates and Phenylpropanoids | 98.70 | Flavonoids | 100.00 | Flavanones | 100.00 |
| Naringin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavanones | 100.00 |
| Cascaroside C/Cascaroside D | Polyketides | 99.60 | Polycyclic aromatic polyketides | 100.00 | Anthraquinones and anthrones | 99.70 |
| Cirsimarin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavones | 100.00 |
| myrciacitrin IV | Shikimates and Phenylpropanoids | 99.80 | Flavonoids | 100.00 | Flavanones | 100.00 |
| Demethyloleuropein | Terpenoids | 100.00 | Monoterpenoids | 100.00 | Secoiridoid monoterpenoids | 100.00 |
| Flavone | Shikimates and Phenylpropanoids | 99.50 | Flavonoids | 100.00 | Flavones | 100.00 |
| 1,8-Dihydroxy-9(10H)-anthracenone | Polyketides | 100.00 | Polycyclic aromatic polyketides | 100.00 | Anthraquinones and anthrones | 100.00 |
| Kaempferol | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavonols | 100.00 |
| Chrysoeriol 7-O-glucoside | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavones | 100.00 |
| Narirutin -O-glucoside | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavanones | 100.00 |
| Narirutin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavanones | 100.00 |
| Chlorogenic acid | Shikimates and Phenylpropanoids | 99.80 | Phenylpropanoids (C6–C3) | 97.10 | Cinnamic acids and derivatives | 99.50 |
| Caffeic acid | Shikimates and Phenylpropanoids | 98.70 | Phenylpropanoids (C6–C3) | 96.30 | Cinnamic acids and derivatives | 98.20 |
| Aloe emodin | Polyketides | 100.00 | Polycyclic aromatic polyketides | 100.00 | Anthraquinones and anthrones | 100.00 |
| 3-Feruloylquinic acid | Shikimates and Phenylpropanoids | 99.70 | Phenylpropanoids (C6–C3) | 98.70 | Cinnamic acids and derivatives | 99.80 |
| Genistein | Shikimates and Phenylpropanoids | 99.80 | Isoflavonoids | 100.00 | Isoflavones | 99.90 |
| Rhoifolin -O-glucoside | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavones | 100.00 |
| Procyanidin dimer B1 | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Proanthocyanins | 99.90 |
| Formononetin | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 100.00 | Isoflavones | 99.90 |
| Luteolin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 100.00 | Flavones | 100.00 |
| Prunin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavanones | 100.00 |
| Kaempferol 3-O-glucosyl-rhamnosyl-galactoside | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavonols | 100.00 |
| Nepetin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavones | 100.00 |
| 3-Caffeoylquinic acid | Shikimates and Phenylpropanoids | 99.80 | Phenylpropanoids (C6–C3) | 97.10 | Cinnamic acids and derivatives | 99.50 |
| -O-Acetylglycitin | Shikimates and Phenylpropanoids | 100.00 | Isoflavonoids | 100.00 | Isoflavones | 100.00 |
| 2,4,6-Trimethoxybenzophenone | Polyketides | 99.60 | Phloroglucinols | 100.00 | Acyl phloroglucinols | 99.90 |
| Daidzin | Shikimates and Phenylpropanoids | 100.00 | Isoflavonoids | 100.00 | Isoflavones | 100.00 |
| Geraldone | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavones | 100.00 |
| 6-Methoxyflavonol | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 100.00 | Flavonols | 100.00 |
| 1-Sinapoyl-2-feruloylgentiobiose | Shikimates and Phenylpropanoids | 94.00 | Phenylpropanoids (C6–C3) | 99.70 | Cinnamic acids and derivatives | 99.90 |
| 3-Methoxynobiletin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavonols | 100.00 |
| 1,2-Diferuloylgentiobiose | Shikimates and Phenylpropanoids | 92.90 | Phenylpropanoids (C6–C3) | 99.80 | Cinnamic acids and derivatives | 100.00 |
| Quercetin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 100.00 | Flavonols | 100.00 |
| Phloretin | Shikimates and Phenylpropanoids | 99.10 | Flavonoids | 100.00 | Chalcones | 100.00 |
| Galangin | Shikimates and Phenylpropanoids | 99.70 | Flavonoids | 100.00 | Flavonols | 100.00 |
| Coumestrol | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 99.80 | Coumestan | 99.80 |
| Apocynin | Shikimates and Phenylpropanoids | 98.80 | Phenylpropanoids (C6–C3) | 56.60 | Simple phenolic acids | 31.00 |
| Phloridzin | Shikimates and Phenylpropanoids | 99.60 | Flavonoids | 100.00 | Chalcones | 99.90 |
| Sinensetin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavones | 100.00 |
| Eriodictyol | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavanones | 99.80 |
| Tectoridin | Shikimates and Phenylpropanoids | 100.00 | Isoflavonoids | 100.00 | Isoflavones | 99.90 |
| Chrysin | Shikimates and Phenylpropanoids | 99.50 | Flavonoids | 100.00 | Flavones | 100.00 |
| Taxifolin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Dihydroflavonols | 99.80 |
| Cyanidin 3-O-glucosyl-rutinoside | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Anthocyanidins | 100.00 |
| Sinapaldehyde | Shikimates and Phenylpropanoids | 98.00 | Phenylpropanoids (C6–C3) | 91.40 | Cinnamic acids and derivatives | 29.20 |
| 3-hydroxyflavanone | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavanones | 99.90 |
| Eugenol | Shikimates and Phenylpropanoids | 100.00 | Phenylpropanoids (C6–C3) | 98.80 | Cinnamic acids and derivatives | 96.30 |
| p-Coumaric acid ethyl ester | Shikimates and Phenylpropanoids | 98.70 | Phenylpropanoids (C6–C3) | 97.00 | Cinnamic acids and derivatives | 95.90 |
| Carnosol | Terpenoids | 100.00 | Diterpenoids | 100.00 | Abietane diterpenoids | 99.90 |
| isoquercetin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavonols | 100.00 |
| 3-p-Coumaroylquinic acid | Shikimates and Phenylpropanoids | 100.00 | Phenylpropanoids (C6–C3) | 94.20 | Cinnamic acids and derivatives | 99.60 |
| Morin | Shikimates and Phenylpropanoids | 99.70 | Flavonoids | 100.00 | Flavonols | 100.00 |
| p-Coumaroyl glycolic acid | Shikimates and Phenylpropanoids | 99.10 | Phenylpropanoids (C6–C3) | 15.60 | Cinnamic acids and derivatives | 57.70 |
| Homoeriodictyol | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 99.90 | Flavanones | 99.80 |
| 4-Vinylguaiacol | Shikimates and Phenylpropanoids | 100.00 | Phenylpropanoids (C6–C3) | 90.00 | Cinnamic acids and derivatives | 60.60 |
| Apigenin 7-O-(-malonyl-apiosyl-glucoside) | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavones | 99.90 |
| Kaempferitrin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavonols | 100.00 |
| olmelin | Shikimates and Phenylpropanoids | 99.80 | Isoflavonoids | 100.00 | Isoflavones | 99.90 |
| Coumarin | Shikimates and Phenylpropanoids | 99.90 | Coumarins | 99.80 | Simple coumarins | 90.80 |
| Hesperetin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 99.90 | Flavanones | 99.80 |
| m-Coumaric acid | Shikimates and Phenylpropanoids | 99.30 | Phenylpropanoids (C6–C3) | 91.90 | Cinnamic acids and derivatives | 99.00 |
| 3,4,7,8-TETRAHYDROXYFLAVONE | Terpenoids | 48.10 | Monoterpenoids | 70.20 | Simple diketopiperazine alkaloids | 7.80 |
| -O-Malonyldaidzin | Shikimates and Phenylpropanoids | 100.00 | Isoflavonoids | 99.90 | Isoflavones | 99.90 |
| Hispidulin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavones | 100.00 |
| Apigenin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 100.00 | Flavones | 100.00 |
| Syringaresinol | Shikimates and Phenylpropanoids | 100.00 | Lignans | 100.00 | Furofuranoid lignans | 100.00 |
| Secoisolariciresinol | Shikimates and Phenylpropanoids | 100.00 | Lignans | 100.00 | Dibenzylbutane lignans | 100.00 |
| Kalambroside B | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavonols | 100.00 |
| Naringenin 7-O-glucoside | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavanones | 100.00 |
| Scopoletin | Shikimates and Phenylpropanoids | 100.00 | Coumarins | 99.80 | Simple coumarins | 95.60 |
| Kaempferol 3-O--rutinoside | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavonols | 100.00 |
| Acetyl eugenol | Shikimates and Phenylpropanoids | 99.90 | Phenylpropanoids (C6–C3) | 99.30 | Cinnamic acids and derivatives | 95.80 |
| Didymin/Poncirin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 99.90 | Flavanones | 99.90 |
| 1,2-Disinapoylgentiobiose | Shikimates and Phenylpropanoids | 95.40 | Phenylpropanoids (C6–C3) | 99.50 | Cinnamic acids and derivatives | 100.00 |
| Carnosic acid | Terpenoids | 100.00 | Diterpenoids | 100.00 | Abietane diterpenoids | 99.70 |
| Dihydroquercetin 3-O-rhamnoside | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 99.90 | Dihydroflavonols | 99.70 |
| Isorhamnetin 3-O-glucoside 7-O-rhamnoside | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavonols | 100.00 |
| Quercetin 3-O-xylosyl-rutinoside | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavonols | 100.00 |
| 2-O-Rhamnosylvitexin | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 100.00 | Flavones | 100.00 |
| 4-Ethylcatechol | Shikimates and Phenylpropanoids | 76.80 | Phenylethanoids (C6–C2) | 9.60 | Phenylethanoids | 11.20 |
| 9,10-Dihydroxyanthracene | Polyketides | 91.50 | Naphthalenes | 98.10 | Naphthoquinones | 85.20 |
| 4-Heptyloxyphenol | Shikimates and Phenylpropanoids | 24.30 | Phenolic acids (C6–C1) | 2.90 | Hydrocarbons | 3.10 |

Table A4.
