Abstract
MicroRNAs (miRNAs) are small noncoding RNAs approximately 22 nucleotides in length. Their main function is to regulate gene expression at the posttranscriptional level by inhibiting the translation of messenger RNAs (mRNAs). miRNAs originate in the cell nucleus from specific genes, where they can perform their function. However, they can also be found in serum, plasma, or other body fluids travelling within vesicles called exosomes and/or bound to proteins or other particles such as lipoproteins. miRNAs can form complexes outside the cell where they are synthesized, mediating paracrine and endocrine communication between different tissues. In this way, they can modulate the gene expression and function of distal cells. It is known that the expression of miRNAs can be affected by multiple factors, such as the nutritional or pathological state of the individual, or even in conditions such as obesity, insulin resistance, or after any dietary intervention. In this review, we will analyse miRNAs whose expression and circulation are affected in conditions of obesity and insulin resistance, as well as the changes generated after a dietary intervention, with the purpose of identifying new possible biomarkers of early response to nutritional treatment in these conditions.
1. Introduction
MicroRNAs (miRNAs) are small RNAs of approximately 22 nucleotides in length that regulate gene expression at the posttranscriptional level by repressing target mRNAs. These small RNAs are responsible for the modulation of translation of most cellular proteins that were destined to be synthesized from target mRNAs, suggesting that miRNAs play an important role in the pathophysiology of many diseases [1]. The distribution and expression patterns of miRNAs are regulated by transcriptional and posttranscriptional mechanisms within the cell. Thus, miRNA profiling can be employed as a cellular identity due to the specificity of their expression in different tissues and conditions [2].
MicroRNAs can perform their function in the same cell where they are synthesized, or they can be taken to the extracellular space to travel in circulation, which constitutes one of the most recently studied cell-cell communication mechanisms in many physiological or pathological conditions.
Several studies have shown an association between the expression levels of different miRNAs and various pathologies, including obesity, type-2 diabetes, cardiovascular diseases, neurodegenerative disorders and cancer. It is also known that other factors, such as diet, exercise, or nutritional status, can modulate their expression [3].
A better understanding of how miRNA expression can be regulated by various pathologies and/or factors such as nutritional status and diet could contribute to the development of new therapeutic targets to control the balance of miRNAs within a cell that may prevent the development of metabolic diseases or be proposed as new biomarkers of disease to determine how subjects respond to different treatments, such as dietary treatments.
Therefore, the purpose of this review was to identify the circulating and cellular miRNAs with upregulated expression under conditions of obesity and insulin resistance that are reported in the literature, as well as the effects of different dietary interventions on their expression, to determine which miRNAs could play a role as biomarkers for early diagnosis, or to indicate response to treatments.
2. Biogenesis and Mechanism of Action of miRNAs
miRNAs synthesis begins in the nucleus through the expression of specific genes that code for MiRNAs. This process starts with RNA polymerase II, which synthesizes a primary miRNA also known as pri-miRNA. After transcription, the pri-miRNA sequence is processed by the complex formed by endoribonucleases called Drosha and DGCR8, resulting in a precursor miRNA sequence called pre-miRNA. This pre-miRNA is a sequence of approximately 60–80 nucleotides, which is exported from the nucleus to the cytoplasm by exportin 5 (XPO5) and Ran GTPases. Subsequently, the pre-miRNA is processed by the type-III endoribonuclease DICER and the RNA-binding proteins TRBP and PACT, giving rise to a double-stranded miRNA, which, after excision from the pre-miRNA hairpin conformation, becomes a mature miRNA [4]. Mature miRNAs then bind to their target mRNA through base-pairing within the RNA-induced silencing complexes (RISC), where mRNA stability and translation are regulated. miRNA-mediated RNA degradation can be achieved via a slicing-competent Argonaute-2 (AGO2) protein that cleaves the target mRNA in the mRNA and miRNA duplex. The function of RISC in translation control is mediated by GW182, which collaborates with AGO2 by recruiting deadenylation/decapping enzymes. This causes the unwinding of the duplex and the stable association of only one of the two strands. This guide strand directs target recognition through base pairing, while the other strand of the original small RNA duplex is discarded and represses translation [5,6].
As mentioned above, miRNAs may perform their function in the cell in which they are synthesized, or they may be present in body fluids or travel in the bloodstream to have a function in distal cells.
3. Circulating miRNAs: Novel Cell-Cell Communication
miRNAs can be exported and imported by cells through different mechanisms, one of the best known being within extracellular vesicles, bound to lipoproteins and carrier proteins, also known as protein-miRNA binding complexes. These are means of transport that make possible the communication of miRNAs with cells and tissues distal to where they were synthesized [7]. Due to this quality, miRNAs can be found in serum, plasma, urine, and other body fluids, making them the subject of recent studies as possible biomarkers in different diseases, since it is known that the amount and profile of miRNAs expressed in these transport media respond to different intracellular transcription factors and extracellular excretion stimuli.
3.1. Extracellular Vesicles: Exosomes
Exosomes are extracellular vesicles with a size < 200 nm originating from the Golgi apparatus. These components present a lipid bilayer due to their subsequent fusion of multivesicular bodies and the plasma membrane of the cell. Exosomes contain diverse molecules, such as RNAs, proteins, DNA fragments, and miRNAs, in different stages of maturation. The lipid bilayer protects those molecules from the extracellular medium during travel, and their degradation is more difficult, ensuring that most of the miRNAs inside them are delivered in good condition to the target cell [2]. Exosomes are detached from multivesicular bodies found in the cytoplasm and attached to the plasma membrane, where they fuse and are released by exocytosis [8]. Regarding what determines the cargo of these exosomes, studies suggest that there are different mechanisms involved in cargo and exosome recognition, such as ubiquitination and glycosylation of exosome membrane proteins, which couple the availability of miRNAs and other molecules within exosomes, the composition of exosomes varies significantly among different tissues and fluctuates based on the metabolic state of the cell [9].
It is known that the cellular release of exosomes also responds to diverse cellular stimuli, and unlike other means of transport of miRNAs, it seems to be a very well-orchestrated mechanism in different pathophysiological conditions or after pharmacological or dietary treatments. On the other hand, various mechanisms of exosome recognition with the target cell for their subsequent internalization have been described; some studies mention the recognition by antigen or membrane receptors of proteins found on the surface of exosomes such as the tetraspanins CD9, CD81, or the most abundant CD63, which also serve as anchors to the receptor cell [10].
Once bound to the surface of the new cell, exosomes can activate intracellular signalling pathways and discharge their contents into the cell by fusion to the membrane or enter the cell by phagocytosis, pinocytosis, or endocytosis [11].
Regarding their lifespan, studies investigating the uptake of exosomes in tissues have shown a half-life of exosomes from 2 to 60 min after being administered intravenously to animal models [12], suggesting not only rapid tissue signalling and communication after a stimulus but also an efficient mechanism of action of the miRNAs that reside in them.
3.2. miRNA-Binding Proteins and Lipoproteins
miRNAs can also be transported in the bloodstream by forming complexes with proteins, and like exosomes, they can enter the target cell through specific receptors for these proteins and release the miRNAs they transport to perform their function. It is suggested that this type of transport is only a fraction of the total number of circulating miRNAs. In miRNA-protein complexes, ribonucleoproteins, and nucleolar proteins such as nucleophosmin 1 (NPM1) have been found, which, once outside the cell, could also facilitate the loading of miRNAs into lipoproteins that also travel in the bloodstream [2]. In addition to exosomes and binding proteins, miRNAs can travel in smaller proportions bound to low-density lipoproteins (LDLs) and high-density lipoproteins (HDLs). HDL-bound miRNAs can be taken up by class B, type I scavenger receptors and subsequently released intracellularly to regulate gene expression. It has been shown that nutritional and metabolic status, e.g., hypercholesterolemia, can alter the abundance of lipoprotein-associated miRNAs, which can be totally distinct from those found within exosomes, thus indicating mechanisms of miRNA transport that are complementary and independent [13].
4. miRNAs in Obesity and Insulin Resistance
Several studies have explored changes in miRNA expression in different metabolic conditions, such as insulin resistance and obesity. Those studies utilized different research models to identify possible biomarkers for the prediction of type-2 diabetes and other cardiovascular diseases. Some of those studies also attempted to determine the response to miRNA treatments aimed at reversing that condition. Other studies have shown that miRNAs may be up- or downregulated in these conditions, and they have identified possible target genes that are influenced by those miRNAs, resulting in the regulation of the synthesis of proteins involved in the pathophysiological processes of obesity and insulin resistance (Table 1 and Table 2).

Table 1.
MiRNAs, exosomal miRNAs, obesity, and diet.

Table 2.
MiRNAs, exosomal miRNAs, insulin resistance, and diet.
4.1. Obesity
Alterations in various circulating miRNAs have been found in human and animal models of obesity. Although most tissues contribute to the pool of circulating miRNAs, adipose tissue is known to play a key role. Alterations in adipose tissue, such as those occurring in various metabolic conditions, can cause significant changes in the expression of circulating miRNAs and thus negatively impact other tissues.
In human and animal models of obesity, an increase in circulating levels of microRNAs that regulate genes involved in metabolic conditions, such as energy expenditure or adipogenesis, has been found. A recent clinical study evaluated the effect of 4 different energy-restricted diets plus 90 min of moderate exercise per week on plasma miR-128-1-5p concentrations in overweight and obese subjects before and after 6 months. The results showed that a high level of miR-128-1-5p was associated with a higher HOMA-IR index (‘Homeostatic Model Assessment of Insulin Resistance’), waist circumference, and body fat percentage and a lower resting energy expenditure (REE), and an interaction analysis revealed that dietary fat and protein intake modify the relationship between the change in circulating levels of this miRNA and REE [28]. Regarding adipogenesis, a study in obese mice showed that after a high-fat diet, there was a significant increase in the exosomal miRNA miR-122, which regulated the expression of adipogenesis-related genes such as PPARγ (peroxisome proliferator-activated receptor γ), ADIPOQ (adiponectin), and the VDR/SREBF1 (vitamin D receptor/sterol regulatory element-binding transcription factor-1) axis, among others [20].
Growing interest exist in understanding how circulating miRNAs in exosomes participate in a very specific manner in the regulation of genes involved in the development of obesity and its comorbidities. One study showed that injection of exosomes containing miRNAs obtained from obese mice (miR-122, miR-192, miR-27a, miR-27b) into lean mice induced the development of insulin resistance and dyslipidaemia, suggesting that these miRNAs play an important role in the pathophysiology of obesity, the development of type-2 diabetes, and possibly other cardiovascular diseases [26]. The conception of microRNAs as possible biomarkers of cardiovascular disease in the presence of obesity has been proposed by different research groups worldwide. A cross-sectional study conducted in adolescents with obesity found an association between circulating levels of at least 10 microRNAs and plasma levels of adipokines such as adiponectin, leptin, and other markers of metabolic syndrome such as glucose, insulin, the HOMA-IR index, peptide-C, and plasma levels of lipids such as triglycerides, HDL cholesterol, and LDL cholesterol [32].
On the other hand, it is important to consider that obesity is a condition in which a proinflammatory state prevails, and this chronic inflammatory environment in turn favours the appearance of various cardiovascular complications, including atherogenesis. Tang and coworkers show that exosomal miR-27b-3p, derived from visceral adipocytes, is involved in endothelial inflammation and atherosclerosis. Mechanistically, miR-27b-3p binds directly to the protein-coding sequence (CDS) region of PPARα mRNA, thereby decreasing the protein expression [14]. Also, Zhang and coworkers evaluated one of the mechanisms involved in the proinflammatory environment orchestrated by macrophages during obesity in an animal model and in vitro, and they found that exosomal miR-1224 participates in the inhibition of M2 macrophages in adipose tissue [18]. Another study that evaluated the conditions that promote an anti-inflammatory environment regulated by M2 macrophages found that exosomes obtained from THP-1 macrophages polarized with IL-4 (anti-inflammatory interleukin) promoted the expression of miR-21, miR-99a, miR-146b, and miR-378a while decreasing miR-33 expression, and it was also found that there was increased expression of PPARγ and GLUT4 (glucose transporter type 4), improving glucose uptake and insulin metabolism, increasing UCP1 (uncoupling protein 1) and OXPHOS (oxidative phosphorylation system) activity, and promoting lipophagy, mitochondrial activity and beiging in adipocytes [21]. Interestingly, it has been shown that M2 macrophages with anti-inflammatory effects are able to release exosomes with miRNAs that also exert an anti-inflammatory function, as is the case for miR-690, which directly regulates NADK (nicotinamide adenine dinucleotide kinase). NADK modulates macrophage-mediated inflammation and participates in insulin signalling as a gene encoding NAD+ kinase, an enzyme that converts nicotinamide adenine dinucleotide (NAD+) to NADP+. The inhibitory effects of NAD+ kinase on cellular insulin action were evidenced by improved in vitro insulin signalling after siRNA-induced depletion of NADK. Previous studies have shown that during obesity, NAD+ levels are reduced through either repression of NAD+ biosynthesis or greater NAD+ utilization. The authors suggested that miR-690 could even become a new insulin sensitizing agent for the treatment of metabolic diseases [22]. The miR-34a released from adipose tissue exosomes inhibits polarization to M2 macrophages possibly by repressing the expression of KLF4 (Krueppel-like factor 4), a transcription factor involved in stem cell differentiation plasticity, thereby promoting fat tissue-induced inflammation in obesity, and the researchers found that miR-34a expression increases proportionally with the development of diet-induced obesity [25].
Other miRNAs that could also participate in the pathophysiology of obesity-related hepatic steatosis have been studied. Castaño and coworkers [19] studied the regulation of the expression of local miRNAs and circulating exosomal miRNAs in an obesity model induced by a high-fat or high-sucrose diet, these miRNAs were found to be associated with the progression of obesity and the regulation of hepatic steatosis related genes, such as PTGDS (prostaglandin D2 synthase), GGT1 (gamma-glutamyltransferase 1), HK3 (hexokinase 3), PFKP (phosphofructokinase), PKM2 (pyruvate kinase M2), SLC2A5 (solute carrier family 2 member 5), and G6PC (glucose-6-phosphatase catalase). These genes are involved in metabolic pathways such as gluconeogenesis, de novo lipogenesis and insulin signalling. Another research group used an animal model and in vitro studies to analyse how Let-7b-5p, a miRNA, could regulate genes such as UCP1, FATP1 (fatty acid transport protein-1), ATP5A (ATP synthase α-subunit), PGC-1α (peroxisome proliferator-activated receptor γ coactivator 1α) for selfregulation of thermogenesis and oxidative phosphorylation, as well as participate in the hepatic TGF-β (transforming growth factor-beta) signalling pathway and inhibit white adipose tissue beiging, which could also promote the development of non-alcoholic fatty liver disease and obesity [17]. A cross-sectional clinical trial conducted in adolescents compared the expression profile of circulating exosomal miRNAs in the serum of those with obesity and those with non-alcoholic fatty liver disease. This study showed a significant increase in three miRNAs in the group of adolescents with non-alcoholic fatty liver disease (miR-122-5p, miR-27a, miR-335-5p), which have been identified as possible biomarkers of this disease. These miRNAs regulate genes such as WNT10B (Wnt family member 10B), PPARγ and SREBP-1c (sterol regulatory element-binding protein-1c), which are involved in hepatic lipid metabolism and adipocyte differentiation [30].