Discordant superclass assignments in Cascara (3/93 compounds; overall concordance 96.8%).
Table A4.
Discordant superclass assignments in Cascara (3/93 compounds; overall concordance 96.8%).
| Component | Validated Superclass | Predicted Superclass | Acc.% |
|---|---|---|---|
| 4-Ethylcatechol | Benzenoids | Phenylethanoids (C6–C2) | 9.6 |
| 9,10-Dihydroxyanthracene | Polycyclic aromatic polyketides | Naphthalenes | 98.10 |
| 4-Heptyloxyphenol | Aromatic polyketides | Phenolic acids (C6–C1) | 2.9 |
The few discordant cases reflect borderline structural features. For instance, 4-ethylcatechol was misclassified as a phenylethanoid due to the presence of a short C2 side chain, although it lacks the typical conjugated system of this class. Similarly, 9,10-dihydroxyanthracene was predicted as a naphthalene, likely because the model emphasized the fused aromatic scaffold rather than the oxidized anthracene core. Finally, 4-heptyloxyphenol was assigned to phenolic acids despite the absence of a carboxyl group, a misclassification probably arising from underrepresentation of long-chain alkylphenols in the training dataset.
Appendix A.3. Frangula
For Frangula, Table A5 lists all identified metabolites with validated superclass assignments, while Table A6 provides the hierarchical classification with model-derived confidence scores. Table A7 highlights the few discordant cases, which are discussed further below. For full-precision m/z values (reported to four decimal places) together with the corresponding retention times (RT) and mass errors (, ppm), readers are referred to Supplementary Table S1.

Table A5.
Frangula—Identified metabolites with validated Superclass. Molecular weights are rounded to two decimals.
Table A5.
Frangula—Identified metabolites with validated Superclass. Molecular weights are rounded to two decimals.
| Component | Formula | Molecular Weight | Superclass |
|---|---|---|---|
| Pavine | C20H23NO4 | 341.16 | Tyrosine alkaloids |
| Emodin | C15H10O5 | 270.05 | Polycyclic aromatic polyketides |
| Liquiritin | C21H22O9 | 418.13 | Flavonoids |
| Genistein | C15H10O5 | 270.05 | Isoflavonoids |
| Glucofrangulin A | C27H30O14 | 578.17 | Polycyclic aromatic polyketides |
| Unknown compound | – | 528.19 | – |
| Unknown compound | – | 484.16 | – |
| Oleuropein | C25H32O13 | 540.19 | Monoterpenoids |
| Demethyloleuropein | C24H30O13 | 526.17 | Monoterpenoids |
| Unknown compound | – | 510.18 | – |
| Unknown compound | – | 496.16 | – |
| Glucofrangulin B | C26H28O14 | 564.15 | Polycyclic aromatic polyketides |
| Isoliquiritigenin | C15H12O4 | 256.07 | Flavonoids |
| 5,6-O--D-diglucopyranosylangelicin | C23H26O15 | 542.13 | Coumarins |
| Kaempferol | C15H10O6 | 286.05 | Flavonoids |
| Frangulin A | C21H20O9 | 416.11 | Polycyclic aromatic polyketides |
| Frangulin B | C20H18O9 | 402.10 | Polycyclic aromatic polyketides |
| Lespedin | C27H30O14 | 578.17 | Flavonoids |
| Phloretin | C15H14O5 | 274.08 | Flavonoids |
| 2-Ethyl-9,10-anthraquinone | C16H12O2 | 236.08 | Polycyclic aromatic polyketides |
| Neohesperidin dihydrochalcone | C28H36O15 | 612.21 | Flavonoids |
| Nepodin | C13H12O3 | 216.08 | Naphthalenes |
| Daidzein | C15H10O4 | 254.06 | Isoflavonoids |
| Piceatannol | C14H12O4 | 244.07 | Stilbenoids |
| trans-Caffeic acid | C9H8O4 | 180.04 | Phenylpropanoids (C6–C3) |
| 6-Methoxyflavonol | C16H12O4 | 268.07 | Flavonoids |
| Gossypetin 3-sophoroside-8-glucoside | C33H40O23 | 804.20 | Flavonoids |
| 3-Caffeoylquinic acid | C16H18O9 | 354.09 | Phenylpropanoids (C6–C3) |
| Laricitrin 3,7,-triglucoside | C34H42O23 | 818.21 | Flavonoids |
| Patuletin 3-(-acetylrhamnoside)-7-(,-diacetylrhamnoside) | C34H38O19 | 750.20 | Flavonoids |
| Acaciin | C28H32O14 | 592.18 | Flavonoids |
| Pinocembrin | C15H12O4 | 256.07 | Flavonoids |
| Quercitrin | C21H20O11 | 448.10 | Flavonoids |
| Isoschaftoside | C26H28O14 | 564.15 | Flavonoids |
| Methyl chlorogenate | C17H20O9 | 368.11 | Phenylpropanoids (C6–C3) |
| Caffeic acid 3-glucoside | C15H18O9 | 342.10 | Phenylpropanoids (C6–C3) |
| Luteolin | C15H10O6 | 286.05 | Flavonoids |
| Kaempferide | C16H12O6 | 300.06 | Flavonoids |
| Methyl 4-methoxycinnamate | C11H12O3 | 192.08 | Phenylpropanoids (C6–C3) |
| Narirutin | C27H32O14 | 580.18 | Flavonoids |
| Loganin | C17H26O10 | 390.15 | Monoterpenoids |
| Apiin | C26H28O14 | 564.15 | Flavonoids |
| Taxifolin | C15H12O7 | 304.06 | Flavonoids |
| Afzelechin | C15H14O5 | 274.08 | Flavonoids |
| Diosmetin 7-O--D-glucoside | C22H22O11 | 462.12 | Flavonoids |
| isorhamnetin 3-O-alpha-L-[-p-coumaroyl--D-glucopyranosyl-(1→2)-rhamnopyranoside] | C37H38O18 | 770.21 | Flavonoids |
| Oleic acid | C18H34O2 | 282.26 | Fatty acids and conjugates |
| ,,5,7-Tetramethyldihydroquercetin | C19H20O7 | 360.12 | Flavonoids |
| 3-Methoxynobiletin | C22H24O9 | 432.14 | Flavonoids |
| Epigallocatechin-(4)--O-methylgallocatechin | C31H28O14 | 624.15 | Flavonoids |
| Primuletin | C15H10O3 | 238.06 | Flavonoids |
| -Acetylpaeonoside | C29H32O17 | 652.16 | Flavonoids |
| 2,3-Dihydro-,-di-O-methylamentoflavone | C32H24O10 | 568.14 | Flavonoids |
| Piceid | C20H22O8 | 390.13 | Stilbenoids |
| Vicenin-2 | C27H30O15 | 594.16 | Flavonoids |
| Chrysoeriol 7-(-feruloylglucuronosyl)-(1→2)-glucuronide | C38H36O21 | 828.17 | Flavonoids |
| ,5,7-Trimethoxyflavone | C18H16O5 | 312.10 | Flavonoids |
| Catechin | C15H14O6 | 290.08 | Flavonoids |
| Rutin | C27H30O16 | 610.15 | Flavonoids |
| Sakuranetin | C16H14O5 | 286.08 | Flavonoids |
| Hesperetin | C16H14O6 | 302.08 | Flavonoids |
| Flavanol | C15H14O6 | 290.08 | Flavonoids |
| Hispidulin | C16H12O6 | 300.06 | Flavonoids |
| 7-Methoxyflavone | C16H12O3 | 252.08 | Flavonoids |
| Diosmin | C28H32O15 | 608.17 | Flavonoids |
| Schaftoside | C26H28O14 | 564.15 | Flavonoids |
| (E)-4-Methoxycinnamic acid | C10H10O3 | 178.06 | Phenylpropanoids (C6–C3) |
| Isoformononetin | C16H12O4 | 268.07 | Isoflavonoids |
| Isoquercetin | C21H20O12 | 464.10 | Flavonoids |
| Malonyldaidzin | C24H22O12 | 502.11 | Isoflavonoids |
| Quercetin 3-O-xylosyl-rutinoside | C32H38O20 | 742.20 | Flavonoids |
| Liquiritin apioside | C26H30O13 | 550.17 | Flavonoids |
| Isovitexin 7-O-[feruloyl]-glucoside | C37H38O18 | 770.20 | Flavonoids |
| Isoquercitrin | C21H20O12 | 464.10 | Flavonoids |
| 1,2,4-Trihydroxyanthraquinone | C14H8O5 | 256.04 | Polycyclic aromatic polyketides |
| Kaempferol 3-(-rhamnosyl--(6-malyl-glucosyl)-glucoside) | C37H44O24 | 872.22 | Flavonoids |
| Quercetin-3-O-(-trans-p-coumaroyl--glucosyl)rhamnoside | C36H36O18 | 756.19 | Flavonoids |
| 1,2,8-Trihydroxyanthraquinone | C14H8O5 | 256.04 | Polycyclic aromatic polyketides |
| 4-Heptyloxyphenol | C13H20O2 | 208.15 | Aromatic polyketides |
| 5,7-DIHYDROXY-,,-TRIMETHOXYFLAVANONE | C18H18O7 | 346.11 | Flavonoids |
| 7-[(6-Deoxy--L-mannopyranosyl)oxy]-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-3-yl 6-O-(6-deoxy--L-mannopyranosyl)--D-glucopyranoside | C33H40O18 | 724.22 | Flavonoids |
| 4-Hydroxy-3-methyl-9,10-dioxo-9,10-dihydro-2-anthracenyl 6-O-acetyl-2-O-(6-deoxy--L-mannopyranosyl)--D-glucopyranoside | C29H32O14 | 604.18 | Polycyclic aromatic polyketides |
| 5-Hydroxy-3,6,7,8,,-hexamethoxyflavone | C21H22O9 | 418.13 | Flavonoids |

Table A6.