A study in an animal model of hepatic steatosis induced by a high-fat diet evaluated the effect of different interventions, such as a low-fat diet, energy restriction, quercetin supplementation and exercise, on miRNAs that act to regulate the synthesis and action of thyroid hormones. The study showed that although the four interventions improved hepatic steatosis to different degrees, energy restriction showed the greatest improvement in the reduction of steatosis, as well as a possible upregulation of NIS (iodine transporter protein) expression mediated by miRNAs miR-200a, miR-28, miR-339, miR-383, and miR-146b. All four interventions increased thyroid hormone action, possibly by differentially regulating miRNAs that upregulate TRB (T-cell receptor beta locus) and DIO1 (iodothyronine deiodinase 1) expression and by maintaining the homeostasis status dependent on Nrf2 (Nuclear factor erythroid 2-related factor 2), a transcription factor involved in redox homeostasis and antioxidant gene expression [23].
Other studies have sought to identify the abilities of adipose tissue exosomal microRNAs in models of obesity, including cellular repair [31]. An in vitro study of mesenchymal stem cells taken from adipose tissue in subjects with and without obesity demonstrated how exosomes isolated from those cells have the capacity to modulate inflammation, apoptosis processes, and the activation of the MAPK (mitogen-activated protein kinase) and Wnt signalling pathways involved in these processes. Moreover, some miRNAs inside those exosomes, such as miR-1291, miR-888-5p, miR-6892, miR-222-5p, miR-8072, miR-4757-5p, miR-769-5p, and miR-4730, were found to be overexpressed in the cells of the participants with obesity (Table 1).
4.2. Insulin Resistance
The persistent inflammatory state present in obesity is closely linked to the development of insulin resistance, and the mechanisms involved in this condition have been studied in various populations. A cross-sectional clinical trial conducted in adolescents with and without obesity aimed to compare plasma levels of exosomal miRNAs, and the results showed that there was a decrease in miR-223-5p, 33a-3p, miR-181a-5p, and miR-199-5p in those who had obesity. These miRNAs regulate genes such as CHD9 (chromodomain helicase DNA binding protein 9), PTEN (phosphatase and tensin homologue), MTMR12 (myotubularin related protein 12), TBL1X (transducin beta-like 1 X-linked), and CPOX (coproporphyrinogen oxidase), which are involved in lipid metabolism, inflammatory status and insulin resistance, suggesting these miRNAs as biomarkers of obesity in adolescents [29]. The levels of miR-27a and its correlation with insulin resistance were assessed within a pediatric population, and the results showed that serum miR-27a levels were positively correlated with obesity and insulin resistance in children with obesity. This same study also used an animal model with C57BL/6J mice with diet-induced obesity and C2C12 cells and found that in addition, incubation of miR-27a obtained from adipocytes of mice with induced insulin resistance caused insulin resistance in C2C12 skeletal muscle cells. Furthermore, they found that miR-27a suppressed the expression of PPARγ, i.e., genes involved in the development of obesity [60].
Inflammation caused by obesity leads to alterations in insulin signalling pathways. Ying et al. [16] demonstrated in vitro and in an animal model how adipose tissue releases into the bloodstream exosomes with microRNAs that travel to insulin-sensitive tissues such as liver and muscle, as mice with obesity treated with exosomes from adipose tissue of lean mice, with a higher proportion of M2 macrophages, decreased insulin resistance. They also found an increase in miR-155 in adipose tissue exosomes from obese models, which could contribute to the development of insulin resistance.
Another study in an animal model showed that mice with obesity and fed a high-fat diet had lower levels of exosomal miR-141-3p than mice with obesity consuming a normal diet, and the decrease in miR-141-3p was associated with reduced Akt (serine/threonine-protein kinase) phosphorylation. They also found that GW4869, a protein involved in the inhibition of exosome biogenesis and release, might participate in the mechanism of action of miR-141-3p in hepatocytes, which is expressed in higher proportions in models of obesity [24].
In the search to identify microRNAs that may be useful as diagnostic biomarkers and therapeutic targets, several research groups have evaluated how their expression is regulated in the presence of insulin resistance. Ali and collaborators studied the serum levels of miRNAs, mRNAs, and lncRNAs (long noncoding RNAs) in subjects with type-2 diabetes, subjects with prediabetes, and healthy control subjects, with the purpose of identifying genes broadly related to the pathogenesis of insulin resistance and prediabetes. The results showed that the expression levels of TMEM173 (Transmembrane Protein 173), CHUK (Conserved Helix-loop-helix Ubiquitous Kinase), and the miRNAs miR-611, miR-5192, and miR-1976 gradually increased from the control group to the prediabetes group and reached their highest levels in the type-2 diabetes group, while the expression of lncRNAs RP4-60503.4 and AC074117.2 gradually decreased from the control group to the prediabetes group and reached their lowest levels in the type-2 diabetes group [55].
In a cross-sectional clinical study, results revealed that women with metabolic syndrome showed decreased levels of miR-16 and miR-363, while men with one or more risk factors displayed elevated levels of miR-Let-7c and miR-30a. Moreover, there was a positive correlation between waist circumference and higher levels of miR-Let-7c, miR-122, miR-30a, miR-146a, miR-15a, miR-30d, and miR-222. Additionally, individuals with higher plasma glucose and/or greater insulin resistance exhibited increased expression of miR-122, miR-139, miR-Let-7c, miR-126, and miR-30a [56].
Insulin resistance may be related to the development of multiple complications in different insulin-sensitive tissues. In fact, an in vitro study showed the mechanism by which adipose tissue can regulate cardiac insulin resistance through miRNAs. For this purpose, exosomes were extracted from 3T3-L adipocytes that were induced to insulin resistance and used to treat rat ventricular myocytes. The findings were that miR-802-5p was highly expressed in models with insulin resistance and that heat shock protein 60 (HSP60) was its target gene. Its depletion was accompanied by a significant increase in C/EBP (CCAAT/enhancer binding protein) expression and increased phosphorylation of JNK (c-Jun N-terminal kinase) and IRS1 (insulin receptor substrate-1) on serine 307. Therefore, exosomal miR-802-5p from hypertrophic adipocytes caused cardiac insulin resistance through downregulation of HSP60 [41] (Table 2).
5. Regulation of miRNAs by Diet
miRNAs have been proposed as biomarkers of disease, which could be regulated in response to strategies and/or treatments for some pathologies. One of these alternatives is dietary changes, which may be a fundamental treatment component, especially in metabolic diseases. Studies have evaluated changes in miRNAs after a nutritional intervention to determine the effect of nutrients, supplements, foods, dietary patterns, or energy restriction in the diet.
5.1. Energy Restriction
Energy restriction is one of the most widely explored dietary interventions to assess the difference in the expression of various miRNAs that may be altered under conditions of obesity in both animal models and humans.
A recent study evaluated the difference in miRNA expression in rhesus monkeys after prolonged caloric restriction. The study found changes in the expression of 24 known miRNAs and 10 novel miRNAs not known until then. They showed correlations between body weight, adiposity and insulin sensitivity, with at least 10 miRNAs upregulated after dietary energy restriction. miR-125a-5p presented the greatest decrease in its expression after the intervention; in addition, a positive correlation was found between miR-125a-5p abundance and adiposity and a negative correlation with insulin sensitivity [61]. Other microRNAs that decreased in expression after intervention were miR-16, miR-20a, miR-21, miR-92a-5p, miR-130 a-5p, miR-143-5p, and miR-224. It is known that miR-92a-5p is synthesized in brown adipose tissue, a metabolically active tissue for energy expenditure, and has been inversely correlated with brown fat activity in humans [62]. Another study in an animal model found that after 4 weeks of intervention, mice fed an energy-restricted diet had a significant decrease in CD8+ T cells as well as an increase in serum miR-16-5p, miR-196b-5p and miR-218-5p expression compared to the control group. Subsequently, miR-16-5p was shown to decrease the mRNA expression of the proinflammatory cytokines IL-1β, IL-6, and TNFα, which have been implicated in the dysregulation of the insulin signalling pathway [63]. Specifically, it has been reported that TNFα inhibits the proper phosphorylation of IRS-1, favouring the development of insulin resistance. This study suggests that miR-16-5p plays an important role in the anti-inflammatory regulation conferred by a low-energy diet, as well as indirectly enhancing insulin signalling [64].