Frangula—Hierarchical classification of identified metabolites with model accuracy expressed as percentage.
Table A6.
Frangula—Hierarchical classification of identified metabolites with model accuracy expressed as percentage.
| Component | Pathway | Acc.% | Superclass | Acc.% | Class | Acc.% |
|---|---|---|---|---|---|---|
| Pavine | Tyrosine alkaloids | 99.90 | Tyrosine alkaloids | 99.90 | Isoquinoline alkaloids | 88.80 |
| Emodin | Polycyclic aromatic polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Liquiritin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.90 |
| Genistein | Isoflavonoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.80 |
| Glucofrangulin A | Polycyclic aromatic polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Unknown compound | – | – | – | |||
| Unknown compound | – | – | – | |||
| Oleuropein | Monoterpenoids | 99.90 | Monoterpenoids | 99.90 | Secoiridoid monoterpenoids | 100.00 |
| Demethyloleuropein | Monoterpenoids | 99.90 | Monoterpenoids | 99.90 | Secoiridoid monoterpenoids | 100.00 |
| Unknown compound | – | – | – | |||
| Unknown compound | – | – | – | |||
| Glucofrangulin B | Polycyclic aromatic polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Isoliquiritigenin | Flavonoids | 99.90 | Flavonoids | 99.90 | Chalcones | 100.00 |
| 5,6-O--D-diglucopyranosylangelicin | Coumarins | 99.90 | Coumarins | 99.90 | Furocoumarins | 99.90 |
| Kaempferol | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Frangulin A | Polycyclic aromatic polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Frangulin B | Polycyclic aromatic polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Lespedin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Phloretin | Flavonoids | 99.90 | Flavonoids | 99.90 | Chalcones | 99.90 |
| 2-Ethyl-9,10-anthraquinone | Polycyclic aromatic polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Neohesperidin dihydrochalcone | Flavonoids | 99.90 | Flavonoids | 99.90 | Chalcones | 99.90 |
| Nepodin | Naphthalenes | 99.90 | Naphthalenes | 99.90 | Naphthalenes and derivatives | 99.90 |
| Daidzein | Isoflavonoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.90 |
| Piceatannol | Stilbenoids | 99.90 | Stilbenoids | 99.90 | Monomeric stilbenes | 98.70 |
| trans-caffeic acid | Phenylpropanoids (C6–C3) | 96.20 | Phenylpropanoids (C6–C3) | 96.20 | Cinnamic acids and derivatives | 98.20 |
| 6-Methoxyflavonol | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Gossypetin 3-sophoroside-8-glucoside | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| 3-Caffeoylquinic acid | Phenylpropanoids (C6–C3) | 97.40 | Phenylpropanoids (C6–C3) | 97.10 | Cinnamic acids and derivatives | 99.90 |
| Laricitrin 3,7,-triglucoside | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Patuletin 3-(-acetylrhamnoside)-7-(,-diacetylrhamnoside) | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Acaciin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Pinocembrin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.70 |
| Quercitrin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Isoschaftoside | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Methyl chlorogenate | Phenylpropanoids (C6–C3) | 98.50 | Phenylpropanoids (C6–C3) | 98.50 | Cinnamic acids and derivatives | 99.50 |
| Caffeic acid 3-glucoside | Phenylpropanoids (C6–C3) | 98.10 | Phenylpropanoids (C6–C3) | 98.10 | Cinnamic acids and derivatives | 99.00 |
| Luteolin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Kaempferide | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Methyl 4-methoxycinnamate | Phenylpropanoids (C6–C3) | 98.50 | Phenylpropanoids (C6–C3) | 98.50 | Cinnamic acids and derivatives | 98.00 |
| Narirutin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.90 |
| Loganin | Monoterpenoids | 99.90 | Monoterpenoids | 99.90 | Iridoids monoterpenoids | 99.90 |
| Apiin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Taxifolin | Flavonoids | 99.90 | Flavonoids | 99.90 | Dihydroflavonols | 99.90 |
| Afzelechin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavan-3-ols | 99.80 |
| Diosmetin 7-O--D-glucoside | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| isorhamnetin 3-O--L-[-p-coumaroyl--D-glucopyranosyl-()-rhamnopyranoside] | Flavonoids | 100.00 | Flavonoids | 100.00 | Flavonols | 99.90 |
| Oleic acid | Fatty Acids and Conjugates | 97.20 | Fatty Acids and Conjugates | 97.20 | Unsaturated fatty acids | 90.30 |
| ,,5,7-tetramethyldihydroquercetin | Flavonoids | 99.90 | Flavonoids | 99.90 | Dihydroflavonols | 99.70 |
| 3-Methoxynobiletin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| epigallocatechin-()--O-methylgallocatechin | Flavonoids | 99.90 | Flavonoids | 99.90 | Proanthocyanins | 99.60 |
| Primuletin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| -acetylpaeonoside | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| 2,3-dihydro-,-di-O-methylamentoflavone | Flavonoids | 100.00 | Flavonoids | 100.00 | Flavanones;Flavones | 99.40 |
| Piceid | Stilbenoids | 99.90 | Stilbenoids | 99.90 | Monomeric stilbenes | 98.90 |
| vicenin 2 | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Chrysoeriol 7- (-feruloylglucuronosyl) - (1→2) -glucuronide | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| ,5,7-TRIMETHOXYFLAVONE | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Catechin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavan-3-ols | 99.80 |
| Rutin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Sakuranetin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.90 |
| Hesperetin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.80 |
| Flavanol | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavanones;Flavans | 74.50 |
| Hispidulin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| 7-methoxyflavone | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Diosmin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| schaftoside | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| (E)-4-Methoxycinnamic acid | Phenylpropanoids (C6–C3) | 97.90 | Phenylpropanoids (C6–C3) | 97.90 | Cinnamic acids and derivatives | 97.30 |
| isoformononetin | Isoflavonoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.90 |
| isoquercetin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| malonyldaidzin | Isoflavonoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.90 |
| Quercetin 3-O-xylosyl-rutinoside | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| LIQUIRITIN APIOSIDE | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.90 |
| isovitexin 7-O-[feruloyl]-glucoside | Flavonoids | 100.00 | Flavonoids | 100.00 | Flavones | 99.90 |
| Isoquercitrin | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| 1,2,4-Trihydroxyanthraquinone | Polycyclic aromatic polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Kaempferol 3- (-rhamnosyl-- (6-malyl-glucosyl) -glucoside) | Flavonoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Quercetin-3-O-(-trans-p-coumaroyl--glucosyl)rhamnoside | Flavonoids | 100.00 | Flavonoids | 100.00 | Flavonols | 99.90 |
| 1,2,8-Trihydroxyanthraquinone | Polycyclic aromatic polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.80 |
| 4-Heptyloxyphenol | Phenolic acids (C6–C1) | 2.90 | Phenolic acids (C6–C1) | 2.90 | Hydrocarbons | 3.00 |
| 5,7-DIHYDROXY-,,-TRIMETHOXYFLAVANONE | Anthranilic acid alkaloids | 1.30 | Anthranilic acid alkaloids | 1.30 | Pyridine alkaloids | 70.70 |
| 7-[(6-Deoxy--L-mannopyranosyl)oxy]-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-3-yl 6-O-(6-deoxy-alpha-L-mannopyranosyl)--D-glucopyranoside | - | |||||
| 4-Hydroxy-3-methyl-9,10-dioxo-9,10-dihydro-2-anthracenyl 6-O-acetyl-2-O-(6-deoxy--L-mannopyranosyl)--D-glucopyranoside | – | – | – | |||
| 5-hydroxy-3,6,7,8,,-hexamethoxyflavone | – | – | – |

Table A7.
Discordant and unassigned superclass assignments in Frangula (5/83 compounds; overall concordance 94.0%).
Table A7.
Discordant and unassigned superclass assignments in Frangula (5/83 compounds; overall concordance 94.0%).
| Component | Validated Superclass | Predicted Superclass | Acc.% |
|---|---|---|---|
| 4-Heptyloxyphenol | Aromatic polyketides | Phenolic acids (C6–C1) | 2.9 |
| 5,7-DIHYDROXY-,,-TRIMETHOXYFLAVANONE | Flavonoids | Anthranilic acid alkaloids | 1.3 |
| 7-[(6-Deoxy--L-mannopyranosyl)oxy]-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-3-yl 6-O-(6-deoxy-alpha-L-mannopyranosyl)--D-glucopyranoside | Flavonoids | – | – |
| 4-Hydroxy-3-methyl-9,10-dioxo-9,10-dihydro-2-anthracenyl 6-O-acetyl-2-O-(6-deoxy--L-mannopyranosyl)--D-glucopyranoside | Anthraquinones and anthrones | – | – |
| 5-hydroxy-3,6,7,8,,-hexamethoxyflavone | Flavonoids | – | – |
The few discordant cases reflect borderline structural features or gaps in the training data. For instance, 4-heptyloxyphenol was predicted as a phenolic acid despite lacking a carboxyl group, likely because the long alkyl side chain was poorly represented in training examples. Similarly, 5,7-dihydroxy-,,-trimethoxyflavanone, a typical flavonoid, was misclassified as an anthranilic acid alkaloid, probably due to the presence of multiple methoxy substituents mimicking alkaloid-like motifs. In addition, three compounds (a glycosylated flavone derivative, an anthracene glycoside, and a highly methoxylated flavone) were not assigned at all, reflecting either insufficient representation of complex glycosylated structures or ambiguous fragmentation patterns. These cases underline typical boundary conditions in automated annotation, but were readily resolved by expert review without altering the main compositional trends.
Appendix A.4. Rhubarb
For Rhubarb, Table A8 lists all identified metabolites with validated superclass assignments, while Table A9 provides the hierarchical classification with model-derived confidence scores. Table A10 highlights the few discordant cases, which are discussed further below. For full-precision m/z values (reported to four decimal places) together with the corresponding retention times (RT) and mass errors (, ppm), readers are referred to Supplementary Table S1.

Table A8.
Rhubarb—Identified metabolites with validated Superclass. Molecular weights are rounded to two decimals.
Table A8.
Rhubarb—Identified metabolites with validated Superclass. Molecular weights are rounded to two decimals.
| Component | Formula | Molecular Weight | Superclass |
|---|---|---|---|
| Emodin | C15H10O5 | 270.05 | Polycyclic aromatic polyketides |
| Piceatannol | C14H12O4 | 244.07 | Stilbenoids |
| Rhein | C15H8O6 | 284.03 | Polycyclic aromatic polyketides |
| Daidzein | C15H10O4 | 254.06 | Isoflavonoids |
| Catechin | C15H14O6 | 290.08 | Flavonoids |
| Rhein-8-glucoside | C21H18O11 | 446.08 | Polycyclic aromatic polyketides |
| Genistein | C15H10O5 | 270.05 | Isoflavonoids |
| (2Z)-6-hydroxy-2-[(4-hydroxy-3-methoxyphenyl) methylidene]-2,3-dihydro-1-benzofuran-3-one | C16H12O5 | 284.07 | Flavonoids |
| 3-O-Methylquercetintetraacetate | C24H20O11 | 484.10 | Flavonoids |
| (+)-Catechin 3-O-gallate | C22H18O10 | 442.09 | Flavonoids |
| Torachrysone 8-O--D-glucoside | C20H24O9 | 408.14 | Naphthalenes |
| spectaflavoside A | C46H42O22 | 946.22 | Flavonoids |
| 1,4-Dihydroxy-5,8-bis(p-toluidino)anthraquinone | C28H22N2O4 | 450.16 | Polycyclic aromatic polyketides |
| 5-Acetonyl-7-hydroxy-2-methylchromone | C13H12O4 | 232.07 | Chromanes |
| aloesin | C19H22O9 | 394.13 | Chromanes |
| Protocatechuic aldehyde | C7H6O3 | 138.03 | Phenolic acids (C6–C1) |
| Kaempferol | C15H10O6 | 286.05 | Flavonoids |
| Limocitrol 3- [ -L-arabinopyranosyl- () [ galactosyl- () ] -galactoside ] | C35H44O23 | 832.23 | Flavonoids |
| Irilone | C16H10O6 | 298.05 | Isoflavonoids |
| Afzelechin | C15H14O5 | 274.08 | Flavonoids |
| Cassialoin | C21H22O9 | 418.13 | Polycyclic aromatic polyketides |
| Isoliquiritigenin | C15H12O4 | 256.07 | Flavonoids |
| Vitexin | C21H20O10 | 432.11 | Flavonoids |
| Eriodictyol | C15H12O6 | 288.06 | Flavonoids |
| ,-DIMETHYLEPIGALLOCATECHIN GALLATE | C24H22O11 | 486.12 | Flavonoids |
| Resveratrol | C14H12O3 | 228.08 | Stilbenoids |
| Chrysin | C15H10O4 | 254.06 | Flavonoids |
| Resveratrol 3-O-glucoside | C20H22O8 | 390.13 | Stilbenoids |
| Kaempferol 3-O-glucosyl-rhamnosyl-galactoside | C33H40O20 | 756.21 | Flavonoids |
| 1,2,4-Trihydroxyanthraquinone | C14H8O5 | 256.04 | Polycyclic aromatic polyketides |
| Gallic acid | C7H6O5 | 170.02 | Phenolic acids (C6–C1) |
| aescuflavoside | C38H48O25 | 904.25 | Flavonoids |
| Hispidulin | C16H12O6 | 300.06 | Flavonoids |
| ,,7-Trihydroxyflavanone | C15H12O5 | 272.07 | Flavonoids |
| Apigenin | C15H10O5 | 270.05 | Flavonoids |
| Pinocembrin | C15H12O4 | 256.07 | Flavonoids |
| HEXAMETHYLQUERCETAGETIN | C21H22O8 | 402.13 | Flavonoids |
| 3-Hydroxyflavone | C15H10O3 | 238.06 | Flavonoids |
| Primuletin | C15H10O3 | 238.06 | Flavonoids |
| Quercetin 3-sambubioside--glucoside | C32H38O21 | 758.19 | Flavonoids |
| Phloretin | C15H14O5 | 274.08 | Flavonoids |
| 3-hydroxyflavanone | C15H12O3 | 240.08 | Flavonoids |
| Taxifolin | C15H12O7 | 304.06 | Flavonoids |
| Quercetin 3-O-xylosyl-rutinoside | C32H38O20 | 742.19 | Flavonoids |
| Asebogenin | C16H16O5 | 288.10 | Flavonoids |
| Formononetin | C16H12O4 | 268.07 | Isoflavonoids |
| calabricoside A | C32H38O20 | 742.20 | Flavonoids |
| tricin 7-O-(-O-malonyl)--D-glucopyranoside | C26H26O15 | 578.13 | Flavonoids |
| Acacetin | C16H12O5 | 284.07 | Flavonoids |
| chrysoobtusin | C19H18O7 | 358.11 | Polycyclic aromatic polyketides |
| 1-AMINO-4-BENZAMIDOANTHRAQUINONE | C21H14N2O3 | 342.10 | Polycyclic aromatic polyketides |
| Quercetin 3-rutinoside-7-glucuronide | C33H38O22 | 786.19 | Flavonoids |
| Kaempferol 3-O-glucosyl-rhamnosyl-galactoside | C33H40O20 | 756.21 | Flavonoids |
| Syringetin-3-glucoside | C23H24O13 | 508.12 | Flavonoids |
| quercetin 3-O-(3-O-p-coumaroyl, 6-O-feruloyl)-glucoside | C40H34O17 | 786.18 | Flavonoids |
| 1-Formyl-4-hydroxyanthraquinone | C15H8O4 | 252.04 | Polycyclic aromatic polyketides |
| quercetin 3-O-gentiobioside-7-O-rhamnoside | C33H40O21 | 772.20 | Flavonoids |
| kaempferol 3-O-gentiobioside-7-O-rhamnoside | C33H40O20 | 756.21 | Flavonoids |
| Quercetin 3-glucosyl-()-rhamnosyl-()-galactoside | C33H40O21 | 772.21 | Flavonoids |
| Quercetin 3-rutinoside-7,-diglucoside | C39H50O26 | 934.26 | Flavonoids |
| Ononin | C22H22O9 | 430.13 | Isoflavonoids |
| Tetramethylscutellarein | C19H18O6 | 342.11 | Flavonoids |
| Carnosol | C20H26O4 | 330.18 | Diterpenoids |
| KAEMPFEROL-3-O-(-TRANS-P-COUMAROYL--GLUCOSYL)RHAMNOSIDE | C36H36O17 | 740.20 | Flavonoids |
| aromadendrin | C15H12O6 | 288.06 | Flavonoids |
| Biochanin A | C16H12O5 | 284.07 | Isoflavonoids |
| Glycitein | C16H12O5 | 284.07 | Flavonoids |
| (-)-L-Chicoric acid | C22H18O12 | 474.08 | Phenylpropanoids (C6–C3) |
| Scutellarin | C21H18O12 | 462.08 | Flavonoids |
| Ellagic acid | C14H6O8 | 302.01 | Phenolic acids (C6–C1) |
| Myricetin | C15H10O8 | 318.04 | Flavonoids |
| Luteolinidin | C15H11O5 | 271.06 | Flavonoids |
| (E)-Ferulic acid | C10H10O4 | 194.06 | Phenylpropanoids (C6–C3) |
| quercetin 3-O-sophoroside-7-O-rhamnoside | C33H40O21 | 772.21 | Flavonoids |
| Glycitin | C22H22O10 | 446.12 | Isoflavonoids |
| 4-Heptyloxyphenol | C13H20O2 | 208.15 | Aromatic polyketides |
| -HYDROXYFLAVONE | C15H10O3 | 238.06 | Flavonoids |
| ,-O-diacetyloninin | C26H26O11 | 514.15 | Isoflavonoids |
| ANTRAQUINONE DERIVATIVE | C14H8O4 | 240.04 | Polycyclic aromatic polyketides |
| 6-(-D-Glucopyranuronosyloxy)-5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-8-yl -D-glucopyranosiduronic acid | C27H26O19 | 654.11 | Flavonoids |
| 2-(3,4-Dihydroxyphenyl)-7- (-D-glucopyranosyloxy)-5-hydroxy-4-oxo-4H-chromen-3-yl 6-deoxy--L-mannopyranosyl-()-[6-deoxy--L-mannopyranosyl-()]--D-galactopyranoside | C39H50O25 | 918.26 | Flavonoids |
| 5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl) -4-oxo-4H-chromen-3-yl -D-glucopyranosyl-()-6-deoxy--L-mannopyranosyl-()--D-glucopyranoside | C34H42O21 | 786.22 | Flavonoids |
| Kaempferol 7-methyl ether 3- (6- (E) -3,5- dimethoxy-4-hydroxycinnamoylglucosyl) - () - [ rhamnosyl- () -glucoside ] | C45H52O24 | 976.29 | Flavonoids |

Table A9.