In addition to interventions, it has been shown that rapid weight loss can affect the regulation of microRNA expression. In fact, a recent study with obese subjects showed that after a severely energy-restricted diet (<800 Kcal/day) for 4 weeks, there was a modulation in circulating levels of 75 miRNAs. Most notably, miR-34a, miR-208, miR-193, miR-320, miR-433, miR-568, and miR-181a have been proposed as biomarkers of beneficial effects in response to weight loss in individuals with obesity [65].
5.2. High-Protein Diet
An increase in protein intake has been associated with beneficial effects on pathologies such as excess weight. There is now evidence that these benefits may be associated with the regulation of miRNAs. A study showed that men with obesity who consumed a high-protein diet (30% of energy intake) during a 12-week weight-loss intervention had a decreasing trend in circulating levels of miR-223 [36]. This miRNA belongs to a subgroup of miRNAs that travel in the circulation bound to high-density lipoproteins (HDLs). As mentioned above, miR-223 has been found to be increased in populations with obesity and insulin resistance; furthermore, this miRNA was found to modulate the inflammatory phenotype and macrophage activation through the FBXW7/TLR4 (F-box and WD repeat domain containing 7/toll-like receptor 4) axis. FBXW7 is one of the proteins of the ubiquitin ligase complex, and it functions in ubiquitination recognition. Recently, the role of FBXW7 in attenuating TLR4 signalling processes in macrophages has been demonstrated [66].
A study involving men older than 70 years (n = 31) and without any diagnosed disease who consumed a high-protein diet (1.6 g/kg/d) for 10 weeks revealed decreased expression of 5 miRNAs (125b-5p, miR-100-5p, miR99a-5p, miR-23b-3p and mir-203a) compared to the normal-protein diet group (0.8 g/kg/d). These miRNAs have been reported to be gene regulators of proteins involved in various proinflammatory cascades, such as interleukin 6/signal transducer and activator of transcription 3 (IL6/STAT3). The authors propose that a high-protein diet may regulate proinflammatory responses [67].
5.3. Ketogenic Diet
In addition, the regulation of energy and protein intake has been used as a strategy for weight loss or to reduce metabolic disturbances. Additionally, diets high in lipids and low in carbohydrates have been reported as a strategy with beneficial effects on metabolic pathologies. A clinical study in obese subjects (18 women and 18 men) evaluated the effect of a 6-week two-step ketogenic diet on body composition and microRNA expression profile in obese subjects. The results showed significant changes in the expression of three miRNAs, miR-Let-7b-5p, miR-143-3p, and miR-504-5p, all related to the regulation of genes linked to nutrient metabolism, such as mTOR (mechanistic target of rapamycin), PPARs (peroxisome proliferator-activated receptors), insulin and proinflammatory cytokine signalling pathways [68].
Cannataro and coworkers evaluated the expression profile of microRNAs in patients with obesity in response to treatment with a ketogenic diet. After consumption of the ketogenic diet, there was a significant upregulation in the expression of miRNAs compared to subjects with obesity who did not consume the diet. In addition, it was observed that the expression of miRNAs let-7b-5, miR-143-3p, miR-148b-3p, miR-590-5p, miR-520 h, miR-644a, and miR-548d-3p in obese subjects was similar to that in lean subjects. Interestingly, bioinformatic analysis showed that 4 miRNAs are linked to antioxidant and anti-inflammatory metabolic pathways, such as Let-7e-5p, which regulates glutathione peroxidase 7 (GPX7) synthesis, miR-520 h, which regulates Tet methylcytosine dioxygenase 3 (TET3), and miR-548d-3p, which is related to the regulation of superoxide dismutase 2 (SOD2), while miR-30a-5p correlated with decreased catalase expression in red blood cells. The authors concluded that the ketogenic diet results in similar regulation of miRNA expression in obese subjects as in lean subjects [33].
5.4. Diet with Supplementation
Dietary supplementation has been utilized to combat obesity, as it can contribute to weight loss and attenuate some related risks, even without changes in macronutrient intake. Due to the changes associated with these types of interventions, there are studies that suggest the possible role of miRNAs in these benefits. A randomized, crossover clinical study in subjects with prediabetes (n = 49) evaluated the effect of a pistachio-enriched diet on the expression profile of microRNAs related to glucose metabolism and insulin resistance. Participants underwent 2 conditions, a pistachio-supplemented diet (57 g/day) and an isocaloric control diet, for 4 months with a 2-week washout period. Pistachio supplementation increased the expression of miR-21, miR-29b, miR-223, and 15a, which was reflected in a decrease in insulin resistance. Subjects who increased the levels of miRNAs 15a and 21 also showed a decrease in IL-6 expression, which may result in a decrease in the inflammatory state [69]. Specifically, miR-223 has been reported to induce the expression of GLUT4 protein to normalize glucose levels [70]. miR-15a regulates insulin biosynthesis by binding to UCP-2 in mice [71]. For miR-21, a previous study showed that this miRNA reversed insulin resistance in 3T3-L1 cells and significantly increased insulin-induced glucose uptake by translocating GLUT4 to the cell membrane [72]. It was also observed that pistachio supplementation decreased the levels of miR-375 and miR-192. It has been suggested that increased levels of miR-375 indicate increased expression from its synthesis in pancreatic islets in subjects with type-2 diabetes, prediabetes, and alterations in glucose or insulin [73]. miR-192 has been reported to be elevated in subjects with prediabetes and in animal models of mice with glucose intolerance [61]. Thus, pistachio consumption in these amounts could regulate some metabolic pathways involved in glucose metabolism by action of these miRNAs.
In an animal model, it was shown that a diet supplemented with soy (25%) for 28 days promoted significant changes in the expression of two miRNAs, miR-145a-5p and miR-455p, compared to the control group. The analysis suggests that these miRNAs may impact nuclear receptor expression, the glutamine/glutathione ratio, catalytic activity and iron metabolism in the liver [74]. Another study evaluated the effect of a diet high in protein, fish oil and omega-3 in NZ10 mice, which are predisposed to develop obesity and type-2 diabetes. After 19 weeks of intervention, an increase in the expression of miR-205 and a decrease in the expression of miR-411, miR-155, miR-335, and miR-21 were observed compared to the control group (<6% fat). Increase levels of these miRNAs have been reported to be involved in the progression of type-2 diabetes or hepatic steatosis, suggesting that polyunsaturated fatty acids such as omega-3 may regulate gene expression at the hepatic level. Other studies have shown that these miRNAs improve insulin sensitivity and decrease liver triglycerides and hepatic steatosis in animal models [75]. In an animal model, the effect of fish oil supplementation on the hepatic expression of miRNAs and its relationship with the development of non-alcoholic fatty liver disease were evaluated. Separate male Sprague-Dawley rats were included in 3 intervention groups: one group was fed a control diet, the second group was fed a high-fat, high-cholesterol diet, and the third group was fed a high-fat, high-cholesterol diet plus fish oil supplementation. The expression of microRNAs in liver tissue was analysed. The analysis showed that there was a difference in the expression of 79 miRNAs between the high-fat diet with fish oil supplementation and the high-fat diet without fish oil supplementation. The miRNAs that were differentially expressed included miR-29c-3p, miR-30d-5p, miR-33-5p, miR-34a, and miR-328a-3p. It has been reported that these miRNAs might interact with hepatic lipid metabolism, as well as important transductional regulation of different genes, such as Pcsk9 (proprotein convertase subtilisin/kexin type 9), Insig2 (insulin-induced gene 2), Per3 (period circadian regulator 3), and Socs1/3 (suppressor of cytokine signalling 1 y 3), which are related to the development of non-alcoholic liver disease [76].
5.5. Other Related Interventions
A recent review aimed to identify the effect of physical training on miRNAs associated with systemic arterial hypertension, type-2 diabetes, and obesity. It describes the increased expression of 8 miRNAs, including miR-21, miR-27a, miR-30d, miR-126, miR-181a, miR-143, miR-222, and miR-223. These miRNAs have been found to participate in the regulation of genes involved in mechanisms of insulin resistance, adipogenesis or angiogenesis, suggesting a regulatory mechanism with metabolic benefits of physical activation [77].
The effects of an exercise program combined with a weight-loss diet in adolescents with obesity have also been evaluated. This program was shown to decrease glucose levels and circulating levels of miR-126, which has been correlated with parameters of microvascular function [34]. miR-126 has also been linked to IRS1 and to mitochondrial dysfunction, causing altered signalling in the insulin pathway and decreased glycogen synthesis [78].
A recent clinical trial analysed the effects of weight-loss bypass surgery on the upregulation of the expression of microRNAs present in circulating exosomes of six African-American women with obesity (BMI 51 ± 8.8 kg/m2). One year after surgery, there was a decrease in BMI (−18.6 ± 5.1 kg/m2), HOMA index (1.94 ± 0.6 to 0.49 ± 0.1) and concentration of branched-chain amino acids. In addition, upregulation was found in the expression of 168 microRNAs, of which 10 were directly linked to the regulation of genes involved in the insulin signalling pathway: miR-155-5p, miR-503-5p, miR-199a-5p, miR-539-5p, miR-874-3p, miR-4664-5p, miR-4747-5p, miR-516b-5p, miR-126-3p, and miR-122-5p [79].
6. miRNAs Related to Obesity, Insulin Resistance and Diet
Obesity is a condition that is usually accompanied by a long list of metabolic ailments, including insulin resistance. Recent studies have estimated that up to 1 in 2 people with obesity have insulin resistance. Insulin resistance is the precursor to the development of type-2 diabetes, which coexists with the presence of cardiovascular disease. The first line of treatment for both obesity and obesity-related insulin resistance is to make dietary changes with the goal of pursuing healthy weight loss, where diet is a central part of the treatment of these patients. As discussed in this review, both obesity and insulin resistance can be identified through the regulation of characteristic miRNAs, and interestingly, after dietary intervention, we seek to reverse the expression profile of these miRNAs.
We identified miRNAs that are up- and downregulated in conditions of obesity and insulin resistance, and after dietary intervention, these could be proposed as possible biomarkers of early response to nutritional treatment of obesity and insulin resistance (Figure 1) (Venn diagram).
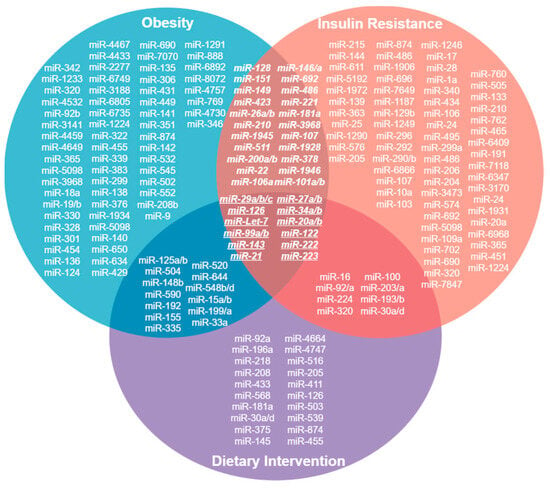
Figure 1.
Venn Diagram. MicroRNAs whose expression is upregulated in conditions of obesity, insulin resistance and after a dietary intervention. 12 of them converge in these conditions and could be proposed as biomarkers of early response in the dietary treatment of obesity and insulin resistance.
7. Discussion
This review summarizes some of the changes in miRNA expression under certain pathologies associated with chronic degenerative diseases. Interestingly, some miRNAs that are upregulated after dietary intervention in conditions of obesity and insulin resistance have been identified (Figure 1). These miRNAs could be proposed as possible biomarkers of early response to nutritional treatment of obesity and insulin resistance. Therefore, it is of great importance to understand the different expression profiles of microRNAs in different pathologies in order to know what is occurring at the cellular level, from the regulation of proteins involved in different signalling pathways to the regulation of proteins that play a decisive role in the development of diseases and their complications. Thus, miRNAs may not only be biomarkers of disease stages but also prognostic biomarkers or biomarkers of response to treatments aimed at preventing or reversing these pathophysiological mechanisms in a timely manner.
This literature review included 26 recent articles that extensively studied the expression of microRNAs in models of obesity in animals and humans. Thus, there is no doubt that there is a wide difference between the circulating levels of microRNAs in obese subjects compared with nonobese subjects, supporting the theory that the disease condition is able to modulate the expression of miRNAs and that their identification in the circulation is an excellent alternative to know what is occurring systemically in various organs. As we could observe in this review, chronic and persistent inflammation, as well as the metabolic consequences that this causes, is one of the most widely studied conditions in models of obesity, and the regulation of genes such as NADK and KLF4, which are mediated by miRNAs such as miR-690 and miR-34a, suggests that microRNAs are directly involved in the polarization of macrophages to an anti-inflammatory or proinflammatory phenotype [22,25].
Studies on the regulation of miRNAs in insulin resistance, which is a pathology closely related to obesity, were also included. Twenty-two articles studying how microRNAs modulate their expression in the presence and development of insulin resistance were included in this review. Animal model and in vitro studies described how miRNAs alter insulin signalling pathways at different times. For example, increased levels of circulating miR-222 were related to the presence of insulin resistance as well as decreased expression of IRS1 in adipose tissue [50].
miRNAs are not only proposed as biomarkers of disease state but also as biomarkers in response to treatment, suggesting the need to search for a suitable biomarker for clinical practice. In this sense, it is important to remember that one quality of biomarkers is that they must be noninvasive, so considering circulating miRNAs, especially in blood, seems to be the best alternative; of those circulating miRNAs, those travelling inside exosomes are the best option, since as mentioned above, exosomes confer protection to miRNAs due to their lipid bilayer, making their travel in the bloodstream safe to ensure their effect on target cells. In addition, according to the literature, research aimed at studying exosomal miRNAs in various pathologies, such as cancer, has grown exponentially, as researchers have sought to elucidate their performance as a biomarker and even as a treatment vehicle.
Interestingly, the regulation of microRNA expression is a reversible epigenetic process that is sensitive to the pathophysiological state of the individual and has also demonstrated its response to nutritional interventions in the different study models, which leads us to believe that the basal nutritional state of the subjects may also be a factor in the regulation of the expression of these miRNAs. Unfortunately, there are very few published studies that discuss the impact of a specific nutritional treatment on the modulation of miRNA expression. In this review, we found only 12 studies that evaluate how diet changes the abundance of miRNAs after a period of treatment compared to baseline levels, and of these, only six are clinical trials conducted in humans.
As shown in this review, there are still few clinical studies in humans that seek to find differences in the expression of miRNAs in metabolic conditions, such as obesity and insulin resistance, together with a particular nutritional intervention. This represents not only a remaining challenge for researchers in the field of nutrigenomics but also a window of opportunity for the innovation of biomarkers of early response to nutritional treatments, which will allow the proposal of new effective strategies for the comprehensive management of patients with cardiovascular diseases, which are the leading causes of death in Mexico and the world.
The possibility of understanding metabolic conditions from the regulation of gene expression makes the study of microRNAs an opportunity to address the future of disease diagnosis, treatment and control.