Rhubarb—Hierarchical classification of identified metabolites with model accuracy expressed as percentage.
Table A9.
Rhubarb—Hierarchical classification of identified metabolites with model accuracy expressed as percentage.
| Component | Pathway | Acc.% | Superclass | Acc.% | Class | Acc.% |
|---|---|---|---|---|---|---|
| Emodin | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Piceatannol | Shikimates and Phenylpropanoids | 99.80 | Stilbenoids | 99.90 | Monomeric stilbenes | 98.70 |
| Rhein | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Daidzein | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.90 |
| Catechin | Shikimates and Phenylpropanoids | 99.80 | Flavonoids | 99.90 | Flavan-3-ols | 99.80 |
| Rhein-8-glucoside | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Genistein | Shikimates and Phenylpropanoids | 99.80 | Isoflavonoids | 99.90 | Isoflavones | 99.80 |
| (2Z)-6-hydroxy-2-[(4-hydroxy-3-methoxyphenyl)methylidene]-2,3-dihydro-1-benzofuran-3-one | Shikimates and Phenylpropanoids | 99.80 | Flavonoids | 99.90 | Aurones | 99.90 |
| 3-O-Methylquercetintetraacetate | Shikimates and Phenylpropanoids | 99.30 | Flavonoids | 99.90 | Flavonols | 99.90 |
| (+)-Catechin 3-O-gallate | Shikimates and Phenylpropanoids | 99.60 | Flavonoids | 99.90 | Flavan-3-ols | 99.90 |
| Torachrysone 8-O--D-glucoside | Polyketides | 99.00 | Naphthalenes | 99.80 | Naphthalenes and derivatives | 99.90 |
| spectaflavoside A | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 99.90 | Flavonols | 100.00 |
| 1,4-Dihydroxy-5,8-bis(p-toluidino)anthraquinone | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 93.40 |
| 5-Acetonyl-7-hydroxy-2-methylchromone | Polyketides | 99.60 | Chromanes | 99.90 | Chromones | 99.90 |
| aloesin | Polyketides | 99.80 | Chromanes | 99.90 | Chromones | 99.90 |
| Protocatechuic aldehyde | Terpenoids | 52.00 | Phenolic acids (C6–C1) | 83.80 | Simple phenolic acids | 45.00 |
| Kaempferol | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Limocitrol 3- [ -L-arabinopyranosyl- () [ galactosyl- () ] -galactoside ] | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Irilone | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.70 |
| Afzelechin | Shikimates and Phenylpropanoids | 99.80 | Flavonoids | 99.90 | Flavan-3-ols | 99.80 |
| Cassialoin | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Isoliquiritigenin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Chalcones | 100.00 |
| Vitexin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Eriodictyol | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.70 |
| ,-DIMETHYL EPIGALLOCATECHIN GALLATE | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavan-3-ols | 99.70 |
| Resveratrol | Shikimates and Phenylpropanoids | 99.80 | Stilbenoids | 99.90 | Monomeric stilbenes | 98.00 |
| Chrysin | Shikimates and Phenylpropanoids | 99.40 | Flavonoids | 99.90 | Flavones | 100.00 |
| Resveratrol 3-O-glucoside | Shikimates and Phenylpropanoids | 99.90 | Stilbenoids | 99.90 | Monomeric stilbenes | 98.90 |
| Kaempferol 3-O-glucosyl-rhamnosyl-galactoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| 1,2,4-Trihydroxyanthraquinone | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Gallic acid | Shikimates and Phenylpropanoids | 97.90 | Phenolic acids (C6–C1) | 97.90 | Simple phenolic acids | 97.00 |
| aescuflavoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Hispidulin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| ,,7-Trihydroxyflavanone | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.80 |
| Apigenin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Pinocembrin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.70 |
| HEXAMETHYL QUERCETAGETIN | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| 3-Hydroxyflavone | Shikimates and Phenylpropanoids | 99.80 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Primuletin | Shikimates and Phenylpropanoids | 98.60 | Flavonoids | 99.90 | Flavones | 99.90 |
| Quercetin 3-sambubioside--glucoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Phloretin | Shikimates and Phenylpropanoids | 99.10 | Flavonoids | 99.90 | Chalcones | 99.90 |
| 3-hydroxyflavanone | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.90 |
| Taxifolin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Dihydroflavonols | 99.90 |
| Quercetin 3-O-xylosyl-rutinoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Asebogenin | Shikimates and Phenylpropanoids | 98.30 | Flavonoids | 99.90 | Chalcones | 99.90 |
| Formononetin | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.90 |
| calabricoside A | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| tricin 7-O-(-O-malonyl)--D-glucopyranoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Acacetin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| chrysoobtusin | Polyketides | 99.60 | Polycyclic aromatic polyketides | 99.80 | Anthraquinones and anthrones | 98.30 |
| 1-AMINO-4-BENZAMIDO ANTHRAQUINONE | Polyketides | 99.80 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Quercetin 3-rutinoside-7-glucuronide | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Kaempferol 3-O-glucosyl-rhamnosyl-galactoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Syringetin-3-glucoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| quercetin 3-O-(3-O-p-coumaroyl, 6-O-feruloyl)-glucoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 100.00 | Flavonols | 99.90 |
| 1-Formyl-4-hydroxyanthraquinone | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| quercetin 3-O-gentiobioside-7-O-rhamnoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| kaempferol 3-O-gentiobioside-7-O-rhamnoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Quercetin 3-glucosyl-()-rhamnosyl-()-galactoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Quercetin 3-rutinoside-7,-diglucoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Ononin | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.90 |
| Tetramethylscutellarein | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Carnosol | Terpenoids | 99.90 | Diterpenoids | 99.90 | Abietane diterpenoids | 99.90 |
| KAEMPFEROL-3-O-(-TRANS-P-COUMAROYL--GLUCOSYL)RHAMNOSIDE | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| aromadendrin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Dihydroflavonols | 99.90 |
| Biochanin A | Shikimates and Phenylpropanoids | 99.70 | Isoflavonoids | 99.90 | Isoflavones | 99.80 |
| Glycitein | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 100.00 | Flavones | 99.90 |
| (-)-L-Chicoric acid | Shikimates and Phenylpropanoids | 97.30 | Phenylpropanoids (C6–C3) | 98.90 | Cinnamic acids and derivatives | 99.90 |
| Scutellarin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Ellagic acid | Shikimates and Phenylpropanoids | 99.90 | Phenolic acids (C6–C1) | 99.90 | Gallotannins | 99.90 |
| Myricetin | Shikimates and Phenylpropanoids | 99.80 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Luteolinidin | Shikimates and Phenylpropanoids | 99.80 | Flavonoids | 99.90 | Anthocyanidins | 99.10 |
| (E)-Ferulic acid | Shikimates and Phenylpropanoids | 99.60 | Phenylpropanoids (C6–C3) | 99.10 | Cinnamic acids and derivatives | 99.20 |
| quercetin 3-O-sophoroside-7-O-rhamnoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Glycitin | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.90 |
| 4-Heptyloxyphenol | Shikimates and Phenylpropanoids | 24.30 | Phenolic acids (C6–C1) | 2.90 | Hydrocarbons | 3.00 |
| -HYDROXYFLAVONE | Polyketides | 93.30 | Monoterpenoids | 13.10 | Oblogolides | 16.70 |
| ,-O-diacetyloninin | – | – | – | |||
| ANTRAQUINONE DERIVATIVE | – | – | – | |||
| 6-(-D-Glucopyranuronosyloxy)-5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-8-yl -D-glucopyranosiduronic acid | – | – | – | |||
| 2-(3,4-Dihydroxyphenyl)-7-(-D-glucopyranosyloxy)-5-hydroxy-4-oxo-4H-chromen-3-yl 6-deoxy--L- mannopyranosyl-()-[6-deoxy--L-mannopyranosyl-()]--D-galactopyranoside | – | – | – | |||
| 5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-4-oxo-4H-chromen-3-yl -D-glucopyranosyl-()-6-deoxy--L-mannopyranosyl-()--D-glucopyranoside | – | – | – | |||
| Kaempferol 7-methyl ether 3- (6- (E) -3,5-dimethoxy-4-hydroxycinnamoylglucosyl) - () - [ rhamnosyl- () -glucoside ] | – | – | – |

Table A10.
Discordant and unassigned superclass assignments in Rhubarb (8/83 compounds; overall concordance 90.0%).
Table A10.
Discordant and unassigned superclass assignments in Rhubarb (8/83 compounds; overall concordance 90.0%).
| Component | Validated Superclass | Predicted Superclass | Acc.% |
|---|---|---|---|
| 4-Heptyloxyphenol | Aromatic polyketides | Phenolic acids (C6–C1) | 2.9 |
| -HYDROXYFLAVONE | Flavonoids | Monoterpenoids | 13.1 |
| ,-O-diacetyloninin | Flavonoids | – | |
| ANTRAQUINONE DERIVATIVE | Polycyclic aromatic polyketides | – | |
| 6-(-D-Glucopyranuronosyloxy)-5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-8-yl -D-glucopyranosiduronic acid | Flavonoids | – | |
| 2-(3,4-Dihydroxyphenyl)-7-(-D-glucopyranosyloxy)-5-hydroxy-4-oxo-4H-chromen-3-yl 6-deoxy--L-mannopyranosyl-()-[6-deoxy--L-mannopyranosyl-()]--D-galactopyranoside | Flavonoids | – | |
| 5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-4-oxo-4H-chromen-3-yl -D-glucopyranosyl-()-6-deoxy--L-mannopyranosyl-()--D-glucopyranoside | Flavonoids | – | |
| Kaempferol 7-methyl ether 3- (6- (E) -3,5-dimethoxy-4-hydroxycinnamoylglucosyl) - () - [ rhamnosyl- () -glucoside ] | Flavonoids | – |
The discordant and unassigned cases in Rhubarb mainly involve borderline or incompletely represented molecular structures. Misclassifications were associated with features that blur the boundaries between distinct chemical families, such as partial overlap between aromatic oxygenated systems and terpenoid-like motifs. The unassigned compounds correspond to highly glycosylated flavonoids and anthraquinone derivatives, for which complex substitution patterns extend beyond the canonical chemical space captured by the model. These instances highlight the model’s limitations in handling extensively conjugated or heavily substituted scaffolds, but were readily resolved through expert review and did not affect the conclusions at the superclass level.
Appendix A.5. Senna
For Senna, Table A11 lists all identified metabolites with validated superclass assignments, while Table A12 provides the hierarchical classification with model-derived confidence scores. Table A13 highlights the few discordant cases, which are discussed further below. For full-precision m/z values (reported to four decimal places) together with the corresponding retention times (RT) and mass errors (, ppm), readers are referred to Supplementary Table S1.

Table A11.
Senna—Identified metabolites with validated Superclass. Molecular weights are rounded to two decimals.
Table A11.
Senna—Identified metabolites with validated Superclass. Molecular weights are rounded to two decimals.
| Component | Formula | Molecular Weight | Superclass |
|---|---|---|---|
| vicenin 2 | C27H30O15 | 594.16 | Flavonoids |
| Rhein | C15H8O6 | 284.03 | Polycyclic aromatic polyketides |
| Guaethol | C8H10O2 | 138.07 | Phenylpropanoids (C6–C3) |
| 1,2,4-Trihydroxyanthraquinone | C14H8O5 | 256.04 | Polycyclic aromatic polyketides |
| Rhein-8-glucoside | C21H18O11 | 446.08 | Polycyclic aromatic polyketides |
| 11-o-Galloylbergenin | C21H20O13 | 480.09 | Coumarins |
| Emodic acid | C15H8O7 | 300.03 | Polycyclic aromatic polyketides |
| demethylwedelolactone | C15H8O7 | 300.03 | Isoflavonoids |
| Sennoside A | C42H38O20 | 862.20 | Isoflavonoids |
| Sennoside B | C42H38O20 | 862.20 | Isoflavonoids |
| Sennidin B | C30H18O10 | 538.10 | Isoflavonoids |
| Aloe emodin | C15H10O5 | 270.05 | Isoflavonoids |
| Sennidin A | C30H18O10 | 538.10 | Isoflavonoids |
| Luteolin | C15H10O6 | 286.05 | Flavonoids |
| Eugenol | C10H12O2 | 164.08 | Phenylpropanoids (C6–C3) |
| creosol | C8H10O2 | 138.07 | Benzenoids |
| Myricetin | C15H10O8 | 318.04 | Flavonoids |
| Coumesterol | C15H8O5 | 268.04 | Isoflavonoids |
| Prodelphinidin T1 | C45H38O20 | 898.20 | Isoflavonoids |
| Quercitrin | C21H20O11 | 448.10 | Isoflavonoids |
| Kaempferol | C15H10O6 | 286.05 | Isoflavonoids |
| Rutin | C27H30O16 | 610.15 | Isoflavonoids |
| (E)-Ferulic acid | C10H10O4 | 194.06 | Isoflavonoids |
| Phloretin | C15H14O5 | 274.08 | Flavonoids |
| Eriodictyol | C15H12O6 | 288.06 | Flavonoids |
| Sinapinic acid | C11H12O5 | 224.07 | Phenylpropanoids (C6–C3) |
| malonyldaidzin | C24H22O12 | 502.11 | Isoflavonoids |
| Cinnamic acid | C9H8O2 | 148.05 | Isoflavonoids |
| Daidzein | C15H10O4 | 254.06 | Isoflavonoids |
| Cyanidin 3-O-sambubioside 5-O-glucoside | C32H39O20 | 743.20 | Isoflavonoids |
| Carnosol | C20H26O4 | 330.18 | Isoflavonoids |
| Irilone | C16H10O6 | 298.05 | Isoflavonoids |
| olmelin | C16H12O5 | 284.07 | Isoflavonoids |
| (±)-Naringenin | C15H12O5 | 272.07 | Isoflavonoids |
| Peonidin 3-O-glucoside | C22H23O11 | 463.12 | Isoflavonoids |
| Secoisolariciresinol | C20H26O6 | 362.17 | Isoflavonoids |
| 3,4-Dicaffeoylquinic acid | C25H24O12 | 516.13 | Isoflavonoids |
| Sennoside C | C42H40O19 | 848.22 | Polycyclic aromatic polyketides |
| Tectochrysin | C16H12O4 | 268.07 | Flavonoids |
| Xanthorin | C16H12O6 | 300.06 | Polycyclic aromatic polyketides |
| Miquelianin | C21H18O13 | 478.08 | Flavonoids |
| Theaflavine | C29H24O12 | 564.13 | Flavonoids |
| Rubrofusarin | C15H12O5 | 272.07 | Naphthalenes |
| Procyanidin C1 | C45H38O18 | 866.21 | Flavonoids |
| Isorhamnetin 3-glucoside | C22H22O12 | 478.11 | Flavonoids |
| (E)-p-coumaric acid | C9H8O3 | 164.05 | Phenylpropanoids (C6–C3) |
| Hispidulin | C16H12O6 | 300.06 | Flavonoids |
| 4-Heptyloxyphenol | C13H20O2 | 208.15 | Aromatic polyketides |
| ,-BISEPIGALLOCATECHIN DIGALLATE | C44H34O22 | 914.15 | Flavonoids |
| Diosmetin-O-glucoside | C22H22O11 | 462.12 | Flavonoids |
| Quercetin 3-O-(2,6-di-O-rhamnosyl) galactoside | C33H40O20 | 756.21 | Isoflavonoids |

Table A12.
Senna—Hierarchical classification of identified metabolites with model accuracy expressed as percentage.
Table A12.