Author Contributions
K.G.H.-G., M.G.-C., L.E.G.-S. and A.A.-N. wrote the first draft; A.R.T., A.E.S.-Z., L.G.N., N.T., R.G.-H., I.M.-V. and A.L.G.-S. contributed to the preparation of the final version of our manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “Convocatoria 2023 para el fondo de apoyo a proyectos de investigación en el campo de la salud del Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán” and Karla Guadalupe Hernández-Gómez is student in the Programa de Doctorado en Ciencias Biomédicas at Universidad Nacional Autónoma de México and has received support through a scholarship from CONACYT (CVU 703851). The APC was funded by Hospital Regional de Alta Especialidad de la Península de Yucatán.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bartel, D.P. Metazoan microRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Safdar, A.; Tarnopolsky, M.A. Exosomes as mediators of the systemic adaptations to endurance exercise. Cold Spring Harb. Perspect. Med. 2018, 8, a029827. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Hannon, G.J. Small RNA sorting: Matchmaking for Argonautes. Nat. Rev. Genet. 2011, 12, 19–31. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of exosome composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef]
- Vallabhajosyula, P.; Korutla, L.; Habertheuer, A.; Yu, M.; Rostami, S.; Yuan, C.X.; Reddy, S.; Liu, C.; Korutla, V.; Koeberlein, B.; et al. Tissue-specific exosome biomarkers for noninvasively monitoring immunologic rejection of transplanted tissue. J. Clin. Investig. 2017, 127, 1375–1391. [Google Scholar] [CrossRef]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, more than markers of extracellular vesicles in reproduction. Int. J. Mol. Sci. 2020, 21, 7568. [Google Scholar] [CrossRef]
- Horibe, S.; Tanahashi, T.; Kawauchi, S.; Murakami, Y.; Rikitake, Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer 2018, 18, 47. [Google Scholar] [CrossRef]
- Lai, C.P.; Mardini, O.; Ericsson, M.; Prabhakar, S.; Maguire, C.A.; Chen, J.W.; Tannous, B.A.; Breakefield, X.O. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 2014, 8, 483–494. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef]
- Tang, Y.; Yang, L.J.; Liu, H.; Song, Y.J.; Yang, Q.Q.; Liu, Y.; Qian, S.W.; Tang, Q.Q. Exosomal miR-27b-3p secreted by visceral adipocytes contributes to endothelial inflammation and atherogenesis. Cell Rep. 2023, 42, 111948. [Google Scholar] [CrossRef] [PubMed]
- Aas, V.; Øvstebø, R.; Brusletto, B.S.; Aspelin, T.; Trøseid, A.M.S.; Qureshi, S.; Eid, D.S.O.; Olstad, O.K.; Nyman, T.A.; Haug, K.B.F. Distinct microRNA and protein profiles of extracellular vesicles secreted from myotubes from morbidly obese donors with type 2 diabetes in response to electrical pulse stimulation. Front. Physiol. 2023, 14, 1143966. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Hu, L.; Gui, W.; Xiao, L.; Wang, W.; Xia, J.; Fan, H.; Li, Z.; Zhu, Q.; Hou, X.; et al. Hepatocyte TGF-β signaling inhibiting WAT browning to promote NAFLD and obesity is associated with let-7b-5p. Hepatol. Commun. 2022, 6, 1301–1321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yao, X.; Teng, Y.; Zhao, T.; Lin, L.; Li, Y.; Shang, H.; Jin, Y.; Jin, Q. Adipocytes-derived exosomal microRNA-1224 inhibits M2 macrophage polarization in obesity-induced adipose tissue inflammation via MSI2-mediated Wnt/β-catenin axis. Mol. Nutr. Food Res. 2022, 66, e2100889. [Google Scholar] [CrossRef] [PubMed]
- Castaño, C.; Novials, A.; Párrizas, M. Exosomes from short-term high-fat or high-sucrose fed mice induce hepatic steatosis through different pathways. Cells 2022, 12, 169. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, J.; Ren, Y.; Fan, L.; Xiang, W.; He, X. Exosomal miR-122 promotes adipogenesis and aggravates obesity through the VDR/SREBF1 axis. Obesity 2022, 30, 666–679. [Google Scholar] [CrossRef]
- Phu, T.A.; Ng, M.; Vu, N.K.; Bouchareychas, L.; Raffai, R.L. IL-4 polarized human macrophage exosomes control cardiometabolic inflammation and diabetes in obesity. Mol. Ther. 2022, 30, 2274–2297. [Google Scholar] [CrossRef]
- Ying, W.; Gao, H.; Dos Reis, F.C.G.; Bandyopadhyay, G.; Ofrecio, J.M.; Luo, Z.; Ji, Y.; Jin, Z.; Ly, C.; Olefsky, J.M. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 2021, 33, 781–790.e5. [Google Scholar] [CrossRef]
- Xia, S.F.; Jiang, Y.Y.; Qiu, Y.Y.; Huang, W.; Wang, J. Role of diets and exercise in ameliorating obesity-related hepatic steatosis: Insights at the microRNA-dependent thyroid hormone synthesis and action. Life Sci. 2020, 242, 117182. [Google Scholar] [CrossRef]
- Dang, S.; Leng, Y.; Wang, Z.; Xiao, X.; Zhang, X.; Wen, T.; Gong, H.; Hong, A.; Ma, Y. Exosomal transfer of obesity adipose tissue for decreased miR-141-3p mediate insulin resistance of hepatocytes. Int. J. Biol. Sci. 2019, 15, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Hui, X.; Hoo, R.L.C.; Ye, D.; Chan, C.Y.C.; Feng, T.; Wang, Y.; Lam, K.S.L.; Xu, A. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Investig. 2019, 129, 834–849. [Google Scholar] [CrossRef] [PubMed]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Q.; Xiao, X.; Wu, C.; Gao, R.; Peng, C.; Li, D.; Zhang, W.; Du, T.; Wang, Y.; et al. miR-1934, downregulated in obesity, protects against low-grade inflammation in adipocytes. Mol. Cell. Endocrinol. 2016, 428, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Heianza, Y.; Xue, Q.; Rood, J.; Bray, G.; Sacks, F.; Qi, L. Circulating thrifty microRNA is related to insulin sensitivity, adiposity, and energy metabolism in adults with overweight and obesity: The POUNDS Lost trial. Am. J. Clin. Nutr. 2023, 117, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Cabiati, M.; Randazzo, E.; Guiducci, L.; Falleni, A.; Cecchettini, A.; Casieri, V.; Federico, G.; Del Ry, S. Evaluation of exosomal coding and non-coding RNA signature in obese adolescents. Int. J. Mol. Sci. 2022, 24, 139. [Google Scholar] [CrossRef]
- Zhang, J.W.; Pan, H.T. microRNA profiles of serum exosomes derived from children with nonalcoholic fatty liver. Genes Genom. 2022, 44, 879–888. [Google Scholar] [CrossRef]
- Eirin, A.; Meng, Y.; Zhu, X.Y.; Li, Y.; Saadiq, I.M.; Jordan, K.L.; Tang, H.; Lerman, A.; Van Wijnen, A.J.; Lerman, L.O. The micro-RNA cargo of extracellular vesicles released by human adipose tissue-derived mesenchymal stem cells is modified by obesity. Front. Cell Dev. Biol. 2021, 9, 660851. [Google Scholar] [CrossRef]