Senna—Hierarchical classification of identified metabolites with model accuracy expressed as percentage.
| Component | Pathway | Acc.% | Superclass | Acc.% | Class | Acc.% |
|---|---|---|---|---|---|---|
| vicenin 2 | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Rhein | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Guaethol | Shikimates and Phenylpropanoids | 66.30 | Phenylpropanoids (C6–C3) | 30.90 | Cinnamic acids and derivatives | 3.06 |
| 1,2,4-Trihydroxyanthraquinone | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Rhein-8-glucoside | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| 11-o-Galloylbergenin | Shikimates and Phenylpropanoids | 96.60 | Coumarins | 96.40 | Isocoumarins | 95.10 |
| Emodic acid | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| demethylwedelolactone | Shikimates and Phenylpropanoids | 99.80 | Isoflavonoids | 98.70 | Coumestan | 99.90 |
| Sennoside A | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Sennoside B | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Sennidin B | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Aloe emodin | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Sennidin A | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Luteolin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Eugenol | Shikimates and Phenylpropanoids | 99.90 | Phenylpropanoids (C6–C3) | 98.80 | Cinnamic acids and derivatives | 96.20 |
| creosol | Shikimates and Phenylpropanoids | 51.10 | Phenylpropanoids (C6–C3) | 11.00 | Simple phenolic acids | 22.90 |
| Myricetin | Shikimates and Phenylpropanoids | 99.80 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Coumesterol | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 99.80 | Coumestan | 99.80 |
| Prodelphinidin T1 | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 99.90 | Proanthocyanins | 99.90 |
| Quercitrin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Kaempferol | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Rutin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| (E)-Ferulic acid | Shikimates and Phenylpropanoids | 99.60 | Phenylpropanoids (C6–C3) | 99.10 | Cinnamic acids and derivatives | 99.20 |
| Phloretin | Shikimates and Phenylpropanoids | 99.10 | Flavonoids | 99.90 | Chalcones | 99.90 |
| Eriodictyol | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavanones | 99.70 |
| Sinapinic acid | Shikimates and Phenylpropanoids | 99.70 | Phenylpropanoids (C6–C3) | 98.00 | Cinnamic acids and derivatives | 97.80 |
| malonyldaidzin | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.90 |
| Cinnamic acid | Shikimates and Phenylpropanoids | 99.00 | Phenylpropanoids (C6–C3) | 95.20 | Cinnamic acids and derivatives | 91.70 |
| Daidzein | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.90 |
| Cyanidin 3-O-sambubioside 5-O-glucoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Anthocyanidins | 99.90 |
| Carnosol | Terpenoids | 99.90 | Diterpenoids | 99.90 | Abietane diterpenoids | 99.90 |
| Irilone | Shikimates and Phenylpropanoids | 99.90 | Isoflavonoids | 99.90 | Isoflavones | 99.70 |
| olmelin | Shikimates and Phenylpropanoids | 99.70 | Isoflavonoids | 99.90 | Isoflavones | 99.80 |
| (±)-Naringenin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| Peonidin 3-O-glucoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Anthocyanidins | 99.90 |
| Secoisolariciresinol | Shikimates and Phenylpropanoids | 99.90 | Lignans | 100.00 | Dibenzylbutane lignans | 99.90 |
| 3,4-Dicaffeoylquinic acid | Shikimates and Phenylpropanoids | 99.30 | Phenylpropanoids (C6–C3) | 99.40 | Cinnamic acids and derivatives | 99.90 |
| Sennoside C | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Tectochrysin | Shikimates and Phenylpropanoids | 99.30 | Flavonoids | 99.90 | Flavones | 99.90 |
| Xanthorin | Polyketides | 99.90 | Polycyclic aromatic polyketides | 99.90 | Anthraquinones and anthrones | 99.90 |
| Miquelianin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| Theaflavine | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavan-3-ols | 99.90 |
| Rubrofusarin | Polyketides | 99.60 | Naphthalenes | 77.90 | Naphthalenes and derivatives | 98.30 |
| Procyanidin C1 | Shikimates and Phenylpropanoids | 100.00 | Flavonoids | 99.90 | Proanthocyanins | 99.90 |
| Isorhamnetin 3-glucoside | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavonols | 99.90 |
| (E)-p-coumaric acid | Shikimates and Phenylpropanoids | 99.00 | Phenylpropanoids (C6–C3) | 95.10 | Cinnamic acids and derivatives | 97.50 |
| Hispidulin | Shikimates and Phenylpropanoids | 99.90 | Flavonoids | 99.90 | Flavones | 99.90 |
| 4-Heptyloxyphenol | Shikimates and Phenylpropanoids | 24.30 | Phenolic acids (C6–C1) | 2.90 | Hydrocarbons | 3.10 |
| ,-BISEPIGALLOCATECHIN DIGALLATE | Shikimates and Phenylpropanoids | 99.90 | Phenolic acids (C6–C1) | 99.70 | Flavan-3-ols | 94.60 |
| Diosmetin-O-glucoside | Shikimates and Phenylpropanoids | 99.70 | Coumarins | 91.40 | Furocoumarins | 21.60 |
| Quercetin 3-O-(2,6-di-O-rhamnosyl) galactoside | – | – | – |

Table A13.
Discordant and unassigned superclass assignments in Senna (4/51 compounds; overall concordance 92.3%).
Table A13.
Discordant and unassigned superclass assignments in Senna (4/51 compounds; overall concordance 92.3%).
| Component | Validated Superclass | Predicted Superclass | Acc.% |
|---|---|---|---|
| 4-Heptyloxyphenol | Aromatic polyketides | Phenolic acids (C6–C1) | 2.9 |
| ,-BISEPIGALLOCATECHIN DIGALLATE | Flavonoids | Phenolic acids (C6–C1) | 99.7 |
| Diosmetin-O-glucoside | Flavonoids | Coumarins | 91.4 |
| Quercetin 3-O-(2,6-di-O-rhamnosyl) galactoside | Isoflavonoids | – |
The discordant and unassigned cases in Senna mainly concern polyphenolic structures with overlapping functional motifs. The misclassification of large flavonoid derivatives such as ,-bisepigallocatechin digallate and diosmetin-O-glucoside reflects partial similarity with phenolic or coumarin-like scaffolds that share conjugated aromatic systems and hydroxylation patterns. The unassigned compounds correspond to highly glycosylated anthraquinone and isoflavonoid derivatives, for which the model could not confidently assign a superclass due to the structural complexity and rare substitution patterns. Overall, these few discrepancies do not affect the reliability of the classification at the superclass level, confirming the robustness of the annotation workflow.
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Gonfa, Y.H.; Beshah, F.; Tadesse, M.G.; Bachheti, A.; Bachheti, R.K. Phytochemical Investigation and Potential Pharmacologically Active Compounds of Rumex Nepalensis: An Appraisal. J. Basic Appl. Sci. 2021, 10, 18. [Google Scholar] [CrossRef]
- Loschi, F.; Faggian, M.; Sut, S.; Ferrarese, I.; Maccari, E.; Peron, G.; Dall’Acqua, S. Development of an LC–DAD–MS-Based method for the analysis of hydroxyanthracene derivatives in food supplements and plant materials. Molecules 2022, 27, 1932. [Google Scholar] [CrossRef]
- Pandith, S.A.; Hussain, A.; Bhat, W.W.; Dhar, N.; Qazi, A.K.; Rana, S.; Razdan, S.; Wani, T.A.; Shah, M.A.; Bedi, Y.; et al. Evaluation of anthraquinones from Himalayan rhubarb (Rheum emodi Wall. ex Meissn.) as antiproliferative agents. S. Afr. J. Bot. 2014, 95, 1–8. [Google Scholar] [CrossRef]
- Huang, Q.; Lu, G.; Shen, H.M.; Chung, M.C.; Ong, C.N. Anti-cancer properties of anthraquinones from rhubarb. Med. Res. Rev. 2007, 27, 609–630. [Google Scholar] [CrossRef]
- Malik, E.M.; Müller, C.E. Anthraquinones as pharmacological tools and drugs. Med. Res. Rev. 2016, 36, 705–748. [Google Scholar] [CrossRef] [PubMed]
- Chien, S.C.; Wu, Y.C.; Chen, Z.W.; Yang, W.C. Naturally Occurring Anthraquinones: Chemistry and Therapeutic Potential in Autoimmune Diabetes. Evid.-Based Complement. Altern. Med. 2015, 2015, 357357. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Mei, N. Aloe vera: A Review of Toxicity and Adverse Clinical Effects. J. Environ. Sci. Health C 2016, 34, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Sonnenberg, A.; Müller, A.D. Constipation and Cathartics as Risk Factors of Colorectal Cancer: A Meta-Analysis. Pharmacology 1993, 47, 224–233. [Google Scholar] [CrossRef]
- Nusko, G.; Schneider, B.; Müller, G.; Kusche, J.; Hahn, E.G. Retrospective Study on Laxative Use and Melanosis Coli as Risk Factors for Colorectal Neoplasma. Pharmacology 1993, 47, 234–241. [Google Scholar] [CrossRef]
- Siegers, C.P.; Siemers, J.; Baretton, G. Sennosides and Aloin Do Not Promote Dimethylhydrazine-Induced Colorectal Tumors in Mice. Pharmacology 1993, 47, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, N.; Bettiol, A.; Crescioli, G.; Maggini, V.; Gallo, E.; Sivelli, F.; Sofi, F.; Gensini, G.F.; Vannacci, A.; Firenzuoli, F. Association between Anthraquinone Laxatives and Colorectal Cancer: Protocol for a Systematic Review and Meta-Analysis. Syst. Rev. 2020, 9, 19. [Google Scholar] [CrossRef]
- EFSA Panel on Food Additives and Nutrient Sources added to Food (ANS); Younes, M.; Aggett, P.; Aguilar, F.; Crebelli, R.; Filipič, M.; Frutos, M.J.; Galtier, P.; Gott, D.; Gundert-Remy, U.; et al. Safety of Hydroxyanthracene Derivatives for Use in Food. EFSA J. 2018, 16, e05090. [Google Scholar] [CrossRef]
- Commission Regulation (EU) 2021/468 of 18 March 2021. 2021. Available online: https://eur-lex.europa.eu/eli/reg/2021/468/oj (accessed on 7 October 2021).