- Al-Rawaf, H.A. Circulating microRNAs and adipokines as markers of metabolic syndrome in adolescents with obesity. Clin. Nutr. 2019, 38, 2231–2238. [Google Scholar] [CrossRef]
- Cannataro, R.; Caroleo, M.C.; Fazio, A.; La Torre, C.; Plastina, P.; Gallelli, L.; Lauria, G.; Cione, E. Ketogenic diet and microRNAs linked to antioxidant biochemical homeostasis. Antioxidants 2019, 8, 269. [Google Scholar] [CrossRef]
- Tang, D.; Bai, S.; Li, X.; Yao, M.; Gong, Y.; Hou, Y.; Li, J.; Yang, D. Improvement of microvascular endothelial dysfunction induced by exercise and diet is associated with microRNA-126 in obese adolescents. Microvasc. Res. 2019, 123, 86–91. [Google Scholar] [CrossRef]
- Parr, E.B.; Camera, D.M.; Burke, L.M.; Phillips, S.M.; Coffey, V.G.; Hawley, J.A. Circulating microRNA responses between ‘high’ and ‘low’ responders to a 16-Wk diet and exercise weight loss intervention. PLoS ONE 2016, 11, e0152545. [Google Scholar] [CrossRef]
- Tabet, F.; Torres, L.F.C.; Ong, K.L.; Shrestha, S.; Choteau, S.A.; Barter, P.J.; Clifton, P.; Rye, K.A. High-density lipoprotein-associated miR-223 is altered after diet-induced weight loss in overweight and obese males. PLoS ONE 2016, 11, e0151061. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Cardona-Alvarado, M.I.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Sabater, M.; Fuentes-Batllevell, N.; Ramírez-Chávez, E.; Ricart, W.; Molina-Torres, J.; et al. Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J. Nutr. Biochem. 2015, 26, 1095–1101. [Google Scholar] [CrossRef]
- Pescador, N.; Pérez-Barba, M.; Ibarra, J.M.; Corbatón, A.; Martínez-Larrad, M.T.; Serrano-Ríos, M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS ONE 2013, 8, e77251. [Google Scholar] [CrossRef]
- Ortega, F.J.; Mercader, J.M.; Catalán, V.; Moreno-Navarrete, J.M.; Pueyo, N.; Sabater, M.; Gómez-Ambrosi, J.; Anglada, R.; Fernández-Formoso, J.A.; Ricart, W.; et al. Targeting the circulating microRNA signature of obesity. Clin. Chem. 2013, 59, 781–792. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Ye, Y.; Li, D.; Liu, Y.; Lee, E.; Zhang, M.; Dai, X.; Zhang, X.; Wang, S.; et al. Pancreatic β cells control glucose homeostasis via the secretion of exosomal miR-29 family. J. Extracell. Vesicles 2021, 10, e12055. [Google Scholar] [CrossRef]
- Wen, Z.; Li, J.; Fu, Y.; Zheng, Y.; Ma, M.; Wang, C. Hypertrophic adipocyte–derived exosomal miR-802-5p contributes to insulin resistance in cardiac myocytes through targeting HSP60. Obesity 2020, 28, 1932–1940. [Google Scholar] [CrossRef]
- Tian, F.; Tang, P.; Sun, Z.; Zhang, R.; Zhu, D.; He, J.; Liao, J.; Wan, Q.; Shen, J. miR-210 in exosomes derived from macrophages under high glucose promotes mouse diabetic obesity pathogenesis by suppressing NDUFA4 expression. J. Diabetes Res. 2020, 2020, 6894684. [Google Scholar] [CrossRef]
- Su, T.; Xiao, Y.; Xiao, Y.; Guo, Q.; Li, C.; Huang, Y.; Luo, X. Bone marrow mesenchymal stem cells-derived exosomal MiR-29b-3p regulates aging-associated insulin resistance. ACS Nano 2019, 13, 2450–2462. [Google Scholar] [CrossRef]
- Xiong, H.; Liu, W.; Song, J.; Gu, X.; Luo, S.; Lu, Z.; Hao, H.; Xiao, X. Adipose tissue macrophage-derived exosomal miR-210-5p in modulating insulin sensitivity in rats born small for gestational age with catch-up growth. Transl. Pediatr. 2023, 12, 587–599. [Google Scholar] [CrossRef]
- Hong, Y.; Wu, J.; Yu, S.; Hui, M.; Lin, S. Serum-derived exosomal microRNAs in lipid metabolism in polycystic ovary syndrome. Reprod. Sci. 2022, 29, 2625–2635. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Chen, L.; Bian, H.; Chen, X.; Zheng, H.; Yang, P.; Chen, Q.; Xu, H. Natural killer cell-derived exosomal miR-1249-3p attenuates insulin resistance and inflammation in mouse models of type 2 diabetes. Signal Transduct. Target. Ther. 2021, 6, 409. [Google Scholar] [CrossRef]
- Li, L.; Zuo, H.; Huang, X.; Shen, T.; Tang, W.; Zhang, X.; An, T.; Dou, L.; Li, J. Bone marrow macrophage-derived exosomal miR-143-5p contributes to insulin resistance in hepatocytes by repressing MKP5. Cell Prolif. 2021, 54, e13140. [Google Scholar] [CrossRef]
- Jalabert, A.; Reininger, L.; Berger, E.; Coute, Y.; Meugnier, E.; Forterre, A.; Errazuriz-Cerda, E.; Geloen, A.; Aouadi, M.; Bouzakri, K.; et al. Profiling of ob/ob mice skeletal muscle exosome-like vesicles demonstrates combined action of miRNAs, proteins and lipids to modulate lipid homeostasis in recipient cells. Sci. Rep. 2021, 11, 21626. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhou, Y.; Shi, Y.; Zhang, Y.; Liu, K.; Liang, R.; Sun, P.; Chang, X.; Tang, W.; Zhang, Y.; et al. Expression of miRNA-29 in pancreatic β cells promotes inflammation and diabetes via TRAF3. Cell Rep. 2021, 34, 108576. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Song, H.; Shuo, L.; Wang, L.; Xie, P.; Li, W.; Liu, J.; Tong, Y.; Zhang, C.Y.; Jiang, X.; et al. Gonadal white adipose tissue-derived exosomal MiR-222 promotes obesity-associated insulin resistance. Aging 2020, 12, 22719–22743. [Google Scholar] [CrossRef]
- Liu, T.; Sun, Y.C.; Cheng, P.; Shao, H.G. Adipose tissue macrophage-derived exosomal miR-29a regulates obesity-associated insulin resistance. Biochem. Biophys. Res. Commun. 2019, 515, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Jalabert, A.; Vial, G.; Guay, C.; Wiklander, O.P.B.; Nordin, J.Z.; Aswad, H.; Forterre, A.; Meugnier, E.; Pesenti, S.; Regazzi, R.; et al. Exosome-like vesicles released from lipid-induced insulin-resistant muscles modulate gene expression and proliferation of beta recipient cells in mice. Diabetologia 2016, 59, 1049–1058. [Google Scholar] [CrossRef]
- Infante-Menéndez, J.; López-Pastor, A.R.; González-Illanes, T.; González-López, P.; Huertas-Lárez, R.; Rey, E.; González-Rodríguez, Á.; García-Monzón, C.; Patil, N.P.; De Céniga, M.V.; et al. Increased let-7d-5p in non-alcoholic fatty liver promotes insulin resistance and is a potential blood biomarker for diagnosis. Liver Int. 2023, 43, 1714–1728. [Google Scholar] [CrossRef]
- Ye, Z.; Cheng, M.; Fan, L.; Ma, J.; Zhang, Y.; Gu, P.; Xie, Y.; You, X.; Zhou, M.; Wang, B.; et al. Plasma microRNA expression profiles associated with zinc exposure and type 2 diabetes mellitus: Exploring potential role of miR-144-3p in zinc-induced insulin resistance. Environ. Int. 2023, 172, 107807. [Google Scholar] [CrossRef]
- Ali, H.S.; Kamel, M.M.; Agwa, S.H.A.; Hakeem, M.S.A.; Meteini, M.S.E.; Matboli, M. Analysis of mRNA-miRNA-lncRNA differential expression in prediabetes/type 2 diabetes mellitus patients as potential players in insulin resistance. Front. Endocrinol. 2023, 14, 1131171. [Google Scholar] [CrossRef]