- Tinti, L.; Cicaloni, V.; Nezi, P.; Isoldi, G.; Etiope, P.; Barlozzini, B.; Pecorari, R.; Salvini, L. Hydroxyanthracene derivates cytotoxicity: A differential evaluation between single molecule and whole plant extract. Front. Plant Sci. 2023, 14, 1166075. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, H.; Qu, L.; He, Y.; Routledge, M.N.; Gong, Y.Y.; Qiao, B. Identification of rhein as the metabolite responsible for toxicity of rhubarb anthraquinones. Food Chem. 2020, 331, 127282. [Google Scholar] [CrossRef]
- Coates, P.M.; Betz, J.M.; Blackman, M.R.; Cragg, G.M.; Levine, M.; Moss, J.; White, J.D. Encyclopedia of Dietary Supplements, Online edition; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Migues, V.H.; Mauricio, J.; Gomes, A.F.; David, J.P. Determination of anthraquinones in Rhamnus purshiana using high-performance liquid chromatography coupled to diode array detector and simple ultraviolet spectroscopic analysis. J. Sep. Sci. 2022, 45, 2588–2596. [Google Scholar] [CrossRef]
- Demarque, D.P.; Pinho, D.R.; Callejon, D.R.; de Oliveira, G.G.; Silva, D.B.; Carollo, C.A.; Lopes, N.P. New cascarosides from Rhamnus purshiana and fragmentation studies of the class by ion trap mass spectrometry. Rapid Commun. Mass Spectrom. 2017, 31, 1185–1193. [Google Scholar] [CrossRef]
- Van den Berg, A.J.J.; Labadie, R.P. Anthraquinones, Anthrones and Dianthrones in Callus Cultures of Rhamnus frangula and Rhamnus purshiana. Planta Medica 1984, 50, 434–437. [Google Scholar] [CrossRef]
- Kremer, D.; Kosalec, I.; Locatelli, M.; Epifano, F.; Genovese, S.; Carlucci, G.; Končić, M.Z. Anthraquinone profiles, antioxidant and antimicrobial properties of Frangula rupestris (Scop.) Schur and Frangula alnus Mill. Bark. Food Chem. 2012, 131, 1174–1180. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, P.; Xu, L.; He, C.; Peng, Y.; Xiao, P. Evaluation of the content variation of anthraquinone glycosides in rhubarb by UPLC-PDA. Chem. Cent. J. 2013, 7, 170. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Yao, W.; Ji, W.; Wei, J.; Peng, S. Qualitative and quantitative analysis of anthraquinones in rhubarbs by high-performance liquid chromatography with diode array detector and mass spectrometry. Food Chem. 2013, 141, 1640–1648. [Google Scholar] [CrossRef] [PubMed]
- Aichner, D.; Ganzera, M. Analysis of anthraquinones in rhubarb (Rheum palmatum and Rheum officinale) by supercritical fluid chromatography. Talanta 2015, 144, 1239–1244. [Google Scholar] [CrossRef] [PubMed]
- Al-Marzoqi, A.H.; Hadi, M.Y.; Hameed, I.H. Determination of metabolites products by Cassia angustifolia and evaluate antimicobial activity. J. Pharmacogn. Phytother. 2016, 8, 25–48. [Google Scholar]
- Farag, M.A.; El Senousy, A.S.; El-Ahmady, S.H.; Porzel, A.; Wessjohann, L.A. Comparative metabolome-based classification of Senna drugs: A prospect for phyto-equivalency of its different commercial products. Metabolomics 2019, 15, 80. [Google Scholar] [CrossRef] [PubMed]
- Cunha, L.F.; Costa, C.M.; Barroso, P.R.; Kato, K.C.; Oliveira, F.d.; Mendonça Filho, C.V.; Grael, C.F.F.; Gregório, L.E.; Campos, F.F.; Oliveira, P.M.d.; et al. Phytochemical screening and biological assays of ethanolic leaf extract of Senna rugosa. Rodriguésia 2020, 71, e00912019. [Google Scholar] [CrossRef]
- de O. Maia, I.R.; Trevisan, M.T.S.; de V. Silva, M.G.; Klika, K.D.; de Brito, E.; Silva, L.M.A.e.; Pinto, F.d.C.L.; Breuer, A.; Owen, R.W. Characterization and quantitation of polyphenolic compounds in Senna splendida from the northeast of Brazil. Nat. Prod. Commun. 2018, 13, 1934578X1801300614. [Google Scholar] [CrossRef]
- Kim, H.W.; Wang, M.; Leber, C.A.; Nothias, L.F.; Reher, R.; Kang, K.B.; Van Der Hooft, J.J.; Dorrestein, P.C.; Gerwick, W.H.; Cottrell, G.W. NPClassifier: A deep neural network-based structural classification tool for natural products. J. Nat. Prod. 2021, 84, 2795–2807. [Google Scholar] [CrossRef]
- Prete, A.L.; Corradini, B.T.; Costanti, F.; Scarselli, F.; Bianchini, M. Leveraging molecular graphs for natural product classification. Comput. Struct. Biotechnol. J. 2025, 27, 3837–3884. [Google Scholar] [CrossRef]
- Q/1765/24; Ricerca, Sviluppo, Progettazione ed Esecuzione di Servizi (Prove Analitiche) Basate su Spettrometria di Massa e Test In Vitro e Relative Elaborazioni di Dati Bioinformatici [Research, Development, Design and Execution of Services (Analytical Tests) Based on Mass Spectrometry and in Vitro Tests and Related Bioinformatics Data Processing], 2024-03-07. Centro Certificazione Qualità S.r.l.: San Felice a Cancello, Italy, 2024.
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, C.Z.; Sawadogo, W.R.; Bian, Z.X.; Yuan, C.S. Herbal medicines for constipation and phytochemical comparison of active components. Am. J. Chin. Med. 2022, 50, 723–732. [Google Scholar] [CrossRef]
- Chen, G.L.; Li, X.; Saleri, F.D.; Guo, M.Q. Analysis of flavonoids in Rhamnus davurica and its antiproliferative activities. Molecules 2016, 21, 1275. [Google Scholar] [CrossRef]
- Chen, G.-L.; Munyao Mutie, F.; Xu, Y.-B.; Saleri, F.D.; Hu, G.-W.; Guo, M.-Q. Antioxidant, anti-inflammatory activities and polyphenol profile of Rhamnus prinoides. Pharmaceuticals 2020, 13, 55. [Google Scholar] [CrossRef]
- Elansary, H.O.; Szopa, A.; Kubica, P.; Ekiert, H.; Al-Mana, F.A.; El-Shafei, A.A. Polyphenols of Frangula alnus and Peganum harmala leaves and associated biological activities. Plants 2020, 9, 1086. [Google Scholar] [CrossRef] [PubMed]
- Omer, H.A.A.; Caprioli, G.; Abouelenein, D.; Mustafa, A.M.; Uba, A.I.; Ak, G.; Ozturk, R.B.; Zengin, G.; Yagi, S. Phenolic profile, antioxidant and enzyme inhibitory activities of leaves from two Cassia and two Senna species. Molecules 2022, 27, 5590. [Google Scholar] [CrossRef]
- Ye, M.; Han, J.; Chen, H.; Zheng, J.; Guo, D. Analysis of phenolic compounds in rhubarbs using liquid chromatography coupled with electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2007, 18, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Nie, X.; Yang, L.; Wang, L.; Li, J.; Zhou, G. Non-target metabolomics revealed the differences between Rh. tanguticum plants growing under canopy and open habitats. BMC Plant Biol. 2021, 21, 119. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, S.; Del Bo’, C.; Marino, M.; Gargari, G.; Cherubini, A.; Andrés-Lacueva, C.; Hidalgo-Liberona, N.; Peron, G.; González-Dominguez, R.; Kroon, P.; et al. Polyphenols and intestinal permeability: Rationale and future perspectives. J. Agric. Food Chem. 2019, 68, 1816–1829. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).