- Brandão-Lima, P.N.; De Carvalho, G.B.; Payolla, T.B.; Sarti, F.M.; Fisberg, R.M.; Malcomson, F.C.; Mathers, J.C.; Rogero, M.M. Circulating microRNAs showed specific responses according to metabolic syndrome components and sex of adults from a population-based study. Metabolites 2022, 13, 2. [Google Scholar] [CrossRef]
- Byun, J.S.; Lee, H.Y.; Tian, J.; Moon, J.S.; Choi, J.; Lee, S.H.; Kim, Y.G.; Yi, H.S. Effect of salivary exosomal miR-25-3p on periodontitis with insulin resistance. Front. Immunol. 2022, 12, 775046. [Google Scholar] [CrossRef]
- Mantilla-Escalante, D.C.; De Las Hazas, M.C.L.; Crespo, M.C.; Martín-Hernández, R.; Tomé-Carneiro, J.; Del Pozo-Acebo, L.; Salas-Salvadó, J.; Bulló, M.; Dávalos, A. Mediterranean diet enriched in extra-virgin olive oil or nuts modulates circulating exosomal non-coding RNAs. Eur. J. Nutr. 2021, 60, 4279–4293. [Google Scholar] [CrossRef]
- Sardu, C.; Modugno, P.; Castellano, G.; Scisciola, L.; Barbieri, M.; Petrella, L.; Fanelli, M.; Macchia, G.; Caradonna, E.; Massetti, M.; et al. Atherosclerotic plaque fissuration and clinical outcomes in pre-diabetics vs. normoglycemics patients affected by asymptomatic significant carotid artery stenosis at 2 years of follow-up: Role of microRNAs modulation: The ATIMIR study. Biomedicines 2021, 9, 401. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-derived exosomal MiR-27a induces insulin resistance in skeletal muscle through repression of PPARγ. Theranostics 2018, 8, 2171–2188. [Google Scholar] [CrossRef] [PubMed]
- Párrizas, M.; Brugnara, L.; Esteban, Y.; González-Franquesa, A.; Canivell, S.; Murillo, S.; Gordillo-Bastidas, E.; Cussó, R.; Cadefau, J.A.; García-Roves, P.M.; et al. Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. J. Clin. Endocrinol. Metab. 2015, 100, E407–E415. [Google Scholar] [CrossRef]
- Nunez Lopez, Y.O.; Garufi, G.; Seyhan, A.A. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol. Biosyst. 2016, 13, 106–121. [Google Scholar] [CrossRef]
- Dandona, P.; Aljada, A.; Bandyopadhyay, A. Inflammation: The link between insulin resistance, obesity and diabetes. Trends Immunol. 2004, 25, 4–7. [Google Scholar] [CrossRef]
- Yamada, K.; Takizawa, S.; Ohgaku, Y.; Asami, T.; Furuya, K.; Yamamoto, K.; Takahashi, F.; Hamajima, C.; Inaba, C.; Endo, K.; et al. MicroRNA 16-5p is upregulated in calorie-restricted mice and modulates inflammatory cytokines of macrophages. Gene 2020, 725, 144191. [Google Scholar] [CrossRef] [PubMed]
- Manning, P.; Munasinghe, P.E.; Papannarao, J.B.; Gray, A.R.; Sutherland, W.; Katare, R. Acute weight loss restores dysregulated circulating microRNAs in individuals who are obese. J. Clin. Endocrinol. Metab. 2019, 104, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Deiuliis, J.A.; Syed, R.; Duggineni, D.; Rutsky, J.; Rengasamy, P.; Zhang, J.; Huang, K.; Needleman, B.; Mikami, D.; Perry, K.; et al. Visceral adipose microRNA 223 is upregulated in human and murine obesity and modulates the inflammatory phenotype of macrophages. PLoS ONE 2016, 11, e0165962. [Google Scholar] [CrossRef]
- Ramzan, F.; Mitchell, C.J.; Milan, A.M.; Schierding, W.; Zeng, N.; Sharma, P.; Mitchell, S.M.; D’Souza, R.F.; Knowles, S.O.; Roy, N.C.; et al. Comprehensive profiling of the circulatory miRNAome response to a high protein diet in elderly men: A potential role in inflammatory response modulation. Mol. Nutr. Food Res. 2019, 63, e1800811. [Google Scholar] [CrossRef] [PubMed]
- Cannataro, R.; Perri, M.; Gallelli, L.; Caroleo, M.C.; De Sarro, G.; Cione, E. Ketogenic diet acts on body remodeling and microRNAs expression profile. MicroRNA 2019, 8, 116–126. [Google Scholar] [CrossRef]
- Hernández-Alonso, P.; Giardina, S.; Salas-Salvadó, J.; Arcelin, P.; Bulló, M. Chronic pistachio intake modulates circulating microRNAs related to glucose metabolism and insulin resistance in prediabetic subjects. Eur. J. Nutr. 2017, 56, 2181–2191. [Google Scholar] [CrossRef]
- Lu, H.; Buchan, R.J.; Cook, S.A. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc. Res. 2010, 86, 410–420. [Google Scholar] [CrossRef]
- Druz, A.; Chen, Y.C.; Guha, R.; Betenbaugh, M.; Martin, S.E.; Shiloach, J. Large-scale screening identifies a novel microRNA, miR-15a-3p, which induces apoptosis in human cancer cell lines. RNA Biol. 2013, 10, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.Y.; Hu, B.; Hu, X.B.; Zhong, J.; Feng, S.D.; Qin, L.; Liu, G.; Wen, G.B.; Liao, D.F. MiRNA-21 reverses high glucose and high insulin induced insulin resistance in 3T3-L1 adipocytes through targeting phosphatase and tensin homologue. Exp. Clin. Endocrinol. Diabetes 2012, 120, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Poy, M.N.; Hausser, J.; Trajkovski, M.; Braun, M.; Collins, S.; Rorsman, P.; Zavolan, M.; Stoffel, M. miR-375 maintains normal pancreatic α- and β-cell mass. Proc. Natl. Acad. Sci. USA 2009, 106, 5813–5818. [Google Scholar] [CrossRef] [PubMed]
- Seclaman, E.; Balacescu, L.; Balacescu, O.; Bejinar, C.; Udrescu, M.; Marian, C.; Sirbu, I.O.; Anghel, A. MicroRNAs mediate liver transcriptome changes upon soy diet intervention in mice. J. Cell. Mol. Med. 2019, 23, 2263–2267. [Google Scholar] [CrossRef] [PubMed]
- Adi, N.; Adi, J.; Lassance-Soares, R.M.; Kurlansky, P.; Yu, H.; Webster, K.A. High protein/fish oil diet prevents hepatic steatosis in NONcNZO10 mice; association with diet/genetics-regulated micro-RNAs. J. Diabetes Metab. 2016, 7, 676. [Google Scholar] [CrossRef]
- Wang, H.; Shao, Y.; Yuan, F.; Feng, H.; Li, N.; Zhang, H.; Wu, C.; Liu, Z. Fish oil feeding modulates the expression of hepatic microRNAs in a western-style diet-induced nonalcoholic fatty liver disease rat model. BioMed Res. Int. 2017, 2017, 2503847. [Google Scholar] [CrossRef]
- Improta Caria, A.C.; Nonaka, C.K.V.; Pereira, C.S.; Soares, M.B.P.; Macambira, S.G.; Souza, B.S.F. Exercise training-induced changes in micrornas: Beneficial regulatory effects in hypertension, type 2 diabetes, and obesity. Int. J. Mol. Sci. 2018, 19, 3608. [Google Scholar] [CrossRef]
- Ryu, H.S.; Park, S.Y.; Ma, D.; Zhang, J.; Lee, W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS ONE 2011, 6, e17343. [Google Scholar] [CrossRef]
- Hubal, M.J.; Nadler, E.P.; Ferrante, S.C.; Barberio, M.D.; Suh, J.H.; Wang, J.; Dohm, G.L.; Pories, W.J.; Mietus-Snyder, M.; Freishtat, R.J. Circulating adipocyte-derived exosomal MicroRNAs associated with decreased insulin resistance after gastric bypass. Obesity 2017, 25, 102–110. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).